Abstract
A rapid in-house mycobacteriophage-based assay to identify multidrug resistance by detecting the rifampin susceptibility of Mycobacterium tuberculosis in a microtiter plate format was evaluated. The sensitivity, specificity, and overall accuracy of the assay were 100%. This test is rapid to perform and suitable for widespread implementation.
Tuberculosis (TB) control programs are being threatened worldwide by drug-resistant Mycobacterium tuberculosis strains. Detection of multidrug-resistant (MDR) strains (resistant to at least rifampin [RIF] and isoniazid) is of major importance, because these two drugs constitute the more efficacious antibiotics included in the standard TB therapy. Since resistance to RIF can be considered a surrogate marker for MDR M. tuberculosis strains (10), detection of RIF resistance could be used to identify those TB patients who are unlikely to respond to the standard treatment regimen and as a consequence are inadequately treated and remain infectious for longer periods. Although early detection of RIF-resistant (Rifr) strains is available by use of rapid molecular methods (3, 9), there is an urgent need for simple, rapid, and inexpensive multidrug susceptibility screening methods appropriate for widespread routine application (especially for those countries presenting less favorable living conditions, where a high incidence of TB is found) (5).
Mycobacteriophage-based techniques have been reported as potentially useful tools for detection of viable bacilli as well as for antimicrobial susceptibility testing (2, 7, 11). We standardized and assessed an in-house assay (in microtiter format) for determining RIF susceptibility based on the mycobacteriophage amplification principle by studying M. tuberculosis clinical isolates. The method is based on the ability of resistant mycobacteria to support mycobacteriophage D29 infection once exposed to RIF, while sensitive mycobacteria will not be able to support phage replication.
(Part of this study was presented at the 11th European Congress of Clinical Microbiology and Infectious Diseases, held in Istanbul, Turkey, in April 2001.)
Standardization of the technique based on the work by Wilson et al. (11) was carried out previous to evaluation of the technique. Reference strains (see below) were used to determine the parameters of the test such as the inoculum of the strains, the concentration and exposure time of RIF, and the mycobacteriophage titer.
Eighty-nine M. tuberculosis clinical isolates were obtained from specimens cultured for routine diagnosis of TB. They were identified by using DNA probes (AccuProbe; GenProbe, Inc., San Diego, Calif.) and conventional biochemical tests. Eighteen were Rifr strains, and 71 were RIF-sensitive (Rifs) isolates, according to the Bactec 460 TB “in vitro” susceptibility testing method (Becton Dickinson Laboratory Systems, Sparks, Md.) (6). Mycobacterial reference strains were used both in the standardization and in the evaluation of the test (the Rifs M. tuberculosis strain H37Ra [ATCC 25177] and the Rifr M. tuberculosis strain H37Rv [ATCC 35838]).
Fresh cultures from Löwenstein-Jensen medium were used as the source of the mycobacterial organisms. The mycobacterial growths were transferred to sterile screw-cap glass tubes containing 6 to 8 glass beads in 2 ml of 7H9 broth (MAIM S.L., Barcelona, Spain) with 10% (vol/vol) oleic acid-albumin-dextrose-catalase (OADC) enrichment (MAIM S.L.). Suspensions were homogenized by using a vortex mixer for 15 to 20 s. Larger clumps were allowed to settle by letting the suspensions stand for 10 min. The supernatants were transferred to sterile tubes and adjusted to 106 CFU/ml by using 7H9 broth with OADC.
Mycobacteriophage D29 (kindly supplied by R. McNerney, London School of Hygiene and Tropical Medicine) was produced as described previously (8). Mycobacteriophages were inoculated onto a lawn of Mycobacterium smegmatis in Middlebrook 7H9 agar (MAIM S.L.) supplemented with OADC and 1 mM CaCl2 (Merck KGaA, Darmstadt, Germany). Phages were harvested after overnight incubation at 37°C with 7H9 broth and 1 mM CaCl2, passed through a 0.45-μm-pore-size filter, and stored at 4°C for as long as 6 months. The mycobacteriophage stock was quantified by pipetting 10-μl aliquots of serial dilutions onto a lawn of M. smegmatis. The phage suspension was diluted to achieve a working titer of 107 PFU/ml prior to the mycobacteriophage test.
RIF was made up as a 25-mg/ml stock solution in N,N-dimethylformamide (Sigma-Aldrich Chemicals GmbH, Steinheim, Germany) and stored at −20°C until use. The working concentration was achieved by dilution with 7H9 broth-OADC-CaCl2 to 10 μg/ml. The final drug concentration, when it was mixed with the mycobacterial suspension in the microtiter plate, was 5 μg/ml.
The mycobacteriophage-based assay (MBA) was performed as follows. A 75-μl volume of M. tuberculosis bacilli (106 CFU/ml) was placed in wells of sterile microtiter plates (Asahi Techno Glass, Funabasi, Japan) containing 75 μl of RIF (10 μg/ml) and incubated for 24 h at 37°C. Fifty microliters of mycobacteriophage D29 (107 PFU/ml) was added, and the microtiter plates were incubated for 90 min at 37°C. The phages that were unable to infect the bacilli were inactivated by addition of 100 μl of ferrous ammonium sulfate (30 mM), while those inside were protected and replicated within the mycobacteria, causing their lysis and the release of new mycobacteriophage progeny. Mycobacteriophages were detected in 10-μl drops by the formation of plaques on the surface of a lawn of the fast-growing host M. smegmatis (7H9 agar-10% OADC-1 mM CaCl2 and M. smegmatis at 108 CFU/ml) after overnight incubation at 37°C. Results were available with a total turnaround time of 48 h. A strain was interpreted as resistant in the presence of plaques formed by the release of mycobacteriophages from viable M. tuberculosis bacilli after treatment with RIF, and it was considered susceptible in the absence of plaques. A positive control for each clinical isolate, consisting of the strain incubated with assay broth (instead of RIF), and a negative control (broth only) were included in each run of the MBA. Mycobacterial reference strains were assayed as long as new batches of reagents or mycobacteriophages were used in order to assess the performance of the test. The researchers performing the assay were blinded as to the RIF sensitivity or resistance of the M. tuberculosis strains in order to avoid predictable results.
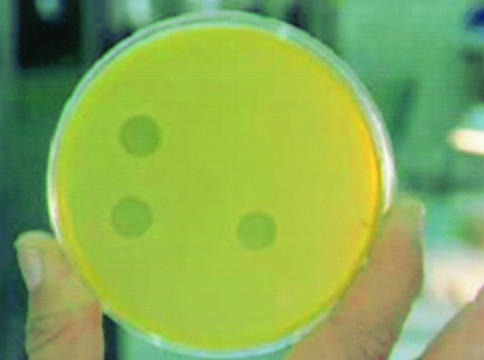
Eighty-nine clinical isolates of M. tuberculosis were assayed in order to compare the performance of the MBA in testing RIF susceptibility with that of the Bactec 460 method. All 89 clinical isolates of M. tuberculosis tested gave concordant results by the standardized MBA and the “gold standard” Bactec 460. The overall accuracy of the MBA relative to the Bactec 460 method was 100% (89 of 89). The sensitivity of the MBA in detecting RIF resistance was 100% (18 of 18), and the specificity achieved was also 100% (71 of 71). The positive predictive value and the negative predictive value were both 100%. The positive and negative process controls as well as the mycobacterial reference strains performed correctly whenever they were tested. Interpretation of an isolate to be scored as susceptible or resistant was easy and clear (Fig. 1). Confluent or total lysis was observed on the 10-μl-drops on the surface of the M. smegmatis lawn in the case of resistant strains, whereas all sensitive isolates showed no plaques.
FIG. 1.
Interpretation of the MBA in the fast-growing M. smegmatis lawn. Bottom left drop, RIFr strain incubated without RIF (drug-free control); bottom right drop, RIFr strain incubated with RIF; upper left drop, RIFs strain incubated without RIF (drug-free control); upper right drop, RIFs strain incubated with RIF.
The MBA demonstrated excellent sensitivity, specificity, and agreement relative to the gold standard Bactec 460 method. All the Rifr strains included in our study were also resistant to isoniazid at least, indicating that RIF resistance can be considered an excellent predictor of MDR in our setting.
Fresh cultures from solid Löwenstein-Jensen medium constituted the source of the mycobacterial organisms in order to facilitate the previous standardization of the bacterial inoculum. Nevertheless, use of liquid cultures would have reduced the time required to report the susceptibility result, as shown in the study performed by Albert et al. (1). In that paper, the authors report the ability to determine RIF susceptibility for clinical strains of M. tuberculosis after growth in the Bactec 460 by using a mycobacteriophage-based test. With 133 clinical isolates studied, the sensitivity, specificity, and overall accuracy of the test were 100, 98.8, and 99.2%, respectively, for detection of RIF resistance (relative to the Bactec 460 system).
Mycobacteriophage-based techniques to screen for resistance to other first- and second-line antituberculosis drugs have also been evaluated, showing excellent and promising results (4, 8).
A technical advantage of the standardized method presented here over those developed in all these previous studies, as presented by McNerney et al. (8), is the use of a microtiter plate-based methodology. Working with a 96-well format reduces the risk of technician exposure to viable M. tuberculosis, because small sample volumes are processed. Moreover, microtiter plates enable one to process a large numbers of strains by following a simple procedure.
This MBA is a rapid test that constitutes an alternative to conventional drug susceptibility testing methods, which take a longer time to achieve results. Several features of the mycobacteriophage-based test make it suitable for widespread application: no requirement for specialized equipment or reagents, low demand for technical skills, low cost, and simplicity of mycobacteriophage use (with regard to production, storage, and safety). Furthermore, it could be particularly useful for laboratories located in areas with limited resources and budgets, where more rapid and expensive molecular methods are unaffordable. Providing reliable tools to detect antituberculosis drug resistance in high-burden regions would benefit public health worldwide.
Acknowledgments
We are indebted to R. McNerney, London School of Hygiene and Tropical Medicine, for her kind collaboration.
This work was supported by a grant from the Comissionat per a Universitats i Recerca (Generalitat de Catalunya, FI/FIAP 1998-2001) and from the Sociedad Española de Neumología y Cirugía Torácica.
REFERENCES
- 1.Albert, H., A. P. Trollip, R. J. Mole, S. J. Hatch, and L. Blumberg. 2002. Rapid identification of multidrug-resistant tuberculosis from liquid cultures using FASTPlaqueTB-RIFTM, a manual phage-based test. Int. J. Tuberc. Lung Dis. 6:523-528. [DOI] [PubMed] [Google Scholar]
- 2.Banaiee, N., M. Bobadilla-Del-Valle, S. Bardarov, Jr., P. F. Riska, P. M. Small, A. Ponce-De-Leon, W. R. Jacobs, Jr., G. F. Hatfull, and J. Sifuentes-Osornio. 2001. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J. Clin. Microbiol. 39:3883-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caws, M., and F. A. Drobniewski. 2001. Molecular techniques in the diagnosis of Mycobacterium tuberculosis and the detection of drug resistance. Ann. N. Y. Acad. Sci. 953:138-145. [DOI] [PubMed] [Google Scholar]
- 4.Eltringham, I. J., S. M. Wilson, and F. A. Drobniewski. 1999. Evaluation of a bacteriophage-based assay (phage amplified biologically assay) as a rapid screen for resistance to isoniazid, ethambutol, streptomycin, pyrazinamide, and ciprofloxacin among clinical isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:3528-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, and M. C. Raviglione. 2001. Global trends in resistance to antituberculosis drugs. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 6.Heifets, L. 1988. Qualitative and quantitative drug susceptibility tests in mycobacteriology. Pulmonary perspective. Am. Rev. Respir. Dis. 137:1217-1222. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs, W. R., Jr., R. G. Barletta, R. Udani, J. Chan, G. Kalkut, G. Sosne, T. Kieser, G. J. Sarkis, G. F. Hatfull, and B. R. Bloom. 1993. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260:819-822. [DOI] [PubMed] [Google Scholar]
- 8.McNerney, R., P. Kiepiela, K. S. Bishop, P. M. Nye, and N. G. Stoker. 2000. Rapid screening of Mycobacterium tuberculosis for susceptibility to rifampicin and streptomycin. Int. J. Tuberc. Lung Dis. 4:69-75. [PubMed] [Google Scholar]
- 9.Soini, H., and J. M. Musser. 2001. Molecular diagnosis of mycobacteria. Clin. Chem. 47:809-814. [PubMed] [Google Scholar]
- 10.Somoskovi, A., L. M. Parsons, and M. Salfinger. 2001. The molecular basis of resistance to isoniazid, rifampicin, and pyrazinamide in Mycobacterium tuberculosis. Respir. Res. 2:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson, S. M., Z. Al-Suwaidi, R. McNerney, J. Porter, and F. A. Drobniewski. 1997. Evaluation of a new rapid bacteriophage-based method for the drug susceptibility testing of Mycobacterium tuberculosis. Nat. Med. 3:465-468. [DOI] [PubMed] [Google Scholar]