Abstract
Genotyping of human rotaviruses was performed on 312 rotavirus-positive samples collected from 2,205 young children with diarrhea in the Upper East District of Ghana, a rural community. Of the 271 (86.9%) rotavirus strains that could be VP7 (G) or VP4 (P) characterized, 73 (26.9%) were of G9 specificity. The predominant G9 genotype was G9P[8], which constituted 79.5% of all G9 strains detected, followed by G9P[6] (12.3%), G9P[10] (2.7%), and G9P[4] (1.3%). G9 strains with mixed P types constituted 2.7% of all G9 strains found in the study. All the G9P[8] strains had a long RNA electrophoretic pattern with VP6 subgroup II specificity. Four G9 isolates, GH1319, GH1416, GH3550, and GH3574, which were selected based on the abundance of stool material and were representative of the three electropherotypes observed, were cloned and sequenced. The Ghanaian isolates shared more than 98% sequence nucleotide homology with other G9 strains from the United States (US1205), Malawi (MW69), Brazil (R160), Japan (95H115), and Nigeria (Bulumkutu). However, they showed only 95% nucleotide homology with the Thai G9 strain Mc345. Phylogenetic analysis of the nucleic acid sequence revealed the existence of at least three clusters, with Ghanaian strains forming one cluster, Nigerian and Brazilian strains forming a second cluster, and U.S., Malawian, and Japanese strains forming a third.
Rotavirus is the most common cause of severe infantile diarrhea worldwide. According to an Institute of Medicine report, 130 million children are estimated to develop diarrhea due to rotavirus each year, resulting in more than 850,000 deaths (18). Rotaviruses are estimated to be responsible for 20% of diarrheal deaths and 6% of all diarrheal episodes in children under the age of 5 years (9).
Seven groups of rotaviruses (A to G) have been isolated and characterized (5). Group A rotaviruses have been identified as the major cause of severe dehydrating gastroenteritis among the young of humans and animals (20) and are responsible for more than 90% of all rotavirus infections. The virus has an inner capsid containing a genome of 11 segmented genes and an outer capsid formed by two structural proteins, VP4 (encoded by the 4th gene) and VP7 (encoded by the 7th, 8th, or 9th gene segment, depending on the strain). The VP7 (designated G because it is a glycoprotein) and VP4 (designated P because it is activated by protease cleavage) proteins are involved in cellular attachment, virus neutralization, and hemagglutination.
Neutralizing antibodies to VP4 and VP7 have been found to correlate with protective immunity. To date, 10 G types and 7 P types have been identified in humans. The most common serotypes found globally are serotypes G1, G2, G3, and G4 (10, 20). Thus, most current formulations of rotavirus vaccines have been designed based on these four most common VP7 serotypes. Recent rotavirus vaccines have shown high efficacy in clinical trials in developed countries, but they have produced conflicting results in a few trials in developing countries. None of these trials have been conducted in Africa or Asia, where such a vaccine will be needed most. Two clinical trials (initiated in Ghana and South Africa) were halted when the RotaShield vaccine was withdrawn because of a suspected possible association with intussusception (4, 6). Thus, questions concerning the immunogenicity and efficacy of rotavirus vaccines in Africa remain unanswered.
Several differences exist in the epidemiology of rotavirus infection between developing and developed countries. For instance, these differences include the earlier age of primary rotavirus infection, the high level of coinfection with other pathogens, and the presence of maternally transferred antibodies in developing countries. In addition, the circulation of atypical rotavirus strains bearing antigenic and genetic diversity may play an important role in the future successes of new rotavirus vaccine candidates. This becomes significant when one examines the increasing number of reports from the developing countries showing a wide range of diversity. For instance, studies in Malawi, Ghana, and South Africa, as well as in Brazil and Bangladesh, have confirmed the existence of significant genetic diversity among rotaviruses (2, 8, 13, 16, 25). This diversity may be due to independent reassortment of the 11 segmented genes (11) or to cross-species transmission, in view of the fact that the host range of rotaviruses is quite wide and hence may further influence variability (21).
After the initial report in 1987, rotavirus strains with G9 specificity seem to have remained dormant for 10 years, emerging as an important epidemiological strain in the last few years. G9 strains have been detected with increasing frequency in India, Brazil, Italy, France, Australia, Nigeria, Japan, and The Netherlands (22-26). The emergence of G9 strains as an important cause of gastroenteritis and the growing global divergence of rotavirus strains point up the need for continuation of surveillance of rotavirus strains in developing countries, especially in Africa, where these vaccines will have the greatest impact. The information generated by rotavirus strain surveillance is important for the development of more efficacious vaccines and also for the evaluation of these vaccines.
While strains with unusual combinations have been detected in previous studies in Ghana (2), strains with G9 serotype specificity were not detected. However, during 1999 and 2000, rotavirus G9 strains were seen to emerge and have been identified as the predominant genotype in diarrheic stools in Ghana, adding to the growing importance of these strains.
This study reports the identification of G9 strains in Ghana, sequence analysis of their VP7 genes, and the phylogenetic relatedness of the VP7 genes with other VP7 G9 genes reported.
MATERIALS AND METHODS
Study site.
A rotavirus surveillance system was set up in three health centers in the Kassena Nankana district of the upper east region of Ghana in August 1998 and was administered until July 2000 to estimate the burden of rotavirus-related diarrhea over a complete 2-year period during a Wyeth Lederle-sponsored rhesus rotavirus tetravalent vaccine trial. At the premature termination of the vaccination component of the study due to concerns over a possible link to intussusception, rotavirus surveillance was continued to the end of the study period and for a further 12 months. During the extension, stool samples were collected from children with diarrhea during the peak rotavirus months of November to February (1). No specimens were collected for the present study from any vaccine recipients.
Stool samples were collected from children below the age of 2 years who had diarrhea and attended the health clinics at Paga and Kassena East or the Navrongo War Memorial Hospital in the Upper East Region of Ghana. Basic demographic and clinical data for patients were collected, and disease outcomes were extracted from patients' hospital or clinic records. In all, 2,537 samples were collected between July 1997 and February 2002. Samples were stored at −20°C until processing.
Rotavirus antigen detection.
An enzyme-linked immunosorbent assay (DAKO, Cambridge, United Kingdom) for rotavirus group A was performed on a 10% suspension of fecal material in phosphate buffer as previously described (3).
Polyacrylamide gel electrophoresis.
Viral double-stranded RNA (dsRNA) was extracted concomitantly from the stool suspensions by the Trizol method (Gibco BRL, Grand Island, N.Y.) and precipitated with isopropanol. The extracted dsRNA was loaded onto a 10% polyacrylamide gel and electrophoresed overnight at 100 V by using a discontinuous Tris buffer system. A 3% stacking gel was employed to enhance the resolution of the gel. Gels were visualized by the silver-staining technique as described by Herring et al. (17).
RT-PCR.
RNA was extracted from 100 μl of 10% fecal suspensions in phosphate-buffered saline and purified with the RNaid kit (Bio 101, La Jolla, Calif.) (11). The extracted RNA was used for a seminested multiplex reverse transcription-PCR (RT-PCR) after specific priming with VP7 and VP4 consensus primer pairs (12, 13) as previously described (2). RT was carried out by incubation at 42°C for 30 min and was followed by 30 cycles of PCR (1 min of denaturing at 94°C, 2 min of annealing at 42°C, and 3 min of extension at 72°C) with a final extension cycle at 72°C for 7 min. The reaction mixture contained 1 μl of RT mixture, 1 μl each of 10 mM dATP, 10 mM dCTP, 10 mM dGTP, and 10 mM dTTP, and 1.5 U of Taq polymerase (Gibco BRL) in a 40-μl volume.
G and P typing PCR.
G typing was performed by using a seminested PCR method as described by Gouvea et al. (12). Briefly, in the first round, the full-length VP7 gene was obtained with primer pair sBeg9-End9 (12). This was followed by a second-round multiplex PCR, which incorporated primer End9, the G-type-specific primer RVG9, and primers aBT1, aCT2, aET3, aDT4, aAT8, and aFT9, which are specific for G types 1, 2, 3, 4, 8, and 9, respectively. For P typing, a similar method using a seminested PCR adapted from the method of Gentsch et al. (11) was used. In the first-round PCR, the full-length VP8* gene products were obtained with the consensus primers Con2 and Con3. This was then followed by a second-round typing PCR which incorporated primer Con3 and primers 1T-1, 2T-1, 3T-1, 4T-1, and 5T-1, which are specific for types P1A[8], P1B[4], P2A[6], P3[9], and P4[10], respectively. All PCR products were examined by gel electrophoresis in 1.2% agarose gels containing 4 μg of ethidium bromide/ml, and the G and P types were determined by the molecular weights of the amplicons.
Nucleotide sequencing.
The RNA electropherotype patterns of the G9 strains were analyzed and grouped. Representatives of each electrophoretic pattern were selected randomly and subjected to sequence analysis of the VP7 genes. The full-length VP7 gene product of the selected isolate was cut out from 2% agarose gels and purified with the QIAquick gel extraction kit (Qiagen, Chatsworth, Calif.) according to the manufacturer's instructions. The purified full-length gene product was cloned into the pGEM-T Easy Vector cloning system (Promega, Southampton, United Kingdom) as recommended by the manufacturers. Plasmids were recovered from the desired selected clones and digested with restriction enzymes to ascertain the size of the insert. Clones with the desired inserts were submitted to the DNA sequencing laboratory at the Department of Microbiology, University of Cape Town, Cape Town, South Africa, for sequencing on the ALFexpress automated sequencer with the M13 forward and reverse primers.
VP6 monoclonal antibodies.
The VP6 subgroup specificity of the rotavirus strains was determined by using the VP6 monoclonal antibodies developed by Greenberg et al. (14). These monoclonal antibodies against subgroup I rotaviruses (255/60) and subgroup II rotaviruses (631/9) have been used extensively in studies worldwide. The methods for their use have been described in detail elsewhere (15). In brief, microtiter plates were coated with the rabbit anti-rotavirus serum in a carbonate buffer and incubated with the rotavirus-positive stools. The monoclonal antibodies were added after overnight incubation and were detected by a horseradish peroxidase conjugate (TMB Enzymatic Kit; Roche, East Sussex, United Kingdom).
Nucleotide sequence accession numbers.
The nucleotide sequences representing the VP7 genes of the Ghanaian G9 strains have been submitted to the GenBank sequence database and assigned accession numbers AY211065 (GH1319), AY211066 (GH1416), AY211067 (GH3550), and AY211068 (GH3574).
RESULTS
Of the total of 2,205 stool and swab samples from young children with diarrhea examined for the presence of rotavirus antigens by enzyme-linked immunosorbent assay, 894 (40.5%) were found to be shedding rotaviruses. Three hundred fourteen of these samples, which were positive by polyacrylamide gel electrophoresis and hence had enough genomic dsRNA, were subjected to genotyping by PCR; of these, 271 samples (86.4%) could be assigned a G or P genotype. However, 43 samples could not be assigned either a G or a P genotype.
Epidemiology of G9 strains.
The distribution of circulating rotavirus VP7 types detected during the study in Navrongo, Paga, and East Kassena is shown in Table 1. Of a total of 271 human rotavirus samples that were assigned a VP7 G genotype, 73 (26.9%) were found to be of G9 specificity. The first rotavirus G9 strain was detected in October 1999 in a 7-month-old boy with diarrhea admitted to the Navrongo War Memorial Hospital. The majority of the G9 isolates were detected in 1999, when 57 samples were characterized as bearing G9 specificity. This represented 78.0% of the total G9 strains detected during the study and 21.0% of rotaviruses genotyped (Table 2). However, rotavirus strains of G9 specificity were observed less commonly in 2000 and 2001, when only 13 and 3 strains, respectively, were detected. This implies that there was a miniepidemic of G9 strains after they were first detected.
TABLE 1.
Rotavirus strains detected in Navrongo, Paga, and East Kassena during surveillance
| Site | No. of strains VP7 genotyped by PCR | Strain distributiona
|
||||||
|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G8 | G9 | UT | ||
| Navrongo | 137 | 6 | 33 | 14 | 0 | 4 | 49 | 31 |
| Paga | 88 | 2 | 28 | 21 | 0 | 3 | 16 | 18 |
| East Kassena | 56 | 4 | 19 | 10 | 0 | 4 | 8 | 11 |
| All sites | 271 | 12 | 67 | 43 | 0 | 8 | 73 | 33 |
No. of strains with each VP7 genotype. UT, untypeable.
TABLE 2.
Human rotavirus G9 genotypes detected in diarrheic stools in the Upper East Region (1998 to 2001)
| Genotype | No. of strains detected
|
Total | |||
|---|---|---|---|---|---|
| 1998 | 1999 | 2000 | 2001 | ||
| G9P[4] | 0 | 0 | 0 | 1 | 1 |
| G9P[6] | 0 | 4 | 4 | 1 | 9 |
| G9P[8] | 0 | 49 | 9 | 0 | 58 |
| G9P[10] | 0 | 2 | 0 | 0 | 2 |
| G9P[mix] | 0 | 2 | 0 | 0 | 2 |
| G9P[?] | 0 | 0 | 0 | 1 | 1 |
| Total | 0 | 57 | 13 | 3 | 73 |
The majority of the G9 strains, 49 of the 73 isolates (67.1%), were detected in stools collected from children attending the Navrongo War Memorial Hospital or from children living in the close vicinity of Navrongo. Sixteen (21.9%) of the children shedding rotavirus G9 strains were from Paga, which is 5 km away and close to the Burkina Faso border, and only 8 (11.0%) children were found to be shedding strains with G9 serotypes in East Kassena, which is 35 km from Navrongo and very rural. This geographic propensity for G9 strains in Navrongo, and the fact that the “outer” strains occurred slightly later in the season, also indicates that a miniepidemic occurred here and spread outwards.
While rotavirus G9 shedding was detected in all age groups, it was particularly common in younger children. Out of the 73 rotavirus strains detected, 38 (52.4%) were found in the stools of children under the age of 12 months, and 20 of these 38 strains (52.6%) were shed by children between the ages of 5 and 8 months. Furthermore, G9 was detected in samples from 30 (41%) children between the ages of 12 and 18 months. Rotavirus G9 shedding was found with less frequency in children above the age of 20 months.
Characterization of G9 strains.
VP7 and VP4 PCR genotyping revealed that the G9 strains were predominantly of the G9P[8] genotype (79.5% of all G9 strains detected), followed by G9P[6] (12.3%), G9P[10] (2.7%), and G9P[4] (1.3%), as shown in Table 2. Strains with mixed P types constituted 2.7% of all G9 strains found.
All the G9P[8] strains shared other characteristics and had a long RNA electrophoretic pattern with VP6 subgroup II specificity (Fig. 1). Strains bearing the G9P[6] genotype exhibited short electrophoretic patterns and belonged to subgroup I. One strain with a mixed G type but bearing a P[6] specificity, G9/1P[6], also had a long RNA electrophoretic pattern, indicating the increased level of genetic diversity present. Furthermore, a single strain with the unusual G9P[4] serotype and a long electrophoretic pattern was detected in 2000, again indicating the potential for genetic reassortment. One interesting aspect was the detection of a high proportion of nontypeable rotavirus strains during the peak of rotavirus G9 infections.
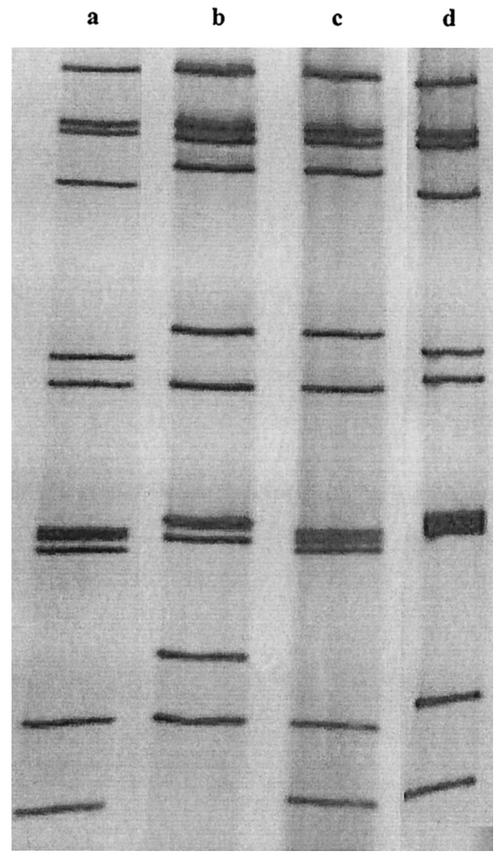
FIG. 1.
Electrophoretic patterns of sequenced Ghanaian G9 strains. Strains in lanes a (GH1416), c (GH3550), and d (GH3574) exhibit long patterns, and the strain in lane b (GH1319) exhibits a short pattern.
In total, three distinct RNA electrophoretic patterns were observed during the study period; most of the strains studied had short electropherotypes. The G9 samples were grouped according to electrophoretic pattern, and four samples, containing strains GH1319, GH1416, GH3550, and GH3574, were selected based on the abundance of stool material and because they were representative of the three electropherotypes.
Sequence analysis of the VP7 gene.
The VP7-encoding genes of these four strains were cloned and sequenced. Strains GH3550, GH1416, and GH3574 had long electropherotypes, while the G9 strain GH1319 had a short electropherotype. The Ghanaian isolates shared more than 98% nucleotide sequence homology with G9 strains from the United States (US1205), Malawi (MW69), Brazil (R160), Japan (95H115), and Nigeria (Bulumkutu). However, the Ghanaian G9 strains showed only 95% nucleotide sequence homology with the Thai strain Mc345, as shown in Table 3.
TABLE 3.
Deduced amino acid and nucleotide homologies of Ghanaian and other selected G9 strains
| Strain | % Homologya
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GH1319 | GH1416 | GH3550 | GH3574 | Bulumkutu | MW69 | R160 | US1205 | 95H115 | Mc345 | |
| GH1319 | 99.8 | 99.9 | 99.9 | 98.5 | 99.1 | 99.2 | 99.0 | 99.2 | 95.3 | |
| GH1416 | 99.4 | 99.7 | 99.9 | 98.3 | 98.9 | 99.1 | 98.8 | 99.0 | 95.1 | |
| GH3550 | 99.7 | 99.1 | 99.8 | 98.4 | 99.0 | 99.2 | 98.9 | 99.7 | 95.2 | |
| GH3574 | 99.7 | 99.7 | 99.4 | 98.4 | 99.0 | 99.2 | 98.9 | 99.1 | 95.2 | |
| Bulumkutu | 99.1 | 98.5 | 98.8 | 98.8 | 98.3 | 98.9 | 98.2 | 98.6 | 94.6 | |
| MW69 | 99.7 | 99.1 | 99.4 | 99.4 | 98.8 | 99.1 | 99.3 | 99.2 | 95.1 | |
| R160 | 99.4 | 98.8 | 99.1 | 99.1 | 99.1 | 99.1 | 99.0 | 99.3 | 95.5 | |
| US1205 | 99.7 | 99.1 | 99.4 | 99.4 | 98.8 | 99.4 | 99.1 | 99.0 | 95.1 | |
| 95H115 | 99.7 | 99.1 | 99.4 | 99.4 | 98.8 | 99.4 | 99.1 | 99.4 | 95.2 | |
| Mc345 | 95.1 | 94.5 | 99.8 | 99.8 | 98.2 | 94.8 | 94.5 | 94.8 | 94.8 | |
Blank rows divide the table diagonally into two sections, with nucleotide homologies in the top right section and amino acid homologies in the bottom left.
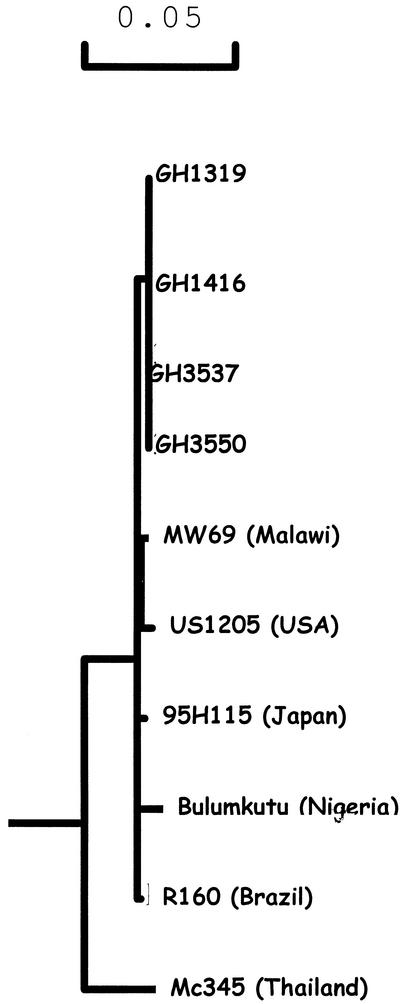
Phylogenetic analysis of the nucleic acid sequence revealed the existence of at least three clusters, with Ghanaian strains forming one cluster, Nigerian and Brazilian strains forming a second cluster, and U.S., Malawian, and Japanese strains forming a third (Fig. 2).
FIG. 2.
Phylogenetic relatedness of Ghanaian G9 strains to other strains. The Mc345, Bulumkutu, R160, MW69, US1205, and 95H115 sequences were obtained from the EMBL gene bank.
DISCUSSION
In this study, G9 strains were identified in Ghana for the first time during a longitudinal study conducted in a rural region. Of course, G9 strains are now recognized as an “emerging” rotavirus strain type globally (22-26) and could soon be ranked among the globally important circulating strains. However, in most locations where G9 strains have been identified, the studies allow limited description of the epidemiology and limited analysis of the strains, because most are point prevalence studies. In this report we identified substantial numbers of G9 strains (73 strains over a 3-year period), and this enables a more thorough investigation of the strains and their epidemiology.
First, because of the longitudinal nature of the surveillance period, we were able to map the emergence of G9 strains in this community. The first G9 strains were recovered from the stool of a 7-month-old female infant admitted to Navrongo War Memorial Hospital in November 1999. The origin of this first isolate is unknown. Within a few weeks, G9 strains were the predominant strains infecting the infants in Navrongo and had begun to spread to outlying towns. By December 1999, G9 strains were recovered from the diarrheal stools of infants in Paga (5 km away from Navrongo), and by early 2000, G9 strains had spread to East Kassena (35 km away).
We hypothesize from this observation that G9 strains emerged in Navrongo from an unknown source and spread by person-to-person transmission to the resident community and hence to the community at large. It is interesting to speculate whether the hospital supplies both (i) a point of origin for the new virus strain, possibly by reassortment of the mixed strains in the environment, facilitated by the increased viral population in a hospital setting, and (ii) the point of amplification of the “new strain” because of the influx of susceptible hosts.
The temporal occurrence of the G9 strains also indicates that the first strain was introduced into the community and was able to establish itself relatively quickly as the predominant strain. The tapering off of the G9 strains in 2001 may reflect the normal periodicity of some rotavirus strains in the community (20) or may indicate that the strains are not biologically competitive against the usual wild-type strains circulating. This remains to be determined and should be made evident by continued surveillance of rotavirus infection in this community in the future. In the United Kingdom, G9 strains were also reported to emerge in this sequential manner (9). The G9 strains were first seen at one center and then spread to other centers throughout the country.
However, the G9 strains infected infants at the normal target age of rotavirus infection in Navrongo (3), showing that they were competing successfully with the more usual circulating rotaviruses and were not targeting a different population group. In Nigeria (24) and in the United Kingdom (7), G9 strains were seen to infect children older than the normal peak age group for rotavirus infection, raising the speculation that the usual rotavirus strains conferred no protection against these strains, thus exposing an older “non-G9-immune” group of children to infection. In this study, with larger numbers of children infected, the G9 strains replaced the other, traditional strains in the primary rotavirus infection episode in young Ghanaian infants.
Characterization of the rotavirus G9 strains showed a variety of strains, although the predominant strain was a G9P[8] strain with VP6 subgroup II and a long RNA electropherotype. The second most common strain was a G9P[6] strain with VP6 subgroup I and a short profile. This is different from the findings of the study recently reported in Nigeria, where the majority of strains revealed a G9P[6]SGI short profile (24). The G9 strains were collected from these two adjacent countries during the same period.
These two constellations of markers seem to reflect the prevailing phenotypes of circulating G9 rotavirus strains (19, 22). In the United Kingdom, G9 strains were also classified into these two profiles, and it was interesting that the G9P[6]SGI short-profile strains were reported for older children (7), whereas the G9P[8]SGII long-profile strains seemed to occur in the usual target group of younger children (19). This epidemiological difference was also seen in West Africa. In the Nigerian study, G9P[6]SGI short-profile strains were identified for older symptomatic children, while in Ghana the G9P[8[SGII long-profile strains were detected in children of the usual target age group (this study). The difference in epidemiology may reflect the suggestion by Iturriza-Gomara and colleagues (19) that the G9P[8] strains have arisen by reassortment of the predecessor G9P[6] strain with the traditional VP7 serotype strains. Thus, the G9P[6] strain is introduced into an immunologically naïve population and is able to symptomatically infect older children. Further work needs to be done to examine this apparent difference in epidemiology.
VP7 cDNA sequence analysis of four of the Ghanaian strains, three with the G9P[8]SGII long pattern and one with the G9P[6]SGI short pattern, indicated >99% homology between strains. While no lineage or sublineage definition antigens, such as were observed in the United Kingdom study (19), were observed in the Ghanaian isolates, some amino acid insertions were seen to have occurred in antigenic region A (amino acids 87 to 101). The full implication of this insertion for serotyping using monoclonal antibodies is not known and will have to be investigated.
There is a high diversity of unusual rotavirus strains circulating in West Africa (A. D. Steele, African Rotavirus Network, personal communication), and strains with unusual G and/or P types continue to be detected with increasing frequency. One of the important emerging types of rotavirus strains is the G9 genotype. While these strains have emerged in miniepidemics, their prevalence continues to grow, as seen in other parts of the world. To effectively monitor the emergence and spread of these strains and of other strains with unusual G and P type combinations, it is imperative that surveillance of rotavirus infections be continued, especially in West Africa. This information will be crucial for the evaluation of future vaccines and will establish the prevalence of these strains.
Acknowledgments
This work was part of an RRTV rotavirus vaccine immunogenicity study and a rotavirus surveillance study supported by the DFID (London, United Kingdom) and WHO. The African Rotavirus Network provided support for the typing and sequencing of the strains isolated. G. E. Armah was supported by grants from the Claude Harris Foundation (Cape Town, South Africa), and M. D. Esona was supported by grants from DAAD, Bonn, Germany.
We thank the staffs of the collaborating laboratories for their support.
REFERENCES
- 1.Armah, G. E., J. A. Mingle, A. K. Dodoo, A. Anyanful, R. Antwi, J. Commey, and F. K. Nkrumah. 1994. Seasonality of rotavirus infection in Ghana. Ann. Trop. Paediatr. 14:223-229. [DOI] [PubMed] [Google Scholar]
- 2.Armah, G. E., C. T. Pager, R. H. Asmah, F. R. Anto, A. R. Oduro, F. Binka, and A. D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 3.Asmah, R. H., J. Green, G. E. Armah, C. I. Gallimore, J. J. Gray, M. Iturriza-Gomara, F. Anto, A. Oduro, F. N. Binka, D. W. Brown, and F. Cutts. 2001. Rotavirus G and P genotypes in rural Ghana. J. Clin. Microbiol. 39:1981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass, D. M. 2000. Rotavirus vaccinology: good news and bad news. J. Pediatr. Gastroenterol. Nutr. 30:10-11. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, R. F. 1996. Natural history of human rotavirus infection. Arch. Virol. Suppl. 12:119-128. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Withdrawal of rotavirus vaccine recommendation. Morb. Mortal. Wkly. Rep. 48:1007. [PubMed] [Google Scholar]
- 7.Cubitt, W. D., A. D. Steele, and M. Iturriza. 2000. Characterization of rotaviruses from children treated at a London hospital during 1996: Emergence of strains G9P2A[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 8.Cunliffe, N. A., J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and C. A. Hart. 1999. Rotavirus G and P types in children with acute diarrhoea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J. Med. Virol. 57:308-312. [PubMed] [Google Scholar]
- 9.de Zoysa, I., and R. G. Feachem. 1985. Interventions for the control of diarrhoeal diseases among young children: rotavirus and cholera immunisation. Bull. W. H. O. 63:569-583. [PMC free article] [PubMed] [Google Scholar]
- 10.Estes, M. 1996. Rotaviruses and their replication, p. 1625-1655. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 11.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouvea, V., and N. Santos. 1999. Rotavirus serotype G5: an emerging cause of epidemic childhood diarrhoea. Vaccine 17:1291-1292. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg, H. B., J. Valdesuso, K. van Wyke, K. Midthun, M. Walsh, V. McAuliffe, R. G. Wyatt, A. R. Kalica, J. Flores, and Y. Hoshino. 1983. Production and preliminary characterisation of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J. Virol. 47:267-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg, H. B., V. McAuliffe, J. Valdesuso, R. G. Wyatt, J. Flores, A. R. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup antigen of rotavirus using monoclonal antibodies. Infect. Immun. 39:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gusmao, R. H., J. D. Mascarenhas, Y. B. Gabbay, Z. Lins-Lainson, F. L. Ramos, T. A. Monteiro, S. A. Valente, U. Fagundes-Neto, and A. C. Linhares. 1999. Rotavirus subgroups, G serotypes, and electrophoretypes in cases of nosocomial infantile diarrhoea in Belem, Brazil. J. Trop. Pediatr. 45:81-86. [DOI] [PubMed] [Google Scholar]
- 17.Herring, A. J., N. F. Inglis, C. K. Ojeh, and D. R. Snodgrass. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Institute of Medicine. 1986. The prospect of immunisation against rotavirus, p. D-13-1-D-13-2. In New vaccines for development: diseases of importance in developing countries, 2nd ed. National Academy Press, Washington, D.C.
- 19.Iturriza-Gomara, M., W. D. Cubitt, A. D. Steele, J. Green, D. G. Brown, U. Desselberger, and J. J. Gray. 2000. Characterisation of rotavirus G9 strains isolated in the UK between 1995 and 1998. J. Med. Virol. 61:510-517. [DOI] [PubMed] [Google Scholar]
- 20.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 21.Nakagomi, O., and T. Nakagomi. 1993. Interspecies transmission of rotaviruses studied from the perspective of genogroup. Microbiol. Immunol. 37:337-348. [DOI] [PubMed] [Google Scholar]
- 22.Oka, T., T. Nakagomi, and O. Nakagomi. 2000. Apparent re-emergence of serotype G9 in 1995 among rotaviruses recovered from Japanese children hospitalised with acute gastroenteritis. Microbiol. Immunol. 44:957-961. [DOI] [PubMed] [Google Scholar]
- 23.Ramachandran, M., C. D. Kirkwood, L. Unicomb, N. A. Cunliffe, R. L. Ward, M. K. Bhan, H. F. Clark, R. I. Glass, and J. R. Gentsch. 2000. Molecular characterization of serotype G9 rotavirus strains from a global collection. Virology 278:436-444. [DOI] [PubMed] [Google Scholar]
- 24.Steele, A. D., L. Nimzing, I. Peenze, M. C. De Beer, A. Geyer, I. Angyo, and N. E. Gomwalk. 2002. Circulation of the novel G8 and G9 rotavirus strains in Nigeria in 1998/1999. J. Med. Virol. 67:608-612. [DOI] [PubMed] [Google Scholar]
- 25.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Widdowson, M. A., G. J. van Doornum, W. H. van der Poel, A. S. de Boer, U. Mahdi, and M. Koopmans. 2000. Emerging group-A rotavirus and a nosocomial outbreak of diarrhoea. Lancet 356:1161-1162. [DOI] [PubMed] [Google Scholar]