Abstract
The human skin papillomaviruses (HPVs) represent a group of ubiquitous viruses detected at a high prevalence in the normal skin of healthy adults. In the present study, we analyzed skin swab samples from babies during their first days of life and from infants at various ages up to age 4 years. Specimens from their parents and, for the newborn babies, environmental samples were also investigated. HPV DNA was already detected on the day of birth in samples from 2 of the 16 babies, and 45% of the samples from the babies were positive for HPV in the days following birth. Seventy-seven percent of the skin samples collected from the mothers were HPV DNA positive. The prevalence of HPV DNA among children from the ages of 1 month to 4 years varied between 50 and 70%. The HPV DNA sequences detected revealed a great diversity of genotypes and putative genotypes. Among 115 samples from 38 infants and 31 parents and 7 environmental samples, a total of 73 HPV types or putative types were isolated. Of these, 26 putative HPV types have not been described before. Our data suggest that asymptomatic HPV infections of normal skin are acquired very early in infancy and are caused by a great multiplicity of HPV types.
The human papillomaviruses (HPVs) can be classified by phylogenetic analysis into specific groups which parallel the tissue tropisms of the viruses (6, 19). Most of the cutaneous HPVs belong to groups B1 and B2, and a few are found in group E. More than 20 of the HPV types in group B1 have originally been isolated from patients suffering from the rare hereditary skin disease epidermodysplasia verruciformis (EV) (12, 13), and the HPV types of this group have therefore been referred to as EV-associated HPVs (16). While some HPV types cause common warts, a few of the HPV types in group B1, mainly HPV type 5 (HPV-5) and HPV-8, have been found in skin cancer lesions from EV patients (13, 16, 17) and immunosuppressed patients (3, 10).
In a previous report (1), we have shown that there is a high prevalence of HPV DNA (80%) in normal skin from healthy adults as well as a surprisingly high multiplicity of HPV genotypes and putative types, with all but one belonging to HPV group B1 or B2. Furthermore, there was an increased prevalence of HPV with age. In order to determine at what time in life skin papillomavirus infections are acquired, we analyzed normal skin samples from children of different age groups.
MATERIALS AND METHODS
Subjects and samples.
This investigation consisted of three parts: part a, a study of 1-month-old, 1-year-old, and 4-year-old children and one of the respective parents; part b, a pilot trial comprising two newborn babies; and part c, a study of 16 newborn babies, their mothers, and their environments.
Part a.
Samples were collected from 20 children in each of three age groups (ages 1 month, 1 year, and 4 years) at a child health clinic as well as from one of the respective parents. The prevalence of warts, eczema, and allergy in the child and the parent, as well as the number of siblings and attendance at nursery school or another school, was assessed by a questionnaire filled out by the parent.
Part b.
A sample was collected from one baby directly after birth and then once a week for 8 weeks. One sample from the mother and one sample from the father were also taken.
Samples were obtained from a second baby on the day of birth and then after 1, 3, 5, 8, 13, and 21 days. Samples were also collected from both parents.
Part c.
Sixteen babies and their mothers were sampled directly after birth (within 1 h) and then daily until the mother and the baby left the hospital (between two and six samples were obtained from each baby). An environmental sample was collected from the mother's bedside table on the maternity ward at the same time as the collection of samples from the child and mother.
All specimens for HPV analysis were collected from the skin on the individual's forehead by using cotton-tipped swabs. The swabs were soaked in saline solution (0.9% NaCl), drawn back and forth five times within a skin area of 3 by 5 cm, and then suspended in 1 ml of 0.9% NaCl. No DNA extraction of the specimens was carried out. The samples were stored at 4°C and analyzed within 72 h.
Informed consent was obtained from all parents in this study. The project was approved by the Ethics Committee of Lund University (LU 106-01).
PCR.
The presence of HPV DNA was determined by PCR. The final volume of 25 μl of PCR solution contained 0.75 μM (each) FAP59 and FAP64 primers (11), 0.2% bovine serum albumin (Sigma-Aldrich, Steinheim, Germany), 0.2 mM (each) deoxynucleoside triphosphate, 0.625 U of AmpliTaq Gold DNA polymerase, GeneAmp PCR buffer II, and 3.5 mM MgCl2 (Applied Biosystems, Warrington, United Kingdom), as well as 5 μl of the sample. Forty-five cycles of amplification were performed after denaturation for 10 min at 94°C. Each cycle consisted of 94°C for 90 s, 50°C for 90 s, and 72°C for 90 s.
In each batch of tests, H2O (without template) was included as a negative control, and samples containing HPV-11 or HPV-20 (both clinical samples) served as positive controls. As a PCR amplification control, all HPV DNA-negative samples were analyzed for human DNA by PCR (8).
HPV type determination.
The HPV types were determined by cloning and sequence analysis. The PCR amplicons were cloned into the pCR-script SK(+) cloning vector (Stratagene, La Jolla, Calif.). Between two and five clones per sample were sequenced with both the forward and the reverse primers (BigDye; Applied Biosystems) with a Perkin-Elmer 373A automated sequencer, and the complementary sequences were aligned with MacMolly computer software (version 3.8).
Comparison of the DNA sequences obtained with those of previously established HPV types and putative types was performed by using the BLAST server (http://www.ncbi.nlm.nih.gov/blast/).
The PCR products of the HPV DNA obtained span the L1 region from nucleotides 6044 to 6480, with the numbering being relative to that for the HPV-20 sequence. Since the PCR products obtained in this study represent only a part of the L1 gene, the isolated DNA sequences with less than 90% homology with any of the fully characterized HPV types or previously described HPV sequences have been designated new putative HPV types. The guidelines from the Papillomavirus Nomenclature Committee 1995 (14th Int. Papillomavirus Conf., Quebec City, Quebec, Canada, 1995) were followed to define the putative new HPV types found in this study (9).
Nucleotide sequence accession numbers.
The sequences of HPV types FA44, FA45, FA47, FA50, FA51, FA54, FA55, FA61, FA65 to FA72, and FA79 to FA88 have been submitted to GenBank with the following accession numbers: FA44, AY009877; FA45, AY009878; FA47, AY009881; FA50, AY009882; FA51, AY009885; FA54, AY009888; FA55, AY009886; FA61, AY040281; FA65, AY049756; FA66, AY049757; FA67, AY049758; FA68, AY049759; FA69, AY049760; FA70, AY049761; FA71, AF411918; FA72, AF411919; FA 79, AF455142; FA80, AF455143; FA81, AF455144; FA82, AF455145; FA83, AF455146; FA84, AF479248; FA85, AF479249; FA86, AF479250; FA87, AF479251; and FA88, AF479252.
RESULTS
Part a: samples from 1-month-, 1-year-, and 4-year-old children.
Twelve of 20 samples, 10 of 20 samples, and 14 of 20 of the samples from the 1-month-, 1-year-, and 4-year old children, respectively, were HPV DNA positive (Table 1). Eighty-five percent (51 of 60) of the samples from the parents were PCR positive for HPV DNA.
TABLE 1.
Prevalence of HPV in skin samples from 1-month-, 1-year-, and 4-year-old infants and their parentsa
| Age group | HPV prevalence (no. [%] of individuals)
|
|
|---|---|---|
| Children | Parents | |
| 1 mo | 12 (60) | 18 (90) |
| 1 yr | 10 (50) | 16 (80) |
| 4 yrs | 14 (70) | 17 (85) |
| All subjects (n = 60) | 36 (60) | 51 (85) |
Twenty samples were collected from each group of children and parents.
For 3 of the 12 HPV DNA-positive 1-month-old babies, the same HPV type or putative type (HPV-5, FA23, and FA55) was detected in the specimen from the child and the respective parent (Table 2). A total of 17 different HPV types or putative types were isolated from the individuals in this age group, and 4 of them (FA55, FA80, FA81 and FA82) have not been described previously. Eleven HPV types or putative types were isolated from the parents, two of which (FA55 and FA83) have not been described before.
TABLE 2.
HPV DNA in skin samples from 1-month-, 1-year-, and 4-year-old children and their parentsa
| Age group and child no. | HPV type or putative type
|
|
|---|---|---|
| Child | Parent | |
| 1 mo | ||
| 4 | HPV-5 | HPV-5 |
| 6 | HPV-9 | NEG |
| 8 | FA23, FA81 | FA23, HPV-vs73-1, HPV-38 |
| 10 | FA6 | HPV-20 |
| 12 | FA80, HPV-21, HPV-49 | NEG |
| 13 | FA31, FA35 | FA83, HPV-49 |
| 14 | FA82, FA20 | HPV-50 |
| 17 | FA10 | NEG |
| 18 | HPV 50 | HPV-12 |
| 19 | FA14 | HPV-37 |
| 20 | HPV-25 | NEG |
| 21 | FA55 | FA55 |
| 1 yr | ||
| 0 | HPV-80 | FA45, FA54 |
| 1 | HPV-12, FA38 | FA20, FA38 |
| 5 | FA70, HPV-vs92-1 | FA47, HPV-vs92-1 |
| 6 | FA65, FA51 | NEG |
| 7 | FA66, FA16.2 | FA54 |
| 10 | FA67 | FA31, FA38 |
| 12 | FA23 | FA23, HPV-vs43-1 |
| 27 | FA1, FA20 | NEG |
| 29 | HPV-8, HPV-38 | HPV-38 |
| 31 | HPV-5 | FA12, FA24 |
| 4 yr | ||
| 1 | FA15, HPV-vs20-4 | FA69, FA16 |
| 5 | FA14 | HPV-5, FA5, FA23 |
| 55 | FA68 | FA24, FA38 |
The putative HPV types not previously described are designated FA45, FA47, FA51, FA54, FA55, FA65 to FA70 and FA80 to FA83. NEG, negative for HPV DNA.
Among the 1-year-old children, four harbored one HPV type or putative HPV type (HPV-38, HPV-vs92-1, and FA23 and FA38), as did the corresponding parent. A total of 16 HPV types and putative types were isolated from the 1-year-old children, and 5 of these (FA51, FA65, FA66, FA67, and FA70) have not been described previously (Table 2). Ten different HPV types and putative types were detected from the parents of these children, and three of them (FA45, FA47, and FA54) have not been described previously.
The HPV types from 3 of the 13 HPV DNA-positive 4-year-old children were determined (Table 2). None of the samples from these children and their respective parent had any HPV type or putative type in common. One new putative HPV type (FA68) was found in a child, and another (FA69) was detected in one of the parents.
The answers to the questionnaires revealed no differences (regarding the appearance of warts, eczema, and allergy in the child and their parent and siblings) between the HPV DNA-positive and -negative children.
Part b: pilot trial with two newborn babies.
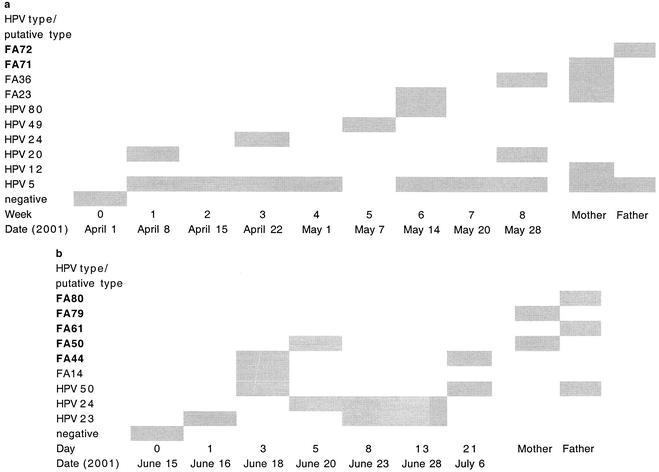
The specimen from the first baby, obtained directly after birth, was HPV DNA negative, while samples collected from weeks 1 to 8 were all HPV DNA positive (Fig. 1a). Altogether, seven HPV types or putative types (HPV-5, HPV-20, HPV-24, HPV-49, HPV-80, and FA23 and FA36) were detected in the samples from the baby. The sample from the mother contained HPV-5, HPV-12, FA23 and FA36, and a new putative type (FA71), while HPV-5 and a new putative type (FA72) were isolated from the father.
FIG. 1.
HPV DNA prevalence and HPV type distribution over time for two newborn babies and both of their parents. New putative types (FA44, FA50, FA61, FA71, FA72, FA79, and FA80) are indicated in boldface. (a) Samples were collected from the baby directly after birth and then once a week for 8 weeks. (b) Samples were obtained on the day of birth and then on days 1, 3, 5, 8, 13, and 21.
The second baby was HPV DNA negative directly after birth and then HPV DNA positive at days 1, 3, 5, 8, 13, and 21 (Fig. 1b). Six different HPV types or putative types (HPV-23, HPV-24, HPV-50, and FA14, FA44, and FA50) were detected in the samples from this baby. The putative types FA50 and FA79 were isolated from the mother; and the sample from the father contained HPV-50 and FA61 and FA80. Five new putative HPV types (FA44, FA50, FA61, FA79, and FA80) were isolated.
Part c: samples from newborn babies and their mothers and environmental samples.
HPV DNA was found in 2 of the 16 specimens from the newborn babies (day 0). Among the samples collected during the babies' second day of life, HPV DNA was detected in those from half of the babies (8 of 16 babies), and among the 17 samples collected from day 2 to day 5, HPV was detected in those from 7 babies (41%) (Table 3). The HPV DNA prevalences were 77% (38 of 49) among the samples from the mothers and 18% (9 of 49) among the environmental specimens. HPV DNA was detected in at least one sample from 11 of the 16 babies (69%; with between two and six samples collected from each baby). Four of the 11 HPV DNA-positive babies harbored at least one HPV type or putative HPV type in common with the mother (baby 2, putative type FA84; baby 12, putative type FA7; baby 13, HPV-65; and baby 15, HPV-12 and HPV-47). For two of these babies (baby 13 and baby 15), HPV-65 and HPV-47, respectively, were detected in the sample taken directly after birth. Two or more specimens from each of three of the babies were positive for DNA of the same HPV type or putative type (baby 6, FA35 and RTRX7; baby 13, HPV-65; and baby 15, HPV-12) (Table 3).
TABLE 3.
Longitudinal HPV DNA findings for skin samples from newborn babies, their mothers, and the environmenta
| Baby no. | Sexb | HPV type recoveredc
|
|||||
|---|---|---|---|---|---|---|---|
| Day 0
|
Day 1
|
||||||
| Baby | Mother | Environment | Baby | Mother | Environment | ||
| 1 | M | − | HPV-38, HPV 76 | − | − | HPV-38 | − |
| 2 | F | − | + | − | FA84 | FA84 | − |
| 3 | F | − | FA20, HPV-20, HPV-38 | − | FA5 | HPV-60 | − |
| 4 | F | − | − | − | − | FA37 | FA11 |
| 5 | F | − | + | − | − | + | − |
| 6 | M | − | FA35, HPV 5 | − | RTRX7 | HPV-5 | − |
| 7 | F | − | − | − | FA20 | − | − |
| 8 | F | − | HPV-8, HPV 20 | − | FA85 | FA86, HPV-8 | FA87, RTRX7, HPV-21 |
| 9 | F | − | + | − | − | + | + |
| 10 | M | − | + | − | − | + | − |
| 11 | M | − | + | + | − | + | − |
| 12 | M | − | FA7 | − | − | + FA7 | − |
| 13 | F | + HPV 65 | FA23, HPV-50 | − | HPV 65 | + HPV 65 | − |
| 14 | M | − | − | − | − | − | − |
| 15 | M | HPV-47 | HPV-12 | HPV-14D | FA88, HPV-12 | − | − |
| 16 | M | − | − | − | FA13 | FA5, HPV-20 | FA28 |
The new putative HPV types are designated FA45, FA61, and FA84 to FA88.
F, female; M, male.
−, no HPV DNA was recovered.
None of the HPV types or putative types found in the environmental samples was detected in the corresponding samples from the baby or the mother. HPV types and putative types HPV-14D, HPV-21, FA11, FA28, FA34, and FA87 were detected only in the environmental samples. HPV-5 was found in two different environmental samples as well as in three different samples from one mother, and RTRX7 was detected in an environmental sample and in two different samples from one baby. However, the environmental samples in which HPV DNA was found were not from the same rooms as the mother and the baby in question.
Altogether, seven new putative HPV types (types FA45, FA61, FA84, FA85, FA86, FA87, and FA88) that have not been described previously were isolated from the newborn babies, mothers, or the environment.
All HPV DNA-negative samples from children and parents were positive for human DNA by PCR, indicating that no PCR-inhibiting substances were present. Eighty-five percent (34 of 40) of the HPV DNA-negative environmental samples were positive for human DNA by PCR, and all nine HPV DNA-positive environmental samples tested positive for human DNA.
In total, the HPV types detected in 115 samples from 38 children and 31 parents plus 7 environmental samples were determined, resulting in the identification of 26 new putative HPV types (FA44, FA45, FA47, FA50, FA51, FA54, FA55, FA61, FA65 to FA72 and FA79 to FA88) together with 20 HPV types (HPV-5, HPV-8, HPV-9, HPV-12, HPV-14D, HPV-19, HPV-20, HPV-21, HPV-23, HPV-24, HPV-25, HPV-37, HPV-38, HPV-47, HPV-49, HPV-50, HPV-60, HPV-65, HPV-76, and HPV-80) and 27 previously characterized putative HPV types (FA1, FA5, FA6, FA7, FA10, FA11, FA12, FA13, FA14, FA15, FA16, FA20, FA23, FA24, FA27, FA28, FA31, FA34, FA35, FA36, FA37, FA38, HPV-vs20-4, HPV-vs43-1, HPV-vs73-1, HPV-vs92-1, and RTRX7).
DISCUSSION
The present study shows that HPV colonization of human skin occurs very early in life and with a great multiplicity of different HPV types. Actually, the acquisition of subclinical HPV infections seems to start the moment humans are born. Thus, the HPV DNA prevalence among children from the age of 1 month to 4 years ranged between 55 and 70%, which is close to the prevalence (about 75%) for their parents and other adults.
The samples obtained just after birth from the two babies in the pilot trial were both negative for HPV DNA, while all of the subsequent samples were HPV DNA positive, and one or more of the HPV types was found repeatedly. More than half of the HPV types detected in the specimens from the babies were also isolated from the respective parents, even although only one sample per parent was obtained and analyzed. It can be assumed that more HPV types or putative types would have been found to be common between the babies and their parents if additional clones had been analyzed or if more samples from the parents had been tested.
The conclusions drawn from the results for the two families in the pilot study were strengthened by the data for the 16 newborn babies and their mothers. Four of the 11 neonate-mother pairs shared the same HPV type or putative HPV type. The detection of HPV DNA in the environmental samples collected from bedside tables in a maternity ward demonstrates the efficiency of shedding, and thus the potential infectiousness of the HPVs from the skin. However, none of the HPV types detected in the environmental samples were detected in the specimens from the respective newborn babies or their mothers, while mothers and babies frequently shared the same HPV type. Thus, it is reasonable to believe that close skin contact with another individual whose skin is infected with HPV, such as between mother and child, is more efficient for transmission of the viruses.
It can be questioned whether the HPV DNA found in the normal skin of a person represents an established infection or if it merely has the character of a contamination from the environment, i.e., HPV produced in other individuals. By the method of analysis used in the present study, it is not possible to determine which of the two alternatives is correct in individual cases.
Another question is how long it takes for a subclinical HPV infection of the skin to establish itself before new virus particles are shed from the surface of the skin. This question is very interesting with regard to the newborn infants, for whom it can be assumed that they were not exposed to HPV of the skin until they were born. Studies with different HPV types and cultured cells showed that uptake and internalization of the virus particles take place within 2 h (14, 15). However, little is known at present about the time that it takes before the HPVs that enter the skin establish a subclinical HPV infection and shed new HPV particles. In our study, HPV DNA was isolated in two samples from babies collected directly after birth. These two samples probably do not represent true infections but, rather, virus shed from the mother, especially since the same HPV type was also isolated in one specimen from the respective mother.
The HPV types of the skin are widely spread among humans and give rise to persistent subclinical infections without causing warts or other lesions of the skin in healthy individuals (1, 2). Papillomaviruses have been found in a wide range of vertebrates (18) and are believed to have codeveloped with different species over long periods of time (4, 5, 7), and an interesting thought is that they have thereby given an evolutionary advantage to their hosts.
Table 3a.
| HPV type recoveredc
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 2
|
Day 3
|
Day 4
|
Day 5
|
||||||||
| Baby | Mother | Environment | Baby | Mother | Environment | Baby | Mother | Environment | Baby | Mother | Environment |
| − | HPV-20, HPV-23 | − | − | FA1, HPV-23, HPV-76 | HPV-5, HPV-14D | FA61 | FA27, HPV-23, HPV-38 | − | − | HPV-76 | − |
| − | FA84 | − | |||||||||
| − | − | FA34 | FA23 | FA37 | − | − | FA37 | − | |||
| − | FA34 | FA23 | FA37 | − | FA37 | − | |||||
| FA35, RTRX7, HPV-80 | HPV-5, HPV-8 | − | |||||||||
| − | − | − | − | − | − | ||||||
| − | HPV-20 | − | |||||||||
| FA7, HPV-19 | FA7, HPV-23 | − | |||||||||
| − | − | − | |||||||||
| − | HPV-12 | HPV-47 | HPV-5 | HPV-49 | FA45 | − | HPV-12 | HPV-12 | − | ||
Acknowledgments
We thank the pilot trial families, William, Helena, and Per in Dösjebro and Carolina, Irini, and Dag in Lund, for bringing us samples. Thanks are also due to Susanne Goldberg-Falck, Ingrid Holmberg, and Carina Prahl, nurses at the child health clinic Familjens Hus, Malmö, and to Kristina Larsson, midwife at the maternity ward at the University Hospital in Malmö, for collecting samples.
This work was supported by the Cancer Foundation of Malmö University Hospital and the Alfred Österlund Foundation.
REFERENCES
- 1.Antonsson, A., O. Forslund, H. Ekberg, G. Sterner, and B. G. Hansson. 2000. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 74:11636-11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astori, G., D. Lavergne, C. Benton, B. Hockmayr, K. Egawa, C. Garbe, and E. M. de Villiers. 1998. Human papillomaviruses are commonly found in normal skin of immunocompetent hosts. J. Investig. Dermatol. 110:752-755. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, R. J., L. M. Tieben, H. L. Smits, J. N. Bavinck, B. J. Vermeer, and J. ter Schegget. 1995. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J. Clin. Microbiol. 33:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard, H. U. 1994. Coevolution of papillomaviruses with human populations. Trends Microbiol. 2:140-143. [DOI] [PubMed] [Google Scholar]
- 5.Chan, S. Y., H. U. Bernard, M. Ratterree, T. A. Birkebak, A. J. Faras, and R. S. Ostrow. 1997. Genomic diversity and evolution of papillomaviruses in rhesus monkeys. J. Virol. 71:4938-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, S. Y., H. Delius, A. L. Halpern, and H. U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, S. Y., R. S. Ostrow, A. J. Faras, and H. U. Bernard. 1997. Genital papillomaviruses (PVs) and epidermodysplasia verruciformis PVs occur in the same monkey species: implications for PV evolution. Virology 228:213-217. [DOI] [PubMed] [Google Scholar]
- 8.Deragon, J. M., D. Sinnett, G. Mitchell, M. Potier, and D. Labuda. 1990. Use of gamma irradiation to eliminate DNA contamination for PCR. Nucleic Acids Res. 18:6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Villiers, E. M. 2001. Taxonomic classification of papillomaviruses. Papillomavirus Rep. 12:57-63.
- 10.de Villiers, E. M., D. Lavergne, K. McLaren, and E. C. Benton. 1997. Prevailing papillomavirus types in non-melanoma carcinomas of the skin in renal allograft recipients. Int. J. Cancer 73:356-361. [DOI] [PubMed] [Google Scholar]
- 11.Forslund, O., A. Antonsson, P. Nordin, B. Stenquist, and B. G. Hansson. 1999. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 80:2437-2443. [DOI] [PubMed] [Google Scholar]
- 12.Jablonska, S., J. Dabrowski, and K. Jakubowicz. 1972. Epidermodysplasia verruciformis as a model in studies on the role of papovaviruses in oncogenesis. Cancer Res. 32:583-589. [PubMed] [Google Scholar]
- 13.Jablonska, S., and S. Majewski. 1994. Epidermodysplasia verruciformis: immunological and clinical aspects. Curr. Top. Microbiol. Immunol. 186:157-175. [DOI] [PubMed] [Google Scholar]
- 14.Liu, W. J., Y. M. Qi, K. N. Zhao, Y. H. Liu, X. S. Liu, and I. H. Frazer. 2001. Association of bovine papillomavirus type 1 with microtubules. Virology 282:237-244. [DOI] [PubMed] [Google Scholar]
- 15.Muller, M., L. Gissmann, R. J. Cristiano, X. Y. Sun, I. H. Frazer, A. B. Jenson, A. Alonso, H. Zentgraf, and J. Zhou. 1995. Papillomavirus capsid binding and uptake by cells from different tissues and species. J. Virol. 69:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orth, G. 1987. Epidermodysplasia verruciformis, p. 199-243. In N. P. Salzman and P. M. Howley (ed.), The Papovaviridae, the Papillomaviruses, vol. 2. Plenum Press, New York, N.Y.
- 17.Orth, G., S. Jablonska, M. Jarzabek-Chorzelska, S. Obalek, G. Rzesa, M. Favre, and O. Croissant. 1979. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 39:1074-1082. [PubMed] [Google Scholar]
- 18.Sundberg, J. P. 1987. Papillomavirus infections in animals, p. 40-103. In K. Syrjänen, L. Gissmann, and L. G. Koss (ed.), Papillomaviruses and human disease. Springer-Verlag, Berlin, Germany.
- 19.van Ranst, M., J. B. Kaplan, and R. D. Burk. 1992. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J. Gen. Virol. 73:2653-2660. [DOI] [PubMed] [Google Scholar]