Abstract
Bacteremia caused by Francisella tularensis is rare and has been reported mainly in the United States and infrequently in Europe. We report herein the first case of bacteremic F. tularensis pneumonia in an immunocompetent individual in southern Europe.
CASE REPORT
A 56-year-old man who was born blind and with cerebral palsy and who lived in a rural area was hospitalized with a cough, fever (39.7°C), and severe dehydration. At admission, a chest radiograph showed alveolar lesions of the right middle lobe. Laboratory studies revealed a white blood cell count of 12,000 cells/mm3 (92% polymorphonuclear leukocytes), a C-reactive protein level of 102 mmol/liter, and a fibrinogen level of 6.43 g/liter. In this human immunodeficiency virus-seronegative patient, further evaluation revealed normal levels of immunoglobulin G (IgG), IgA, and IgM in serum.
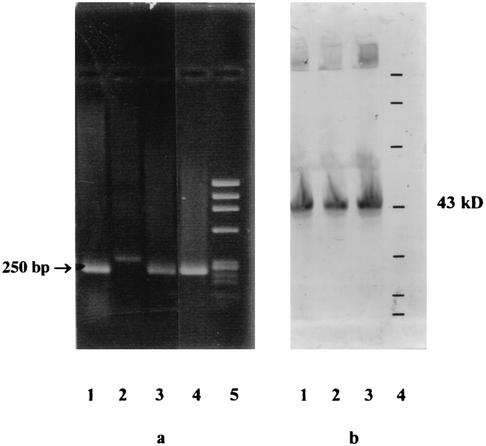
Two sets of blood culture samples were collected in BACTEC Plus Aerobic/F medium and BACTEC Plus Anaerobic/F medium (BACTEC 9050 system; BBL Microbiology Systems, Cockeysville, Md.). Because of the inability of the patient to cooperate and his altered clinical status, no sample could be obtained from the lower respiratory tract. The patient was treated empirically with cefotaxime (1 g, three times per day) and netilmicin (100 mg, twice per day). He became apyretic within 48 h. The netilmicin was stopped after 7 days, and the cefotaxime was continued for three additional days. After 6 days of incubation, bacterial growth was detected (based on spectrophotometric CO2 detection) in one blood culture vial (aerobic culture) of the two sample sets by the BACTEC system. Gram staining revealed very small gram-negative bacilli. Subculture was performed with chocolate agar plates in a humidified atmosphere at 35°C with 5% CO2 and yielded transparent and mucoid colonies within 48 h. No growth was noted on bromocresol purple lactose agar or MacConkey agar plates. The organism was strictly aerobic and was negative in tests for oxidase, urease, and nitrate reductase and positive in tests for γ-glutamyl transpeptidase and tributyrine. There was no satellitism around X- and V-factor-impregnated disks on Mueller-Hinton agar. Cultural and biochemical characterization permitted us to differentiate the isolate from species belonging to the Haemophilus-Actinobacillus-Cardiobacterium-Eikenella-Kingella group. Latex agglutination tests were negative for Brucella but positive for Francisella tularensis (F. tularensis slide agglutination test; Difco). The identity of the organism as F. tularensis was subsequently confirmed by the detection of the 17-kDa lipoprotein gene of F. tularensis by PCR (27) (Fig. 1a) and the 43-kDa protein of F. tularensis by Western blotting (Fig. 1b) (2). Francisella-specific F5 and F11 primers (9) were used to PCR amplify the 16S ribosomal DNA fragment containing the nucleotide signature that differentiates F. tularensis biovar palaearctica (biovar B) from all the other members of the genus (1, 9). DNA sequences were determined with an ABI PRISM 377 DNA sequencer (Applied Biosystems dye terminator cycle sequencing kit). Alignment of these sequences with those of F. tularensis biovar palaearctica (strain L26086) demonstrated the presence of an A at nucleotide position 1153, indicating that our isolate belongs to biovar B (1, 9). In vitro susceptibility testing was performed using a disk diffusion technique on chocolate agar. Because of the absence of Francisella-specific standards, results were interpreted according to the guidelines of the Comité de l'Antibiogramme de la Société Française de Microbiologie for susceptibility testing of Haemophilus spp. (3). According to these criteria, our isolate was susceptible to aminoglycosides (gentamicin, netilmicin), tetracycline, and fluoroquinolones (pefloxacin, ciprofloxacin) and resistant to β-lactams (amoxicillin, piperacillin, cefotaxime). β-Lactamase was detected by using a nitrocefin-impregnated paper disk (Cefinase; BBL).
FIG. 1.
(a) PCR products of the F. tularensis 17-kDa lipoprotein gene (250 bp) (27). PCR products that migrated in a 1% agarose gel were stained with ethidium bromide. Lanes: 1 and 4, strain isolated from our patient; 2, negative control (Escherichia coli K-12); 3, positive control (F. tularensis clinical isolate; Institut Pasteur); 5, molecular size markers (φX174 RF DNA/HaeIII fragments of 1,358, 1,078, 872, 603, 310, 281, 271, 234, 194, 118, and 72 bp [from top to bottom]). (b) Western blots of whole-cell lysates from F. tularensis strains with antiserum against F. tularensis 43-kDa protein (2). Samples were separated by sodium dodecyl sulfate-polyacrylamide electrophoresis in a 12.5% gel, blotted onto nitrocellulose, probed with rabbit anti-43-kDa antibodies, and incubated with an anti-rabbit IgG conjugated with peroxidase. Lanes 1, F. tularensis clinical isolate (Institut Pasteur); 2, F. tularensis strain isolated from a tick (Institut Pasteur); 3, strain isolated from our patient; 4, molecular size protein standards (212, 107, 69, 43, 28.6, 19, and 15.5 kDa [from top to bottom]; Gibco).
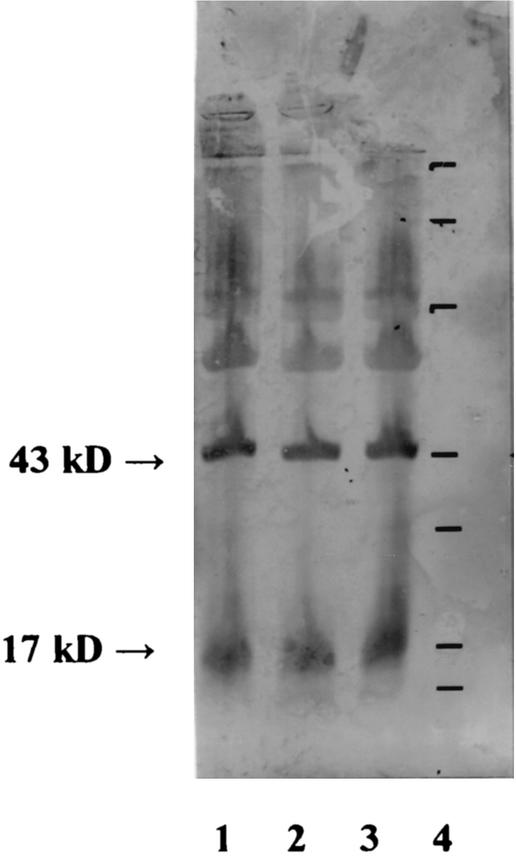
Two weeks after the completion of treatment, the patient remained asymptomatic and was discharged to return home. Two months later, at follow-up, the patient was well and a serum sample was collected. An agglutination serologic test (F. tularensis antigen; Difco) showed antibodies against F. tularensis at a titer of 1:1,280. Moreover, antibodies against the specific 17- and 43-kDa proteins were detected by Western blot analysis (2) by using cell lysates of the patient's isolate as well as of two other F. tularensis strains isolated from a human with adenitis and from a tick at the Institut Pasteur (Fig. 2).
FIG. 2.
Western blot analysis showing the antibody responses to 17- and 43-kDa F. tularensis proteins in the serum from our patient 2 months after the onset of infection. Supernatants of whole-cell sonicates from F. tularensis (lane 1, F. tularensis clinical isolate [Institut Pasteur]; lane 2, F. tularensis strain isolated from a tick [Institut Pasteur]; lane 3, strain isolated from our patient) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted with the patient's serum, and incubated with peroxidase-conjugated anti-human antibodies. Lane 4 contains molecular size protein standards (212, 107, 69, 43, 28.6, 19, and 15.5 kDa [from top to bottom]; Gibco).
Tularemia occurs primarily in the northern hemisphere. Three main biovars of F. tularensis are responsible for this zoonosis: the highly virulent biovar A (biovar tularensis), which is found mostly in North America and exceptionally in Europe (12); biovar B (palaearctica), which is not as virulent as biovar A and which is present in Europe, Asia, and, rarely, in North America; and biovar novicida, which has recently been considered a third biovar of F. tularensis (14, 30). The organism has been recovered from most animal species, including wild and domestic mammals and invertebrates (ticks, mosquitoes), and from their environments (dust, water). The clinical presentation depends on the organism and the route of transmission. It is almost entirely a rural disease. In Europe, hares, rabbits, and rodents are important in the ecology of the disease. Humans are usually contaminated by direct contact with ill or dead animals; thus, the most frequent clinical presentations are the ulceroglandular, the glandular, and the oculoglandular forms, which occur after the individual has handled affected animals. Pharynx infections after the ingestion of inadequately cooked meat (30) or contaminated water (13) have also been reported. Infections following tick bites are less common in Europe. F. tularensis can also cause lung infections after inhalation of infectious aerosols or systemic illness, such as typhoidal tularemia. Only 20 documented cases of bacteremia caused by F. tularensis have been reported worldwide, mainly in the United States (16 cases) (7, 10, 11, 14, 17, 19, 20, 22-25, 28) and more rarely in northern Europe (2 cases in Norway and 2 cases in Sweden) (13, 29). The blood isolates obtained in the United States belong mainly to biovar A and rarely to biovar B, while all the isolates obtained in northern Europe belong to biovar B. The higher virulence of the strains belonging to biovar A may also explain the severity of the cases of bacteremic tularemia reported in the United States (rhabdomyolysis, n = 5; renal failure, n = 4; shock, n = 3; death, n = 5). The presence of underlying immunodeficiencies (e.g., AIDS or liver transplantation) may favor the occurrence of tularemic bacteremia caused by strains belonging to biovar B, as in the two cases in the United States reported by Limaye and Hooper (20). However, in our case, as in all the other cases reported in northern Europe, there was no evidence of underlying disease. In 16 of the 20 published case reports, F. tularensis bacteremia was associated, as in our case, with underlying pneumonia (7, 11, 13, 17, 19, 20, 23, 24, 25, 28, 29). Among the other reported cases, two were characteristic of typhoidal tularemia (11, 14) and another was an oropharyngeal form (13). In 10 cases, a history of previous exposure to ticks or wild animals was found (10, 11, 13, 17, 20, 22-25). In four of the reported cases, neither decreased host defense factors nor a previous history of direct or indirect animal contact was noted (19, 25, 29). In our case, there was no history of direct animal contact. However, such contamination cannot be ruled out since the patient was blind and lived in a rural area. He might have had exposure to an animal and could not remember it. Nevertheless, occasional naturally occurring cases of tularemic pneumonia often arise after inhalation of contaminated dust. Thus, in our patient, the source of infection might also be environmental.
In the present case, the diagnosis of tularemia was fortuitous. This may be explained, at least partially, by the fact that tularemia occurs infrequently, resulting in a low index of diagnostic suspicion among clinicians and microbiologists. However, a diagnosis of bacteremic tularemia is important to consider because of the potential severity of this infection and because F. tularensis is also considered a dangerous potential biological weapon (4). Because of the high virulence of this microorganism, culture techniques are usually avoided for the diagnosis of F. tularensis infections, particularly in areas where biovar A is the most prevalent. Detection of F. tularensis in clinical specimens may also be achieved by using fluorescence-labeled antibodies. PCR-based methods have been proposed for use in the detection of F. tularensis DNA in human skin lesions or in the blood of experimentally infected mice (18, 21). These procedures permit a more rapid diagnosis but are usually performed in reference laboratories. Thus, in most clinical laboratories a diagnosis of tularemia relies on serological tests except for patients with bacteremia in whom tularemia may be diagnosed more or less fortuitously by the growth of F. tularensis in blood cultures. Serology permits a diagnosis only after 10 or more days from the onset of illness. F. tularensis may be isolated by culture in less than 10 days, but the sensitivity of this method may be low in the absence of a suggestive exposure history since the culture conditions may be inappropriate for this fastidious organism (5). Thus, culture of the organism, although growth may be delayed, remains a valuable means of diagnosis and is necessary for antibiotic susceptibility testing, biovar characterization (1, 8, 9, 16), and molecular epidemiological typing (6, 15). Isolation of F. tularensis from blood necessitates the use of highly nutritive culture media and a longer incubation time than the usual 5-day cycle used in many laboratories with continuous-monitoring devices. In our case, the blood culture was positive after 6 days of incubation. However, in other cases, an incubation period of 12 or even 21 days was necessary before positive blood cultures could be obtained (11, 20). A final blind subculture has also been recommended for cases for which there is a high index of suspicion (5, 26).
Thus, our case underlines the fact that bacteremic pneumonia due to F. tularensis may be unpredictable, and therefore prolonged incubation times for blood cultures in cases of unexplained pneumonia are indicated. Moreover, this is the first European case of bacteremic pneumonia due to F. tularensis occurring outside northern Europe.
REFERENCES
- 1.Anda, P., J. S. Del Pozo, J. M. Diaz Garcia, R. Escudero, F. J. Garcia Pena, M. C. Lopez Velasco, R. E. Sellek, M. R. Jimenez Chillaron, L. P. Sanchez Serrano, and J. F. Martinez Navarro. 2001. Waterborne outbreak of tularemia associated with crayfish fishing. Emerg. Infect. Dis. 7(Suppl. 3):575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevanger, L., J. A. Maeland, and A. I. Naess. 1989. Competitive enzyme immunoassay for antibodies to a 43,000-molecular-weight Francisella tularensis outer membrane protein for the diagnosis of tularemia. J. Clin. Microbiol. 27:922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Comité de l'Antibiogramme de la Société Française de Microbiologie. 2002. Communiqué 2002. Société Française de Microbiologie, Paris, France.
- 4.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russel, and K. Thonat. 2001. Tularemia as a biological weapon. Medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 5.Doern, G. V. 2000. Detection of selected fastidious bacteria. Clin. Infect. Dis. 30:166-173. [DOI] [PubMed] [Google Scholar]
- 6.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman, K. A., R. E. Enscore, S. L. Lathrop, B. T. Matyas, M. McGuill, M. E. Schriefer, D. Stiles-Enos, D. T. Dennis, L. R. Petersen, and E. B. Hayes. 2001. An outbreak of primary pneumonic tularemia on Martha's Vineyard. N. Engl. J. Med. 345:1601-1606. [DOI] [PubMed] [Google Scholar]
- 8.Forsman, M., G. Sandström, and B. Jaurin. 1990. Identification of Francisella species and discrimination of type A and type B strains of F. tularensis by 16S rRNA analysis. Appl. Environ. Microbiol. 56:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forsman, M., G. Sandström, and A. Sjöstedt. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38-46. [DOI] [PubMed] [Google Scholar]
- 10.Fulkerson, W., A. Spickard, and B. W. Davis. 1979. Opportunistic infections in chronic lymphocytic leukemia. South. Med. J. 72:1487-1488. [DOI] [PubMed] [Google Scholar]
- 11.Gries, D. M., and M. P. Fairchok. 1996. Typhoidal tularemia in a human immunodeficiency virus-infected adolescent. Pediatr. Infect. Dis. J. 15:838-840. [DOI] [PubMed] [Google Scholar]
- 12.Gurycova, D. 1998. First isolation of Francisella tularensis subsp. tularensis in Europe. Eur. J. Epidemiol. 14:797-802. [DOI] [PubMed] [Google Scholar]
- 13.Hoel, T., O. Scheel, S. H. G. Nordahl, and T. Sandvik. 1991. Water- and airborne Francisella tularensis biovar palaearctica isolated from human blood. Infection 19:348-350. [DOI] [PubMed] [Google Scholar]
- 14.Hollis, D. G., R. E. Weaver, A. G. Steigerwalt, J. D. Wenger, C. W. Moss, and D. J. Brenner. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson, A., I. Göransson, P. Larsson, and A. Sjöstedt. 2001. Extensive allelic variation among Francisella tularensis strains in a short-sequence tandem repeat region. J. Clin. Microbiol. 39:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson, A., A. Ibrahim, I. Göransson, U. Eriksson, D. Gurycova, J. E. Clarridge III, and A. Sjöstedt. 2000. Evaluation of PCR-based methods for discrimination of Francisella species and subspecies and development of a specific PCR that distinguishes the two major subspecies of Francisella tularensis. J. Clin. Microbiol. 38:4180-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser, A. B., D. Rieves, A. H. Price, M. R. Gelfand, R. E. Parrish, M. D. Decker, and M. E. Evans. 1985. Tularemia and rhabdomyolysis. JAMA 253:241-243. [PubMed] [Google Scholar]
- 18.Karhukorpi, E.-K., and J. Karhukorpi. 2001. Rapid laboratory diagnosis of ulceroglandular tularemia with polymerase chain reaction. Scand. J. Infect. Dis. 33:383-385. [DOI] [PubMed] [Google Scholar]
- 19.Ledoux, M. S. 2000. Tularemia presenting with ataxia. Clin. Infect. Dis. 30:211-212. [DOI] [PubMed] [Google Scholar]
- 20.Limaye, A. P., and C. J. Hooper. 1999. Treatment of tularemia with fluoroquinolones: two cases and review. Clin. Infect. Dis. 29:922-924. [DOI] [PubMed] [Google Scholar]
- 21.Long, G. W., J. J. Oprandy, R. B. Narayanan, A. H. Fortier, K. R. Porter, and C. A. Nacy. 1993. Detection of Francisella tularensis in blood by polymerase chain reaction. J. Clin. Microbiol. 31:152-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludmerer, K. M., and J. M. Kissane. 1984. Fever, leukopenia, acute renal failure and death in a 65-year-old man. Am. J. Med. 77:117-124. [DOI] [PubMed] [Google Scholar]
- 23.Overholt, E. L., W. D. Tigertt, P. J. Kadull, M. K. Ward, N. D. Charkes, R. M. Rene, T. E. Salzman, and M. Stephens. 1961. An analysis of forty-two cases of laboratory-acquired tularemia. Am. J. Med. 30:785-805. [DOI] [PubMed] [Google Scholar]
- 24.Pittmann, B., E. B. Shaw, Jr., and W. B. Cherry. 1977. Isolation of Francisella tularensis from infected frozen human blood. J. Clin. Microbiol. 5:621-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Provenza, J. M., S. A. Klotz, and R. L. Penn. 1986. Isolation of Francisella tularensis from blood. J. Clin. Microbiol. 24:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reary, B. W., and S. A. Klotz. 1988. Enhancing recovery of Francisella tularensis from blood. Diagn. Microbiol. Infect. Dis. 11:117-119. [DOI] [PubMed] [Google Scholar]
- 27.Sjöstedt, A., K. Kuoppa, T. Johansson, and G. Sandström. 1992. The 17-kDa lipoprotein and encoding gene of Francisella tularensis LVS are conserved in strains of Francisella tularensis. Microb. Pathog. 13:243-249. [DOI] [PubMed] [Google Scholar]
- 28.Tancik, C. A., and J. A. Dillaha. 2000. Francisella tularensis endocarditis. Clin. Infect. Dis. 30:399-400. [DOI] [PubMed] [Google Scholar]
- 29.Tärnvik, A., C. Henning, E. Falsen, and G. Sandström. 1989. Isolation of Francisella tularensis biovar palaearctica from human blood. Eur. J. Clin. Microbiol. Infect. Dis. 8:146-150. [DOI] [PubMed] [Google Scholar]
- 30.Wong, J. D., and D. S. Shapiro. 1999. Francisella, p. 647-651. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.