Abstract
To study humoral and cellular immunity against human parechovirus type 1 (HPEV1), the viral capsid proteins VP0, VP1, and VP3 were expressed and purified as glutathione S-transferase (GST)-tagged recombinant proteins. The fusion proteins were used to raise antisera in rabbits. VP0 and VP1 antisera specifically detected HPEV1-infected cells in culture by immunoperoxidase staining and immunofluorescence. Furthermore, antisera against the VP0 and VP1 proteins had neutralizing effects against HPEV1 infection. When the HPEV1 antibody titers of 20 adults and 55 children were determined by a microneutralization test, the prevalence of HPEV1 antibodies in the adult population was 96%, while 50% of children were seropositive. Selected sera were used to evaluate HPEV1 fusion proteins as antigens in an enzyme immunoassay. The VP3 capsid protein appeared to be suitable for the purpose, with specificity of 100% and sensitivity of 96% compared to the neutralization test. Furthermore, T-cell responses to the purified HPEV1 and HPEV1 capsid fusion proteins were studied in 20 adults. Sixty percent of the subjects had T-cell proliferation responses to purified HPEV1, and 90% of the subjects also had positive T-cell responses to at least one of the GST capsid proteins.
Human parechovirus type 1 (HPEV1), the type member of the genus Parechovirus, is a common pathogen with a worldwide distribution (5, 9). Infections occur early in life, and seropositivity increases rapidly after 1 year of age. The most common clinical manifestations are gastroenteritis and respiratory infections, while central nervous system symptoms are more rare than in enterovirus infections. Parechoviruses form one of the nine genera in the family Picornaviridae and include two human pathogens, HPEV1 and HPEV2, and a rodent virus, Ljungan virus (12). Like other picornaviruses, parechoviruses are small nonenveloped particles containing an RNA genome with positive polarity and a protein capsid consisting of 60 copies of each of the capsid proteins VP1 to VP4 (8). However, parechoviruses have distinctive molecular properties compared to other picornaviruses, including differences in capsid protein processing (VP0 is not cleaved to VP2 and VP4) and functions of the nonstructural proteins (3, 7, 21).
B-cell antigenic epitopes of some picornaviruses, like polioviruses and rhinoviruses, are relatively well known (22). The major antigenic sites in polioviruses exist in VP1 and, to a lesser extent, in VP2 and VP3 (15). There is a linear epitope in VP1, while amino acids participating in two conformational epitopes are scattered along the capsid proteins. It has been shown by using rabbit antibodies and synthetic peptides that in HPEV1 one immunogenic region resides in VP0 and another resides at the C terminus of VP1 (10). The latter region contains an RGD motif which participates in recognition of αVβ3 integrin on the host cell surface (21).
Cellular immunity and T-cell epitopes of enteroviruses, like coxsackie B viruses (CBV) and polioviruses, have been studied by proliferation tests, and most T-cell epitopes in enteroviruses tend to localize in the VP1 and VP3 proteins (6, 13). T cells can recognize common epitopes in enteroviruses (2), and these cross-reactive epitopes localize in the VP2 and VP3 proteins and, to a lesser extent, in VP1. It was recently shown that T-cell recognition sites on CBV type 4 (CBV4) mainly locate in the conserved regions of the VP2, VP3, and VP4 proteins (14). T-cell lines originally selected by CBV4 antigen also recognized equivalent conserved regions in other enteroviruses.
In order to cast light on the immunogenicity of HPEV1, the capsid proteins of the virus were expressed as fusion polypeptides in bacteria and their characteristics as B- and T-cell antigens were tested using different assays. Furthermore, the diagnostic potentials of the reagents obtained were evaluated.
MATERIALS AND METHODS
Molecular cloning.
HPEV1 (strain Harris) cDNA (8) was used for amplification of HPEV1 capsid protein VP0, VP1, and VP3 genes by PCR. The specific primers included additional restriction enzyme cleavage sites and stop codons for cloning and expression. The part of the genome coding for VP0 was amplified with the sense primer 5′-GGGGAATTCATGGAGACAATTAAG-3′ and the antisense primer 5′-GGGCTCGAGTCAATTATCATATATGTT-3′, and the gene coding for VP3 was amplified with the primers 5′-GGGGAATTCGCACCAAATGGTAAA-3′ and 5′-GGGCTCGAGTCACTGGAATGTAACAAC-3′ (the stop codons are shown in boldface, and the EcoRI and XhoI cloning sites are underlined). To generate the cDNA encoding VP1, the primers 5′-GGGATCCAATTCATGGGGTTCACAG-3′ and 5′-GCGGCCGCTCAATATGGACTCTGATTTGT-3′ were used (the stop codon is shown in boldface, and the BamHI and NotI sites are underlined). The lengths of the amplicons VP0, VP3, and VP1 were 866, 758, and 701 bp, respectively (Fig. 1A). The PCR products of expected sizes were excised from agarose gels and eluted with a QIAquick Gel Extraction kit (Qiagen). The amplicons were cloned into the pGEX 4T-1 (VP0 and VP3) or pGEX 4T-2 (VP1) expression vector (Pharmacia) and subsequently analyzed by restriction enzyme digestion and sequencing to confirm their identities.
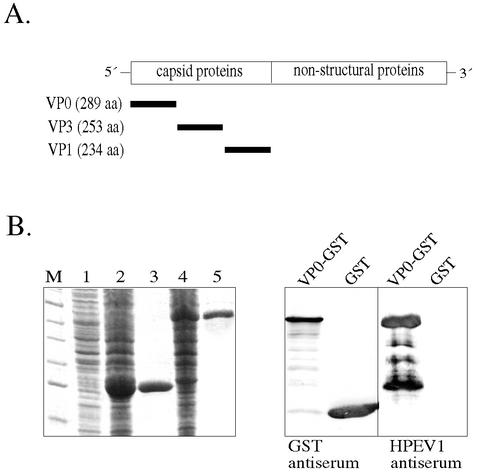
FIG. 1.
(A) Genome localization of the genes for the HPEV1 capsid proteins (VP0, VP3, and VP1) expressed as GST fusion polypeptides. (B) SDS-PAGE (left) and immunoblotting (right) analysis of the VP0-GST fusion protein. Lanes: M, molecular size markers; 1, uninduced E. coli cells; 2, induced E. coli cells expressing GST; 3, purified GST (26 kDa); 4, induced E. coli cells expressing VP0-GST; 5, purified VP0-GST (58 kDa). After separation by SDS-PAGE, the proteins were blotted onto a nitrocellulose filter and detected by primary antibodies (GST antiserum diluted 1:3,000 and HPEV1 antiserum diluted 1:1,000) and secondary antibody (alkaline phosphatase-conjugated anti-rabbit IgG). The samples analyzed in the right panel are VP0-GST and GST.
Expression and purification of recombinant HPEV1 proteins.
The recombinant plasmids and control pGEX vectors were transformed into Escherichia coli BL21 cells. The transformed bacteria were grown to an optical density (OD) of 0.6 in Luria-Bertani medium, and protein expression was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside; Promega). After induction for 5 h at 25°C (VP0 and VP3) or at 37°C (VP1), the cells were collected and stored at −20°C until protein purification was performed. The bacterial pellets were first suspended in a buffer containing 0.5 mM phenylmethylsulfonyl fluoride (Sigma), 10 μg of aprotinin (Sigma)/ml, and 0.5 mg of lysozyme (Sigma)/ml and then incubated on ice for 30 min. Lysis was completed by sonication (Soniprep 150; Sanyo), and the lysate was stirred in 1% Triton X-100 (BDH Laboratory Supplies) for 30 min. The supernatant fraction of the lysate was separated by centrifugation (Sorvall ss-34; 13,000 rpm; 30 min) and incubated with Glutathione Sepharose 4 Fast Flow-Matrix at 4°C overnight. The proteins were eluted with 10 mM glutathione in 50 mM Tris-HCl, pH 8.0, and finally dialyzed against phosphate-buffered saline (PBS) in Slide-A-Lyzer cassettes (Pierce) at 4°C overnight.
Production of rabbit antisera.
The capsid fusion proteins were used to raise polyclonal antisera in rabbits. Rabbits were immunized subcutaneously with 100-μg doses of protein in Freund's complete adjuvant followed by two booster doses in incomplete adjuvant at 30- and 14-day intervals. The sera were collected 2 weeks after the last immunization. HPEV1 antiserum against purified virus was previously produced in rabbits (10). Glutathione S-transferase (GST) antiserum was a kind gift from V. Hukkanen, Department of Virology, University of Turku.
Immunostaining of HPEV1-infected cells.
A method used by Joki-Korpela et al. (10) was applied for immunoperoxidase staining. A-549 cells were infected with HPEV1, and the cells were fixed with cold methanol at 12 and 16 h postinfection Primary capsid antibodies were added at 1:100 dilution, and antisera against whole viruses were added at 1:400 dilution. Peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) (Dako) was used as a secondary antibody at 1:200 dilution.
For immunofluorescence staining, A549-4A cells (cloned A549 cells originally obtained from the American Type Culture Collection) were infected with HPEV1 and fixed at 16 and 24 h postinfection. Primary antibodies were used at 1:100 dilution, and fluorescein isothiocyanate-conjugated donkey antibodies against mouse (Jackson Immunoresearch) or tetramethyl rhodamine isothiocyanate-conjugated donkey antibodies against rabbit (Jackson Immunoresearch) were applied as secondary antibodies at a dilution of 1:250.
Plaque neutralization assay.
Rabbit antisera against the fusion proteins were diluted 1:100 and anti-HPEV1 antiserum was diluted 1:1,000 in Hank's balanced salt solution containing 0.6% fetal calf serum. The antisera were incubated with 100 PFU of HPEV1 for 1 h at 37°C and added to confluent monolayers of A549-4A cells on six-well plates. After 30 min of incubation at 37°C in a 5% CO2 atmosphere, the virus-antibody solution was removed and the cells were overlaid with 0.5% carboxymethyl cellulose. After 2 days of incubation at 37°C, the carboxymethyl cellulose was removed and the cells were stained with crystal violet. The neutralization effect was calculated as the proportion between the number of plaques in the neutralized cells and the number of plaques in the unneutralized control cells.
Determination of antibody levels in human sera by microneutralization.
HPEV1 antibody levels in the serum samples of 20 adults and 55 young children (≤6 years) were determined by a microneutralization test. Fourfold dilutions starting from 1:10 on microtiter plates (Nunclon 96-well Microtest plate) were tested by adding 150 PFU of HPEV1 to the wells and incubating them for 2 h at 37°C; 30,000 A-549 cells per well were then added, and the plates were incubated for 6 days before the cells were stained with crystal violet and the virus growth was observed. Titers were determined as the highest dilutions of the serum samples capable of neutralizing the virus.
IgG enzyme immunoassay (EIA).
Ninety-six-well plates (Maxisorp; Nunc International) were coated with 100 ng of purified fusion proteins or HPEV1 in 50 mM NaHCO3, pH 9.6, overnight at room temperature and blocked with 0.1% bovine serum albumin in PBST (PBS containing 0.1% Tween) for 1 h at room temperature. The control wells were similarly coated with the control cell lysate (expressing GST). Human sera were diluted in a sample buffer containing 2% control cell lysate in 1% bovine serum albumin and 1% fetal calf serum-PBST and preincubated at 37°C for 1 h prior to their application to the coated plates in triplicate. As a secondary antibody, horseradish peroxidase-conjugated anti-human IgG (Dako) was used at a dilution of 1:6,000. Reactivity was measured by adding the substrate chromogen solution for 20 min at room temperature and measuring the OD at 492 nm in a spectrophotometer (Multiscan EX; Labsystems). The net OD was obtained by subtracting the OD for the E. coli lysate control from that of the test with the recombinant antigen.
Lymphocyte proliferation assay.
The method described by Juhela et al. (11) was used in the proliferation studies. Peripheral blood mononuclear cells were incubated in quadruplicate wells with HPEV1 capsid antigens (VP0, VP3, and VP1) or purified HPEV1 in 96-well microtiter plates for 6 days. The GST protein, prepared similarly to the capsid antigens, was included as a control antigen. Furthermore, CBV4 (strain J.V.B. Benschoten) and poliovirus type 1 (strain Sabin) were used. Tritiated thymidine (2 μCi/ml; Amersham) was added 18 h before the cells were harvested on glass fiber filters using a Tomtec (Orange, Conn.) 93 Mach III manual harvester. The incorporated radioactivity was measured with a Micro-Beta scintillation counter (Wallac, Turku, Finland). Stimulation indices (SI) for purified viruses were calculated by dividing the counts per minute of the virus-stimulated cells by the counts per minute of the unstimulated control cells. Similarly, the counts per minute of the fusion protein-stimulated cells were compared to the counts per minute of the GST-stimulated control cells. An SI value of ≥3 was considered a positive response.
RESULTS
Production of recombinant parechovirus proteins.
HPEV1 capsid proteins were expressed as GST fusion proteins in E. coli and affinity purified using glutathione-Sepharose. The expression levels of HPEV1 fusion proteins in bacterial cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The VP0 (Fig. 1B) and VP1 fusion proteins were the predominant products in the bacterial cells, whereas the level of VP3-GST expression was lower under the conditions used. A major fraction of the capsid proteins could be purified in soluble form from the supernatants (yield, 2 to 4 mg of protein from 400 ml of E. coli culture). The GST protein appeared exclusively in soluble form when expressed alone. The solubility of the capsid proteins could be further improved in pilot optimization procedures by decreasing the induction temperature (e.g., to 25°C) and by avoiding oversonication of the cells. The purity of the proteins was confirmed by visualizing single bands in SDS-PAGE. The identities of the recombinant antigens were further tested by immunoblotting (e.g., VP0 [Fig. 1B]). All the recombinant capsid proteins were similarly recognized by the rabbit anti-HPEV1 antiserum and by antiserum against GST. Furthermore, antisera against VP0-GST, VP1-GST, and VP3-GST recognized the respective antigens (data not shown).
However, some partial degradation of the proteins was observed in immunoblotting when rabbit anti-HPEV1 (Fig. 1B) or specific capsid protein antiserum was used. An additional band of 50 kDa was copurified with VP1-GST, but it was not reactive in immunoblotting with rabbit anti-HPEV1 or anti-VP1-GST antiserum. Purified proteins were easily degraded after freezing-thawing, and therefore, they were divided into aliquots and stored at −70°C.
Immunostaining of HPEV1-infected cells.
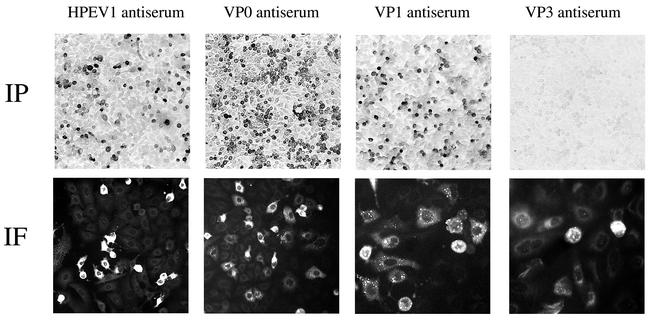
Capsid protein antisera were assayed by immunofluorescence and immunoperoxidase staining for identification of HPEV1 in cell culture. Rabbit antisera against VP0 and VP1 detected HPEV1-infected cells, as did the antisera against purified HPEV1 virus in immunoperoxidase staining (Fig. 2), whereas antisera against VP3 did not. None of the capsid protein antisera reacted with the two enteroviruses tested, CBV3 and echovirus type 1 (EV1) (data not shown). Similarly, no reactivity was observed when antiserum against CBV3 or EV1 was tested against HPEV1-infected cells. Again, in immunofluorescence staining, antisera against VP0 and VP1 showed strong signals whereas the anti-VP3 antiserum detected fewer cells with weaker signals. The specificity of the fluorescence staining was verified by double staining of the cells with HPEV1 antiserum produced in mice and the capsid protein antisera. The green fluorescein staining of the whole virus and the red rhodamine signal showed specific colocalization in the cells.
FIG. 2.
Detection of HPEV1 antigens in virus-infected A549-4A cells by immunoperoxidase (IP) staining and immunofluorescence (IF). The cell monolayers were infected with ∼200 PFU of HPEV1, and HPEV1 antiserum and capsid protein antisera were used at a dilution of 1:300.
Neutralization of HPEV1 infection.
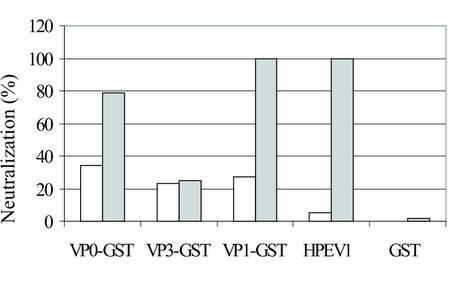
The neutralization effects of the rabbit capsid protein antisera in HPEV1 infection were tested by plaque assay. The antisera against the VP1 protein completely inhibited HPEV1 infection at 1:100 dilution (Fig. 3) and almost as efficiently at 1:1,000 dilution (80% inhibition). The effects with the corresponding preimmune sera were 12 and 27%, respectively. The antisera against the VP0 protein, used at 1:100 dilution, neutralized HPEV1 infection by 79%, whereas 34% neutralization with the preimmune serum was observed. No neutralizing effect of the antisera against VP3 was detected compared to the preimmune serum. In comparison, the 1:1,000 dilution of the HPEV1 antiserum completely neutralized the infection in the cell culture. The background effect of anti-GST antibodies in the capsid fusion protein antisera was excluded by showing that virtually no neutralization effect by the GST antiserum existed. When the specificity of the neutralization of HPEV1 by the capsid protein sera was studied by investigating reactivity against enteroviruses, no neutralization of CBV3 or EV1 was observed (data not shown).
FIG. 3.
Neutralization of HPEV1 infection with rabbit antisera against the GST fusion proteins (VP0-GST, VP3-GST, and VP1-GST) and against GST (dilution, 1:100) and purified HPEV1 (dilution, 1:1,000). The open bars indicate the effects of the preimmune serum, and the shaded bars show the effects of the specific antisera.
HPEV1 capsid proteins as antigens in EIA.
First, the HPEV1 antibody titers of the sera of 20 adults (Table 1) and 55 children were determined by a microneutralization test. In adults, 19 of 20 (95%) samples were positive for HPEV1 antibodies (titer ≥ 10), and the mean antibody titer was 1,400. The antibody levels in children were significantly lower: the mean antibody titer was 160, and the prevalence was 47%. The HPEV1 antibody titers in children increased after 1 year of age but were still low compared to those of the adults.
TABLE 1.
T-cell reactivities of 20 adults to HPEV1 GST fusion proteins (VP0, VP3, and VP1) and GSTa
| Subject | Sex/ageb | HPEV1 antibody titer | SI (4.0 μg/ml of Ag)c
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| HPEV1 | VP0 | VP3 | VP1 | GST | CBV4 | PV1 | |||
| 1 | F/31 | 40 | 3 | 2 | 4 | 2 | 1 | 3 | 3 |
| 2 | M/49 | 10 | 1 | 1 | 2 | 3 | 1 | 10 | 14 |
| 3 | F/26 | 10 | 25 | 5 | 48 | 22 | 1 | 62 | 28 |
| 4 | F/27 | 0 | 10 | 2 | 3 | 5 | 1 | 33 | 19 |
| 5 | F/60 | 160 | 8 | 2 | 2 | 3 | 1 | 5 | 4 |
| 6 | F/51 | 40 | 3 | 4 | 4 | 6 | 1 | 9 | 8 |
| 7 | F/46 | 640 | 3 | 5 | 2 | 4 | 1 | 49 | 12 |
| 8 | F/38 | 10,240 | 41 | 14 | 8 | 7 | 1 | 65 | 67 |
| 9 | M/53 | 2,560 | 2 | 2 | 16 | 4 | 2 | 4 | 19 |
| 10 | F/55 | 160 | 10 | 19 | 10 | 5 | 1 | 3 | 2 |
| 11 | F/27 | 160 | 48 | 3 | 21 | 13 | 2 | 16 | 13 |
| 12 | F/51 | 160 | 1 | 2 | 2 | 1 | 1 | 9 | 1 |
| 13 | M/43 | 160 | 15 | 3 | 13 | 3 | 1 | 3 | 4 |
| 14 | F/33 | 640 | 1 | 1 | 2 | 1 | 1 | 1 | 2 |
| 15 | F/62 | 40 | 2 | 1 | 4 | 4 | 1 | 3 | 2 |
| 16 | M/48 | 40 | 1 | 2 | 18 | 7 | 1 | 4 | 16 |
| 17 | F/23 | 2,560 | 12 | 7 | 8 | 2 | 2 | 22 | 63 |
| 18 | F/55 | 10,240 | 1 | 2 | 21 | 16 | 2 | 12 | 3 |
| 19 | F/26 | 640 | 6 | 1 | 9 | 2 | 1 | 1 | 1 |
| 20 | F/27 | 160 | 1 | 2 | 5 | 2 | 1 | 20 | 6 |
Reactivities against CBV4 and poliovirus 1 (PV1) were also measured. Antibody titers were determined by the microneutralization test.
F, female; M, male.
Mean counts of stimulated cells divided by mean counts of unstimulated cells. SI values of ≥3 were considered to represent positive responses. Ag, antigen.
To test the HPEV1 capsid proteins as antigens in EIA, preliminary EIAs with rabbit hyperimmune sera were performed. All the capsid protein immune sera reacted with the corresponding proteins, whereas the preimmune sera did not (data not shown). Some reactivity of the fusion protein antigens with the GST antiserum was detected, but the signals were lower than those with the specific HPEV1 fusion protein antisera. When the HPEV1 virion antigen was used, it reacted strongly with VP1-GST and VP0-GST rabbit antisera at 10−3 dilution. Preliminary immunoblotting experiments suggested that the VP3 fusion protein would be the most promising candidate for HPEV1 antibody detection. The results showed that 18 of the 20 (90%) HPEV1-seropositive sera reacted with the VP3-GST protein, 7 sera reacted with the VP0-GST protein, and none of the sera reacted with the VP1-GST protein or GST (data not shown).
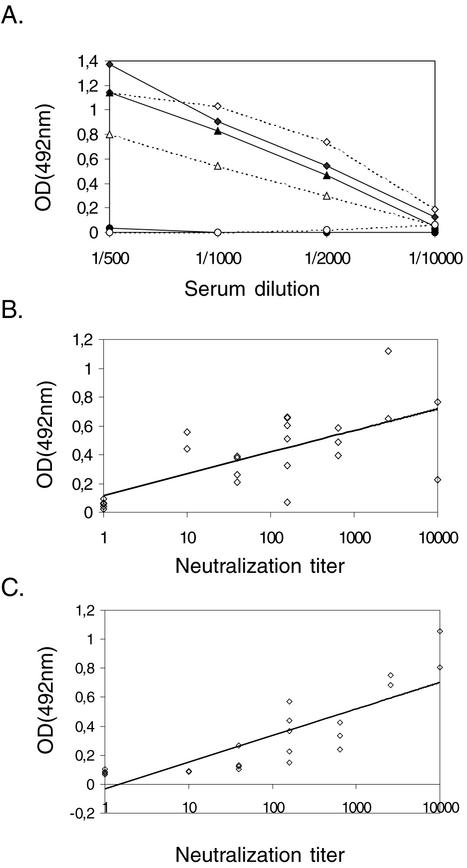
The signals of the seropositive human sera in EIA correlated with the dilution level of the serum, indicating specific reactivity against the VP3 antigen (Fig. 4A). The reactivity for the VP3 fusion protein antigen followed that for the purified HPEV1 antigen. To further evaluate the efficacy of the VP3 protein as an antigen in EIA, human serum samples known to be positive or negative for HPEV1 antibodies, based on microneutralization, were tested at a dilution of 1:1,000. The threshold in the EIA was set as the mean of the negative control sera plus 2 standard deviations. This threshold indicated 100% specificity, as the reactivities of all of the seronegative sera were under the threshold, and 96% sensitivity, with 1 of 19 seropositive sera under the threshold. The results of 18 positive and 5 negative sera in EIA and the microneutralization test are shown in Fig. 4B. The difference between the mean ODs of the positive (0.388) and negative (0.055) samples was approximately ninefold. The correlation between the HPEV1 antibody titer by neutralization and reactivity in EIA was slightly weaker, with the VP3-GST antigen (R2 = 0.46) (Fig. 4B) than with the purified HPEV1 antigen (R2 = 0.69) (Fig. 4C). These results suggest that the VP3 protein specifically and efficiently reacts with HPEV1 antibodies in human sera.
FIG. 4.
(A) Reactivities of the VP3 fusion protein (dotted lines) and purified HPEV1 (solid lines) as antigens in EIA. The microtiter wells were coated with 100 ng of the VP3-GST protein or HPEV1, and the reactivities of two positive sera (diamonds and triangles) and one negative serum (circles) for HPEV1 antibodies in the microneutralization assay were measured. (B) Correlation of the neutralization titer with the VP3-GST antigen in EIA in human serum samples known to be positive (n = 19) or negative (n = 5) for HPEV1 antibodies as determined by the microneutralization test. (C) Correlation of the HPEV1 neutralization antibody titer with the reactivity with purified HPEV1 antigen in EIA in human serum samples known to be positive (n = 19) or negative (n = 5) for HPEV1 antibodies.
T-cell proliferation.
T-cell responses to the purified HPEV1 and GST capsid proteins were studied in 20 adults (Table 1). Twelve out of the 20 adults (60%) had positive T-cell responses (SI ≥ 3) to purified HPEV1. T-cell responses to purified CBV4 and purified poliovirus type 1 were more frequent (90 and 75%, respectively) than those to purified HPEV1. HPEV1 capsid proteins elicited responses as follows: 8 out of 20 adults (40%) had positive responses to VP0 protein, 15 subjects (75%) had positive responses to VP3 protein, and 14 subjects (70%) had positive responses to VP1 protein. Ninety percent of the subjects had positive responses to at least one of the capsid proteins. No positive T-cell responses to the GST protein were detected. All eight subjects who had positive T-cell responses to VP0 proteins also had positive responses to purified HPEV1, whereas responses to VP3 and VP1 proteins were often found without a response to HPEV1. No direct correlation was seen between the level of T-cell proliferation and titers of neutralizing antibody to HPEV1.
DISCUSSION
HPEV1 is a frequent human pathogen, and HPEV1 infections are particularly common during the first years of life (5). The virus was first isolated in 1956 from children suffering from diarrhea (24), and it is still mostly detected in gastroenteritis and respiratory tract infections, but other manifestations have also been reported (22). Parechoviruses are most commonly diagnosed by virus culture in cells followed by neutralization typing, which is both laborious and time-consuming. The use of cell lysates and whole virus as antigens in serological assays is also inconvenient. Therefore, new tools for HPEV1 diagnosis are required. In order to improve specific HPEV1 diagnosis and to study immunity against the virus, we expressed viral capsid proteins in bacteria and produced antisera against them in rabbits.
We were able to purify all of the fusion proteins in soluble form, although we encountered some solubility problems. The formation of insoluble aggregates could be overcome by lowering the induction temperature (e.g., to 25°C), avoiding oversonication, and adjusting the amount of IPTG and the length of induction. Although proteins can be solubilized from aggregates with denaturants, this procedure may lead to misfolding of proteins, and there is no guarantee that the proteins will retain their native antigenicities through this procedure. Thrombin cleavage of the capsid proteins from the GST moiety was performed, but the cleavage remained inefficient regardless of the optimization of the digestion time and temperature. In addition, the cleaved fusion proteins became insoluble and bound to the Sepharose matrix.
The rabbit antisera against HPEV1 capsid proteins were used to detect infected cells in culture. Detection of HPEV1-infected cells was efficient with VP0 and VP1 antisera, and reactivity for human enteroviruses, CBV3 and EV1, was not observed. Thus, the antisera targeted against capsid proteins specifically reacted with HPEV1, allowing the detection of HPEV1-infected cells in culture as described previously with the antisera against purified HPEV1 (9). This procedure accelerates the routine diagnostics, because the virus can be specifically identified before visible signs of infection (cytopathic effect) appear, and there is no need for passaging the virus further in culture for neutralization typing. The VP0 and VP1 antisera are therefore suitable tools for detection and typing of HPEV1 in immunoperoxidase and immunofluorescence staining.
In the neutralization assay, the rabbit antiserum against the VP1 capsid protein neutralized HPEV1 infection efficiently, reducing 80% of infectivity at 1:1,000 dilution and totally neutralizing the infection at 1:100 dilution. Previously, peptide antisera targeted against an antigenic epitope at the C terminus of VP1 has been shown to reduce HPEV1 infectivity by 50% at 1:30 dilution (10). The antiserum against the whole VP1 protein was significantly more efficient, which might be due to additional immunological epitopes in VP1 outside the C-terminal epitope. Our neutralization result with the VP0 antiserum (79% inhibition at 1:100 dilution) resembles the neutralization result with peptide antiserum against the antigenic region of VP0 (76% inhibition at 1:30 dilution) (10).
Virus infection induces both humoral and cellular immunity. B-cell epitopes of picornaviruses have been studied by using synthetic peptides, peptide scanning, and neutralization escape mutants. In polioviruses, coxsackie A virus type 9 and CBV4, and human rhinovirus type 14, the major antigenic regions are mostly concentrated in the capsid protein VP1, whereas VP2 and VP3 are less involved in the antigenic structures (15, 16, 17, 18, 20). In this light, the VP1 protein of HPEV1 was surprisingly poorly immunogenic in immunoblotting and in EIA, although it induced neutralizing antibodies in rabbits. Because B-cell epitopes in general are thought to be conformational, the immunoblotting results with the VP1 and VP0 antigens may reflect the structural nature of their epitopes, since proteins in the blotting procedure are displayed in denatured form. Differences in conformation of the VP3-GST protein may also have affected its immunogenicity and the function of the corresponding antiserum in immunostaining assays compared to the results in EIA.
The EIAs available for enterovirus serodiagnostics are mainly based on heat-treated virus antigens or peptides (19, 23). The sensitivities of the enterovirus EIA methods vary depending on the antigen and the class of antibodies detected. HPEV1 serological diagnosis is presently based on the complement fixation test, and to our knowledge, no EIAs have been described previously. With 100% specificity and 96% sensitivity, although studied with a restricted number of samples, the VP3 protein is a promising candidate for clinical diagnostics. Extending the current IgG detection to IgM class antibody detection could allow rapid diagnosis of acute HPEV1 infections and further extend the usefulness of the test.
When T-cell reactivities to HPEV1 capsid proteins were studied, the VP3 fusion protein elicited responses most often (75%), and the response was also strongest against this antigen in 55% of the subjects. However, the fact that responses to the VP3 and VP1 proteins were often found without a response to HPEV1 and the fact that the proliferation responses did not have a direct correlation with the HPEV1 antibody titers of the sera may indicate that there are some impurities in these preparations causing nonspecific responses. Additionally, there is a possibility for cross-recognition of the protein antigens by T cells. Such cross-reactivity among enterovirus epitopes has been described (1, 2, 4, 13). In these studies, the enteroviral capsid proteins VP1 and VP4 especially have been reported to contain cross-reactive epitopes.
In this study, we successfully produced and purified HPEV1 capsid fusion proteins in soluble form. The capsid protein antisera against the VP0 and VP1 proteins were favorably used for immunostaining, and these antisera also exhibited neutralizing effects in HPEV1 infection. We also gained proof of the immunogenic epitopes in the VP3 protein, as it was successfully assayed as an antigen in EIA. In addition to the humoral immune responses against VP3, T-cell reactivity to the protein was detected. A systematic epitope mapping of the capsid proteins with HPEV1-specific T-cell lines would provide exhaustive information about the antigenic epitopes in the HPEV1 capsid. Although parechoviruses form a biological and molecular group distinct from other picornaviruses, the possibility of common picornavirus antigenic epitopes needs to be carefully analyzed.
Acknowledgments
This study was supported by grants from the Academy of Finland and the Sigrid Juselius Foundation.
We thank Maaria Vainio for technical assistance and Vilja Pietiäinen and Tuija Pöyry for helpful discussions.
REFERENCES
- 1.Cello, J., A. Samuelson, P. Stalhandske, B. Svennerholm, S. Jeansson, and M. Forsgren. 1993. Identification of group-common linear epitopes in structural and nonstructural proteins of enteroviruses by using synthetic peptides. J. Clin. Microbiol. 31:911-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cello, J., O. Strannegard, and B. Svennerholm. 1996. A study of the cellular immune response to enteroviruses in humans: identification of cross-reactive T cell epitopes on the structural proteins of enteroviruses. J. Gen. Virol. 77:2097-2108. [DOI] [PubMed] [Google Scholar]
- 3.Ghazi, F., P. Hughes, T. Hyypiä, and G. Stanway. 1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79:2641-2650. [DOI] [PubMed] [Google Scholar]
- 4.Graham, S., E. Wang, O. Jenkins, and L. Borysiewicz. 1996. Analysis of the human T-cell response to picornaviruses: identification of T-cell epitopes close to B-cell epitopes in poliovirus. J. Virol. 67:1627-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grist, N., E. J. Bell, and F. Assaad. 1978. Enteroviruses in human disease. Prog. Med. Virol. 24:114-157. [PubMed] [Google Scholar]
- 6.Huber, S., J. Polgar, A. Moraska, M. Cunningham, P. Schwimmbeck, and P. Schultheiss. 1993. T lymphocyte responses in CBV3-induced murine myocarditis. Scand. J. Infect. Dis. Suppl. 88:67-78. [PubMed] [Google Scholar]
- 7.Hughes, P., and G. Stanway. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 81:201-207. [DOI] [PubMed] [Google Scholar]
- 8.Hyypiä, T., C. Horsnell, M. Maaronen, M. Khan, N. Kalkkinen, P. Auvinen, L. Kinnunen, and G. Stanway. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. USA 89:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joki-Korpela, P., and T. Hyypiä. 1998. Diagnosis and epidemiology of echovirus 22 infections. Clin. Infect. Dis. 26:129-136. [DOI] [PubMed] [Google Scholar]
- 10.Joki-Korpela, P., M. Roivainen, H. Lankinen, T. Pöyry, and T. Hyypiä. 2000. Antigenic properties of human parechovirus 1. J. Gen. Virol. 81:1709-1718. [DOI] [PubMed] [Google Scholar]
- 11.Juhela, S., H. Hyöty, R. Uibo, S. Meriste, O. Uibo, M. Lönnrot, M. Halminen, O. Simell, and J. Ilonen. 1999. Comparison of enterovirus-specific cellular immunity in two populations of young children vaccinated with inactivated or live poliovirus vaccines. Clin. Exp. Immunol. 117:100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. Knowles, S. Lemon, P. Minor, A. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. Van Regenmortel, C. M. Fauguet, D. H. L. Bishop, C. H. Calisher, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 13.Mahon, B., K. Katrak, and K. Mills. 1992. Antigenic sequences of poliovirus recognized by T cells: serotype-specific epitopes on VP1 and VP3 and cross-reactive epitopes on VP4 defined by using CD4+ T-cell clones. J. Virol. 66:7012-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marttila, J., H. Hyöty, P. Vilja, T. Härkönen, A. Alho, M. Roivainen, T. Hyypiä, and J. Ilonen. 2001. T cell epitopes in coxsackievirus B4 proteins concentrate in regions conserved between enteroviruses. Virology 293:217-224. [DOI] [PubMed] [Google Scholar]
- 15.Minor, P., M. Ferguson, D. Evans, J. Almond, and J. Icenogle. 1986. Antigenic structure of polioviruses of serotypes 1, 2 and 3. J. Gen. Virol. 67:1283-1291. [DOI] [PubMed] [Google Scholar]
- 16.Prabhakar, B., M. Menegus, and A. Notkins. 1985. Detection of conserved and nonconserved epitopes on coxsackievirus B4: frequency of antigenic change. Virology 146:302-306. [DOI] [PubMed] [Google Scholar]
- 17.Pulli, T., H. Lankinen, M. Roivainen, and T. Hyypiä. 1998. Antigenic sites of coxsackievirus A9. Virology 240:202-212. [DOI] [PubMed] [Google Scholar]
- 18.Roivainen, M., A. Närvänen, M. Korkolainen, M.-L. Huhtala, and T. Hovi. 1991. Antigenic regions of poliovirus type 3/Sabin capsid proteins recognized by human sera in the peptide scanning technique. Virology 180:99-107. [DOI] [PubMed] [Google Scholar]
- 19.Samuelson, A., M. Glimaker, E. Skoog, J. Cello, and M. Forsgren. 1993. Diagnosis of enteroviral meningitis with IgG-EIA using heat-treated virions and synthetic peptides as antigen. J. Med. Virol. 40:271-277. [DOI] [PubMed] [Google Scholar]
- 20.Sherry, B., A. Mosser, R. Colonno, and R. Rueckert. 1986. Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J. Virol. 57:246-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanway, G., N. Kalkkinen, M. Roivainen, F. Ghazi, M. Khan, M. Smyth, O. Meurman, and T. Hyypiä. 1994. Molecular and biological characteristics of echovirus 22: a representative of a new picornavirus group. J. Virol. 68:8232-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanway, G., P. Joki-Korpela, and T. Hyypiä. 2000. Human parechoviruses—biology and clinical significance. Rev. Med. Virol. 10:57-69. [DOI] [PubMed] [Google Scholar]
- 23.Swanink, C., L. Veenstra, Y. Poort, J. Kaan, and J. Galama. 1993. Coxsackievirus B1-based antibody-capture enzyme-linked immunosorbent assay for detection of immunoglobulin G (IgG), IgM and IgA with broad specificity for enteroviruses. J. Clin. Microbiol. 31:3240-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wigand, R., and A. Sabin. 1961. Properties of ECHO types 22, 23, and 24 viruses. Arch. Gesamte Virusforsch. 11:224-247. [DOI] [PubMed] [Google Scholar]