Abstract
Three rare human G12 strains were detected from diarrheic clinical samples of children (<8 months of age) in Calcutta during a routine surveillance study of rotaviral diarrhea in India. The VP7 genes of G12 strains and their products showed maximum homology (97 to 99% at the nucleotide level and 98% at the amino acid level, respectively) with those of two recently reported G12 strains (from the United States and Thailand) but lesser homology with those of prototype G12 strain L26.
Rotaviruses are the major cause of acute gastroenteritis in infants and young of a wide variety of mammalian and avian species (11). In developing countries, approximately 130 million infants are infected with rotaviruses and there are 800,000 annual deaths (5, 12). In group A rotaviruses, 15 VP7 G serotypes and 21 VP4 P genotypes have been reported for humans, animals, and birds (17). Though 10 G types and 10 P types from humans have been reported (15), clinically and epidemiologically the most important strains belong to the G1 to G4 serotypes with P[8] and P[4] genotypes (6, 14). On the other hand, some unusual types (G5, G8, and G9) and rare combinations of G and P types from different countries have also been reported (1, 2, 3, 4, 13, 16, 20). The human rotavirus G12 strains (L26 and L27) were first detected from Philippines in 1990 (19). After more than a decade, in 2002, human G12 strains Se585 (9) and T152 (15) from the United States and Thailand, respectively, were reported. In this study, we report the detection of three rare G12 strains of human rotaviruses in India.
As part of a routine surveillance study for diarrheal diseases, stool samples were collected from children below 4 years of age from B. C. Roy Children's Hospital and also from patients of all age groups admitted to the Infectious Diseases Hospital, Calcutta, India, in 2001. Stool samples were also collected from children with diarrhea from Assam Medical College, Dibrugarh, Assam, India; Capital Hospital and Municipality Hospital, Bhubaneshwar, Orissa, eastern India; and Post-Graduate Institute of Medical Education and Research, Chandigarh, northern India. A total of 454 samples were screened for rotaviruses by RNA electrophoresis as described earlier by Herring et al. (10). The fecal specimens were processed for extraction of rotavirus double-stranded RNA suitable for reverse transcription and amplification of the VP4 and VP7 genes as described previously (4). G and P typing of positive samples was carried out by nested and multiplex PCR with consensus and type-specific primers as described previously (4, 7, 8, 18, 21). The amplified products were purified with either the QIAquick gel extraction kit or the PCR purification kit (Qiagen, GmBH) in accordance with the manufacturer's instructions. Direct sequencing was carried out by using ABI PRISM BigDye terminator cycle sequencing kits (Applied Biosystems) with an automated DNA sequencer, the ABI PRISM 310 genetic analyzer (Applied Biosystems). The sequencing of the VP7 genes of all three isolates was repeated three times to eliminate sequencing error. Nucleotide sequences of the VP7 genes of group A rotaviruses deposited at the GenBank database along with the VP7 gene sequences of ISO 1, ISO 2, and ISO 5 were analyzed for the construction of a phylogenetic tree by the Bootstrap neighbor-joining tree method, with a random number generator seed of 500 and 5,000 bootstrap trials, using the CLUSTAL X software program.
In 2001, a total of 454 stool samples were screened for rotaviruses by RNA electrophoresis; there were 326, 104, 98, and 26 samples from diarrheic patients in Calcutta, Dibrugarh, Bhubaneshwar, and Chandigarh, respectively. Out of 161 rotavirus-positive samples, 126 samples were available in adequate quantity for VP7 genotyping. Reverse transcription-PCR results showed that the G types or P types of 21 samples could not be determined (data not shown). The full-length VP7 gene was amplified in the first PCR; however, there was no visible DNA band after the second round of multiplex PCR. A few such untypeable strains were selected for VP7 gene sequencing. After the sequencing, the partial sequences (32 to 1,034 bp) from three strains (ISO 1, ISO 2, and ISO 5) showed maximum homology with that from human prototype G12 strain L26 (19). The VP7 gene of group A rotaviruses is 1,065 nucleotides long and codes for a 326-amino-acid protein and showed maximum identity with that from the L26 rotavirus strain (Table 1). Low identities at the nucleotide and amino acid levels were observed (65 to 75% at the nucleotide level and 67 to 81% at the amino acid level) compared with all sequences from G1 to G15 prototype strains other than L26 (Table 1). These three G12 strains (ISO-1, ISO-2, and ISO-5) were collected from children less than 8 months of age admitted to the B. C. Roy Children's Hospital between January and April 2001. ISO-1 and ISO-2 rotaviruses were long-electropherotype strains, whereas ISO-5 was a short-electropherotype strain. The VP4 genotype of ISO-2 and ISO-5 was P[6]; on the other hand, ISO-1 was untypeable.
TABLE 1.
Nucleotide and amino acid homology of VP7 sequences of group A rotaviruses (G1 to G15 genotype-specific prototype strains) with three G12 strains
| VP7 genotype | Prototype strain | Homology (%)a with:
|
|||||
|---|---|---|---|---|---|---|---|
| ISO 1
|
ISO 2
|
ISO 5
|
|||||
| nt | aa | nt | aa | nt | aa | ||
| G1 | Wa | 75 | 77 | 75 | 77 | 75 | 77 |
| G2 | S2 | 72 | 75 | 72 | 76 | 73 | 76 |
| G3 | AU1 | 75 | 80 | 75 | 81 | 74 | 81 |
| G4 | ST3 | 72 | 75 | 72 | 75 | 73 | 75 |
| G5 | OSU | 75 | 79 | 75 | 79 | 75 | 79 |
| G6 | PA151 | 75 | 81 | 75 | 81 | 75 | 81 |
| G7 | Ty-1 | 65 | 67 | 65 | 67 | 66 | 68 |
| G8 | 69M | 72 | 77 | 73 | 78 | 72 | 78 |
| G9 | WI61 | 76 | 81 | 76 | 81 | 76 | 81 |
| G10 | Mc35 | 73 | 79 | 73 | 78 | 73 | 79 |
| G11 | YM | 73 | 80 | 73 | 80 | 74 | 80 |
| G13 | L338 | 75 | 78 | 75 | 78 | 75 | 78 |
| G14 | F123 | 74 | 79 | 75 | 79 | 75 | 79 |
| G15 | Hg18 | 70 | 73 | 70 | 73 | 71 | 73 |
| G12 | L26 | 90 | 92 | 90 | 92 | 90 | 92 |
| G12 | T152 | 97 | 98 | 97 | 98 | 97 | 98 |
| G12 | Se585 | 99 | 99 | 99 | 99 | 98 | 99 |
nt, nucleotide level; aa, amino acid level. Values ≥90% are in boldface.
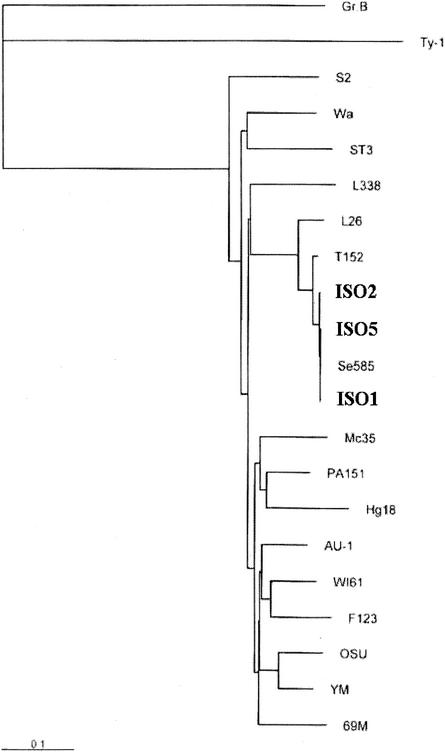
To understand the potential implication of these findings for the epidemiology of rotavirus infection, we sequenced the VP7 genes of some untypeable strains. Sequencing three untypeable rotavirus strains resolved them as G12. G12 was first detected in Philippines in 1990 (19) and in the United States (9) and Thailand (15) in 2002. So this is the first report of the isolation of three G12 strains in India. The percentages of homology at the nucleotide (90%) and amino acid (92%) levels with prototype G12 strain L26 were low. Similarly, the percentages of homology with other G1 to G15 type-specific strains of group A rotaviruses were very low. On the other hand, the percentages of identity with U.S. (Se585) and Thailand (T152) strains, the recently reported G12 strains, were very high (97 to 99%). The percentages of identity within the strains of same type are usually very high, i.e., >95% in rotaviruses (data not shown), but in this case the identity with the prototype L26 G12 strain was very low (92%). The phylogenetic analysis of VP7 gene sequences supports the above observation (Fig. 1). The L26 strain was in one branch, and all the other G12 strains clustered together in a separate branch. Though the strains detected and sequenced were very few, genetic variability within the G12 strain was very high in comparison to that for other type-specific strains of group A rotaviruses. Therefore, these strains may belong to two different subtypes of the same G12 type.
FIG. 1.
Phylogenetic tree of VP7 gene sequences representing different serotype-specific rotaviruses. The bar indicates the variation scale.
Detection of three strains with G12 specificity within 4 months from one hospital in Calcutta is very significant, as only four such strains were detected from all over the world prior to this detection, and this observation implies that G12 may be an emerging strain.
A complete molecular characterization of these untypeable strains and routine surveillance for detection of rotaviruses are essential to have a clear picture of strain diversity in different populations. This information is essential for designing a suitable rotavirus vaccine for the control of childhood diarrhea.
Nucleotide sequence accession numbers.
Nucleotide sequences of the VP7 genes of three G12 rotavirus strains were submitted to the GenBank database, and the accession numbers for ISO-1, ISO-2, and ISO-5 are AY206861, AY098669, and AF508734, respectively.
Acknowledgments
We thank S. C. Bhunia for excellent technical assistance for the detection of rotaviruses by RNA polyacrylamide gel electrophoresis.
We acknowledge the financial support of the Indian Council of Medical Research [ICMR], the Government of India and Japan International Cooperation Agency, Government of Japan. Soma Das was supported by a senior research fellowship from the Council of Scientific and Industrial Research, Government of India. V. Varghese, S. Chaudhuri, and P. Barman were supported by junior research fellowships from ICMR.
REFERENCES
- 1.Bon, F., C. Fromantin, S. Aho, P. Pothier, E. Kohli, and The AZAY Group. 2000. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J. Clin. Microbiol. 38:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cubitt, W. D., A. D. Steele, and M. Iturriza. 2000. Characterization of rotaviruses from children treated at London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 3.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. M. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das, S., A. Sen, G. Uma, V. Varghese, S. Chaudhuri, S. K. Bhattarcharya, T. Krishnan, P. Dutta, D. Dutta, M. K. Bhattarcharya, U. Mitra, N. Kobayashi, and T. N. Naik. 2002. Genomic diversity of group A rotavirus strains infecting humans in eastern India. J. Clin. Microbiol. 40:146-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Zoysa, I., and R. G. Feachem. 1985. Interventions for the control of diarrhoeal diseases among young children: rotavirus and cholera immunization. Bull. W. H. O. 63:569-583. [PMC free article] [PubMed] [Google Scholar]
- 6.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. I):S30-S36. [DOI] [PubMed] [Google Scholar]
- 7.Gouvea, V., N. Santos, and M. D. C. Timenetsky. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 32:1338-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parassar, R. I. Glass, J. R. Gentsch, and the National Rotavirus Strain Surveillance System. 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant strain (P2A[6], G12) and rare P3[9] strains related to bovine rotaviruses. Virology 294:256-269. [DOI] [PubMed] [Google Scholar]
- 10.Herring, A. J., N. F. Inglis, C. K. Ojeh, D. R. Snodgrass, and J. D. Menzies. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 12.Miller, M. A., and L. McCann. 2000. Policy analysis of the use of hepatitis B, Haemophilus influenzae type B, Streptococcus pneumoniae conjugate and rotavirus vaccine in national immunization schedules. Health Econ. 9:19-35. [DOI] [PubMed] [Google Scholar]
- 13.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palombo, E. A. 1999. Genetic and antigenic diversity of human rotaviruses: potential impact on the success of candidate vaccine. FEMS Microbiol. Lett. 181:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Pongsuwanna, Y., R. Guntapong, M. Chiwakul, R. Tacharoenmuang, N. Onvimala, M. Wakuda, N. Kobayashi, and K. Taniguchi. 2002. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. J. Clin. Microbiol. 40:1390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramachandran, M., B. K. Das, A. Vij, R. Kumar, S. S. Shambal, N. Kesari, H. Rawat, L. Bahl, S. Thakur, P. A. Woods, R. I. Glass, M. K. Bhan, and J. R. Gentsch. 1996. Unusual diversity of human rotavirus G and P genotypes in India. J. Clin. Microbiol. 34:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao, C. D., K. Gowda, and B. S. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi, K., F. Wakasugi, Y. Pongsuwanna, T. Urasawa, S. Ukae, S. Chiba, and S. Urasawa.1992. Identification of human and bovine rotavirus serotypes by polymerase chain reaction. Epidemiol. Infect. 109:303-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi, K., T. Urasawa, N. Kobayashi, M. Gorziglia, and S. Urasawa. 1990. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J. Virol. 64:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unicomb, L. E., G. Poddar, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, H., K. Taniguchi, F. Wakasugi, S. Ukae, S. Chiba, M. Ohseto, A. Hasegawa, T. Urasawa, and S. Urasawa. 1994. Survey on the distribution of the gene 4 alleles of human rotaviruses by polymerase chain reaction. Epidemiol. Infect. 112:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]