Abstract
A longitudinal study of the infection dynamics of Salmonella enterica was carried out with three Danish farrow-to-finish swine herds. To account for variations in Salmonella shedding over time, litters from each herd were divided into two cohorts. Each cohort consisted of 30 pigs, for a total of 180 pigs. Pigs were individually monitored by monthly bacteriologic and serologic examinations from weaning to slaughter. At weaning, individual sows were examined bacteriologically and serologically. At slaughter, cecal contents, ileocecal lymph nodes, and carcass swab samples were obtained from 131 pigs. A total of 88 pigs were found to be shedding Salmonella on one or more occasions. Only the Salmonella serotype Typhimurium was detected during the study period. At weaning, no sows or piglets were found to be shedding, but a serological reaction was detected in 11 sows. The prevalence in culture peaked in the nursery and subsequently declined to undetectable levels before slaughter. The seroprevalence peaked approximately 60 days after the peak prevalence in culture. Salmonella was detected in individual fecal samples at least once in 53% of the pigs, and 62% of the pigs were seropositive more than once. Only 3.7% of all pigs were found to be culture positive on more than one occasion. Piglets from seroreacting sows had a significantly (P = 0.0339) lower probability of shedding in the nursery. Under the assumption that shedding lasted at least 1 or 2 weeks, the average shedding time was estimated to have been 18 or 26 days. An association between serology, on-farm bacteriology, and Salmonella prevalence in culture at slaughter was shown. Marked differences in prevalence in sera and prevalence in culture between cohorts and within herds were observed. These differences emphasize the need for caution when using point estimates in on-farm interventions and surveillance in subclinically infected swine herds.
Salmonella enterica does not normally cause clinical disease in pigs, but subclinical Salmonella infections constitute an important food safety problem throughout the world. From a consumer viewpoint, continuing efforts are needed to reduce the occurrence of Salmonella in pork. In order to achieve this, information about the dynamics of Salmonella infections in swine herds over time (e.g., duration of infection and disease transmission patterns) can be a useful tool. However, so far only limited information in this field is available.
Results from a comprehensive longitudinal study in two multiple-site pig production systems in the United States revealed considerable temporal variability in Salmonella prevalence between cohorts of pigs (11). In a Danish study, the Salmonella status of sow herds was measured based upon (i) the prevalence of serotype Typhimurium bacteria among weaners and (ii) the seroprevalence among sows. Both factors were shown to constitute important risk factors for Salmonella infection in finisher herds, as measured by the high seroprevalence found by examining meat juice samples at slaughter (15).
We decided to conduct an investigation to study the complex nature of subclinical Salmonella infections at the levels of the herd and the individual pig. The objectives of the study were the following: (i) to describe the time of onset and duration of Salmonella shedding; (ii) to study the patterns of bacterial transmission between individual pigs until slaughter; (iii) to investigate the transmission between the different age groups, as well as the association between bacteriological shedding and serological response in cohorts of pigs from weaning to slaughter; (iv) to compare antemortem and postmortem findings in individual pigs and thereby focus on food safety; and (v) to provide input estimates for a quantitative risk analysis model, simulating Salmonella prevalence from the growing pig to the slaughtered carcass (1).
MATERIALS AND METHODS
Selection of herds.
Three Danish farrow-to-finish swine herds with moderate to high levels of Salmonella serotype Typhimurium infection were selected for the study. The selection was based on serological and bacteriological data from the national Salmonella surveillance program in Denmark. According to the program, all herds in which pigs are delivered for slaughter are classified into three levels, based on the results of monthly monitoring of Salmonella antibodies in meat juice samples. Level 1 herds are those that have samples with a low (<40%) prevalence, level 2 herds are those that have samples a moderate (40 to 70%) prevalence, and level 3 herds are those that have samples with a high (>70%) prevalence (2). In herds classified as levels 2 and 3, mandatory representative microbiological surveys are carried out. Based on results from these surveys, potential participants for the present study were identified by using the central Zoonosis Register (16). Only herds in which Salmonella serotype Typhimurium bacteria were found were considered for inclusion in the study. Salmonella serotype Typhimurium was chosen because in Denmark this serotype is predominant in pigs (4) and serotype Typhimurium is the second most common serotype isolated in human salmonellosis (3).
The participating farmers agreed not to introduce Salmonella-reducing measures during the 6-month study period. Among five farms potentially suited for the study, pooled pen fecal samples were collected from various rooms to ensure that Salmonella bacteria were present, preferably in most of these rooms. Three farms were chosen based on results from these preliminary samples and based on geographical convenience. Two of the farms, with 650 and 440 sows, were two-site operations. The remaining farm was a three-site, 300-sow operation. All three herds were self-supplying.
Sampling scheme.
In each herd, 10 litters were randomly selected, and in each litter, the ears of six randomly selected piglets were tagged. To account for variations in Salmonella shedding over time, litters from each herd were divided into two groups of five litters that were raised at approximately 1-month intervals. Thus, on each farm there were two cohorts consisting of 30 pigs each, yielding a total of 180 piglets at the start of the study. All ear-tagged pigs from a given cohort were supposed to be raised together for the entire observation period.
Individual blood and fecal samples were collected from the sows at weaning. In addition, individual blood and fecal samples were collected from pigs on the following occasions: (i) just prior to weaning (feces only), (ii) midway through the period in the nursery, (iii) just before leaving the nursery, (iv) monthly in the finishing unit, and (v) just prior to slaughter. Due to different management practices, one herd was sampled on seven occasions and the other two herds were sampled on six occasions. The intervals between sampling occasions varied from 20 to 29 days between sampling occasions just prior to weaning, midway through the nursery period, and just before leaving the nursery. The intervals between sampling occasions in the finishing unit ranged from 21 to 35 days, except with cohort 1B, for which the interval was only 14 days because of an earlier slaughter than expected. The maximum difference between sampling occasions between cohorts within herds was 7 days.
All pigs within a cohort were slaughtered on the same day. At slaughter, the individual carcasses were identified and samples were collected from ileocecal lymph nodes, cecal contents, and the carcass surface. The carcass samples were taken by swabbing 100 cm2 of the ham, chest, and jaw regions, respectively. A template was placed on the surface of the carcass, and one cotton gauze swab was used to swab all three sites (8). Pigs were processed within 1 h after arrival. Abattoir data were collected to investigate the association between antemortem shedding, carrier state, and possible cross-contamination with Salmonella bacteria at slaughter.
Bacteriological analyses were carried out by using standard microbiological culturing methods, including nonselective preenrichment, selective enrichment, and serotyping. Blood samples were analyzed by using the Danish mix-enzyme-linked immunosorbent assay with a cutoff optical density percentage (OD%) of 20%.
Statistics.
To assess the relationship between serology in sows and Salmonella shedding by their offspring, the GENMOD procedure in SAS was used (SAS/STAT user's guide, version 8, SAS Institute, Inc., Cary, N.C.). The proportion of culture-positive piglets per sow on the second sampling occasion was used as the observation or response variable. Herd and cohort were used as explanatory variables. The p-scale option in SAS was used to control for overdispersion originating from the herd and the cohort.
RESULTS
Preliminary sampling.
Salmonella serotype Typhimurium was found in all three herds in the preliminary sampling prior to the start of the study. On each farm, Salmonella was primarily found in the nursery and finisher units and it was only occasionally found in gilts and sows. All Salmonella isolates from preliminary samplings were serotype Typhimurium except one isolate, which was serotype 4.5.12:i.
Bacteriological and serological results.
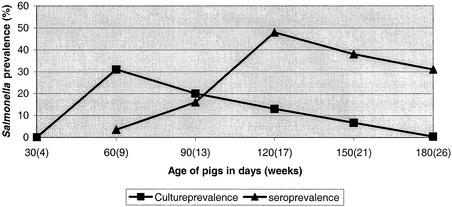
At the time of weaning, none of the 30 sows were culture positive, but one sow in cohort 1B and all sows in cohort 3A were seropositive. In all three herds, only Salmonella serotype Typhimurium was isolated during the entire study period. At weaning, none of the piglets were found to be culture positive. Overall, Salmonella shedding reached a peak in the nursery (i.e., after 60 days, which corresponds to 9 weeks of age), and subsequently declined during the finishing period (Fig. 1). The serological response was observed approximately 30 days later and, on average, reached its peak in the mid-finishing period (i.e., after 120 days, which corresponds to 17 weeks of age) (Fig. 1).
FIG. 1.
Average prevalence of Salmonella in blood and fecal samples from 160 to 180 pigs originating from six cohorts in three Danish farrow-to-finish swine herds participating in a study on Salmonella dynamics in 2001. Pigs were slaughtered between days 150 and 180.
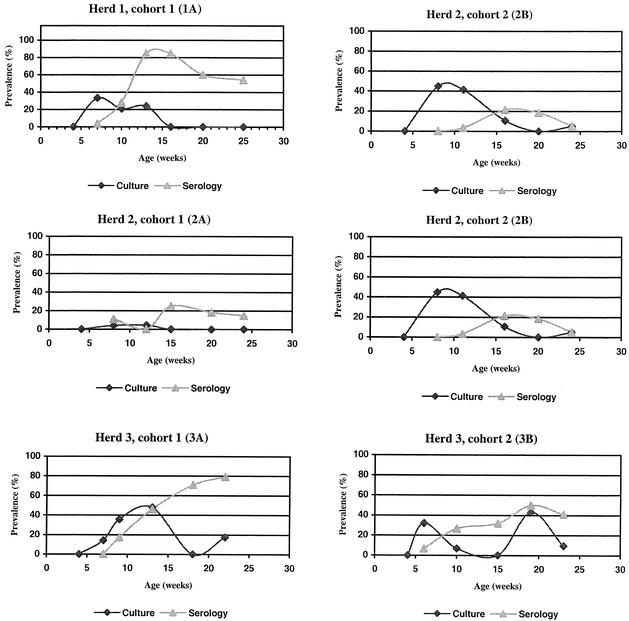
Marked variation in both shedding and seroreaction over time was observed between herds and within herds between cohorts (Fig. 2). All cohorts except cohort 3A showed similar trends in prevalence of Salmonella in culture, i.e., a mid-nursery period peak in the organism's prevalence in culture followed by a decline to undetectable levels before slaughter, regardless of the magnitude of the peak prevalence in culture. The peak prevalence in culture seemed to occur approximately 4 weeks later in cohort 3A than in the five other cohorts. In herd 3, the prevalence in culture started rising towards the end of the study period in both cohorts. This rise coincided with a slurry overflow prior to the farm visit, when the pigs were approximately 19 (cohort 3A) and 16 (cohort 3B) weeks of age.
FIG. 2.
Prevalence of Salmonella in blood and fecal samples from 160 to 180 pigs originating from six cohorts in three Danish farrow-to-finish swine herds participating in a study on Salmonella dynamics in 2001.
Cohorts in herd 3 had an increasing seroprevalence throughout the entire study period. In the other cohorts, the seroprevalence peaked in the early finishing stage (around the age of 14 weeks) and thereafter declined during the late finishing stage; in three cohorts the seroprevalence fell to <40% just before slaughter, while in the remaining cohorts the seroprevalence was still moderate to high (40 to 80% of samples). A conspicuous difference in seroprevalence between cohorts was the presence of antibodies among animals in cohort 2A midway through the nursery period.
Relationship between Salmonella shedding and serology.
The relationship between Salmonella shedding and serological response across all six cohorts is shown in Table 1. Only those pigs sampled on four or more occasions (n = 160) were included. As can be seen in Table 1, Salmonella was detected at least once in 53.1% of the pigs, while 62.4% of the pigs were seropositive more than once. Interestingly, 30.5% [(10.0% + 5.6% + 0.6%)/(35.6% + 13.8% + 3.1% + 0.6%)] of Salmonella-shedding pigs did not show an antibody response at any time during the study. In addition, Salmonella was never isolated from 40.9% [(11.0% + 6.3% + 4.4% + 3.8%)/(23.5% + 18.9% + 8.8% + 11.2%)] of the seropositive animals. Salmonella bacteria were found more than twice in only 3.7% of all pigs tested on at least four occasions.
TABLE 1.
Association between bacteriology and serology in 160 pigs originating from six cohorts in three Danish farrow-to-finish swine herdsa
| No. of times individual pigs were seropositive | % of pigs shedding Salmonella Typhimurium the indicated no. of times
|
Total | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||
| 0 | 22.0 | 10.0 | 5.6 | 0.6 | 0.0 | 38.2 |
| 1 | 11.0 | 10.0 | 1.9 | 0.6 | 0.0 | 23.5 |
| 2 | 6.3 | 6.9 | 4.4 | 1.3 | 0.0 | 18.9 |
| 3 | 4.4 | 3.1 | 1.3 | 0.0 | 0.0 | 8.8 |
| >4 | 3.8 | 5.6 | 0.6 | 0.6 | 0.6 | 11.2 |
| Total | 47.5 | 35.6 | 13.8 | 3.1 | 0.6 | ∼100 |
All pigs were from herds included in our 2001 study of Salmonella dynamics. Only pigs sampled on at least four occasions were included.
Relationship between serology in sows and Salmonella shedding by piglets.
As mentioned earlier, one sow from cohort 1B and all sows from cohort 3A were seropositive when an OD% of 20% was used as a cutoff value. In addition, two sows from cohort 1B and three sows from cohort 2A had seroreactions ranging between 3 and 18 OD%. The remaining 19 sows were nonreactors. Piglets from seroreacting sows had a significantly (P = 0.0339) lower probability of shedding in the mid-nursery period. The odds ratio (OR) for a rise of 1 OD% was 0.94. With this OR, it can be calculated that there was an approximately 50% reduction in the risk of an individual piglet being culture positive in the mid-nursery period with a 10 OD% increase in a sow's seroreaction.
Correlations between sampling results.
The correlations between results from individual pigs on different sampling occasions were assessed by calculating the ORs for bacteriological and serological results (Table 2). Regarding the bacteriological results, we found that the earlier in the study period and the shorter the time interval between sampling occasions, the greater the magnitude of the correlation (assessed by OR). For example, an individual pig that was culture positive on the second sampling occasion had a threefold-higher chance (OR = 3.22) of being culture positive on the third sampling occasion than that of a pig that was culture negative on the second sampling occasion and had a chance (OR = 1.02) of being culture positive on the fifth sampling occasion that was equal to that of a pig that was culture negative on the fifth sampling occasion. As all fecal samples were culture negative on the first sampling occasion, these samples were not included. Sampling on the seventh sampling occasion was carried out only with herd 1, and as none of these fecal samples were positive, the correlation between any other sampling occasion and the seventh sampling occasion was zero. The serological correlations (assessed with ORs) were higher than the bacteriological correlations. We found that there were significant correlations not only between subsequent sample results but also between sampling occasions at larger time intervals, especially late in the study during the finishing period.
TABLE 2.
Correlation between bacteriological and serological findings of Salmonella among pigs on different sampling occasionsa
| Test sample | Sampling occasion | OR on sampling occasion:
|
||||
|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | ||
| Culture | 2 | 3.22* | 0.94 | 1.02 | 0.36 | ND |
| 3 | 2.86* | 1.09 | 3.33 | ND | ||
| 4 | 0.69 | 0.82 | ND | |||
| 5 | 5.95* | ND | ||||
| 6 | ND | |||||
| Serum | 2 | 7.71* | 0.45 | ND | ND | ND |
| 3 | 8.86 | 2.24* | 4.02 | 2.87 | ||
| 4 | 4.99* | 4.76* | 7.65* | |||
| 5 | 21.16* | 7.12* | ||||
| 6 | 10.88* | |||||
ORs were calculated for 160 to 180 pigs originating from six cohorts in three Danish farrow-to-finish swine herds participating in a study on Salmonella dynamics in 2001. *, significant association (P < 0.05) between the results of bacteriological and serological examinations at different sampling occasions, with respect to the detection of Salmonella bacteria and antibodies. ND, not done, as no individuals met both criteria.
Estimating duration of shedding.
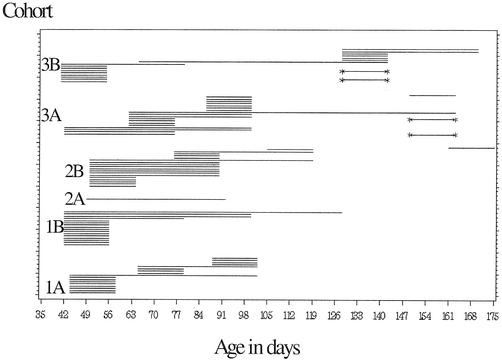
A total of 88 pigs were found to be shedding Salmonella on more than one occasion. To estimate the shedding time, we assumed that shedding began at least 3.5 days or 1 week prior to the first isolation and lasted at least until 3 days or 1 week after the last isolation. An individual pig was considered at risk (i) for as long as it was culture negative or (ii) if it was culture negative on two or more successive occasions. As a consequence, if an individual was found to be culture negative or one sample was missing between two culture-positive events, the pig was considered to have been shedding for the entire time period. This approach was taken because of the known low sensitivity of the culture technique. Only four Salmonella-shedding individuals were found to be culture negative on one sampling occasion between two positive culture findings. A pig that was culture negative on multiple occasions between two culture-positive events was considered to be reinfected. Based on these assumptions, only 4 out of 88 pigs (4.5%) were found to be shedding in two distinct time periods. These four pigs all originated from herd 3 and became culture positive again after a slurry overflow. Figure 3 shows shedding patterns for individual pigs based on the assumption that shedding occurred at least 1 week prior to and after sampling. The overall mean time of shedding was estimated to be 18 or 26 days, depending on the assumptions made. The shortest shedding time period was 7 or 14 days and the longest shedding time period was 94 or 101 days, depending on the assumptions made (Fig. 3).
FIG. 3.
Estimated shedding times of Salmonella serotype Typhimurium in 88 individual pigs originating from six cohorts from three Danish farrow-to-finish swine herds participating in a study on Salmonella dynamics in 2001. One bar represents one individual. Individuals represented by bars flanked by asterisks were considered reinfected.
Slaughter results.
Samples from ileocecal lymph nodes and cecal contents were obtained from 122 finishers, and swab samples were collected from 117 carcasses (Table 3). From 14 finishers, only the ileocecal and cecal contents were obtained, and from nine finishers, only the carcass swab samples were obtained. Overall, microbiological samples of at least one sample type were obtained from 131 finishers. Although Salmonella was isolated from only 7 pigs on the last farm visit prior to slaughter, 31 pigs were found at slaughter to have Salmonella-positive lymph nodes, cecal contents, or carcass surface samples. One pig from cohort 1A and four pigs from cohort 3B had culture-positive lymph nodes and cecal contents. Five out of six pigs that had culture-positive carcass swab samples had culture-negative lymph nodes and cecal contents. The remaining pig, whose carcass was culture positive, also had culture-positive lymph nodes. Two isolates from carcass swab samples in cohort 3B were nontypeable. Among the seven pigs shedding Salmonella on the last sampling occasion at the farm (Fig. 3), only two, from cohort 3B, were found to have culture-positive cecal contents at slaughter.
TABLE 3.
Detection of Salmonella bacteria and serological findings pre- and postslaughter for 131 pigs originating from six cohorts in three Danish farrow-to-finish swine herdse
| Cohort | Fecesa | Lymph nodes | Cecal contents | Carcass swabs | Overall findings at slaughter | Seruma |
|---|---|---|---|---|---|---|
| 1A | 0/24 | 1/24b | 3/24b | 2/24 | 5/24 | 13/24 |
| 1B | 0/29 | 1/26 | 3/26 | 0/22 | 4/26 | 3/29 |
| 2A | 0/20 | 0/16 | 0/16 | 0/15 | 0/17 | 3/20 |
| 2B | 1/20 | 0/19 | 1/19 | 0/20 | 1/20 | 1/20 |
| 3A | 4/23 | 2/19d | 4/19 | 4/20d | 8/24 | 19/24 |
| 3B | 2/21 | 6/18c | 11/18c | 0/16 | 13/20 | 9/22 |
| Total | 7/137 | 10/122 | 22/122 | 6/117 | 31/131 | 48/139 |
Samples were collected at the final farm visit.
One individual had culture-positive ileocecal lymph node and cecal content samples.
Four individuals had culture-positive ileocecal lymph node and cecal content samples.
One individual had culture-positive ileocecal lymph node and carcass swab samples.
All pigs were from a study on Salmonella dynamics in 2001. Values are numbers of animals with positive samples/total numbers of animals.
DISCUSSION
The present study shows that Salmonella occurrence varies between and within age groups within herds, even in herds with an apparent moderate-to-high infection level. Salmonella was predominant in weaners, growers, and finishers, and was only occasionally detected in sows and gilts, prior to the start of the study. These results correspond closely to results from mandatory bacteriological examinations carried out in the Danish Salmonella control program, in which Salmonella typically is detected in the nursery but rarely in the sow unit (13; D. L. Baggesen, J. Dahl, A. Wingstrand, and B. Nielsen, Proc. 14th Int. Pig Vet. Soc. Congr. 1996, abstr., p. 171, 1996). This correspondence of results suggests that the typical infection level among sows is low, which is contrary to the results in a report from the United States (7), where Salmonella shedding was found to be common in sows. Davies et al. (7) found that sows shed several exotic serotypes but found only Salmonella serotype Typhimurium in finishers. In Denmark, all feedstuffs are heat treated, which reduces the prevalence of exotic serotypes. It could be argued that the (less invasive) exotic serotypes are passively transported along the gut or that differences in the microbiological environment of the gastrointestinal tracts of sows and swine of other age groups are of significance.
The decision to monitor more than one cohort per herd was inspired by experiences with bacteriological examinations in the Danish Salmonella control program. Repeated examinations of the same herd might produce variable results, e.g., between batches (within-herd variation) or monthly examinations (true variation over time). Our results show considerable variation in both Salmonella occurrence and shedding patterns between cohorts in the same herds, which shows that inclusion of more than one cohort per herd was justified.
Salmonella in the farrowing unit.
Except for a few sows examined prior to the start of the study, none of the sows and none of the piglets in the cohorts were shedding Salmonella prior to weaning. This finding suggests that sows in subclinically infected herds play a less important role in Salmonella transmission, although the presence of some seroreacting sows indicated a certain level of Salmonella exposure in the sow units. The fact that none of the 30 sows shed Salmonella bacteria at weaning does not exclude the possibility that some sows shed intermittently in the farrowing pens or in the dry sow unit. However, this specific issue was not investigated in this study. Furthermore, other sows in the farrowing unit might have been shedding, which would enable transmission to the nursery through their piglets. This supposition seems to be supported by the results of Funk et al. (11), who found low levels of Salmonella shedding in both sows and piglets before weaning in three of five cohorts examined. Finally, the amount of feces collected from piglets at weaning was in many cases limited, which might result in an underestimation of the actual prevalence (10).
The observation that piglets nursed by seroreacting sows had a significantly lower prevalence of Salmonella in culture after weaning suggests a role for passively transferred protection against Salmonella infection. This possibility was unfortunately not investigated, but it deserves further study.
Transmission in the nursery.
A few studies have shown that it is possible to raise pigs free of Salmonella by strategic movement at or shortly after weaning (6, 9). These studies suggest that sow-offspring transmission plays a minor role and focus attention on the transmission occurring in the finishing unit. On the other hand, Kranker et al. (15) demonstrated that seropositivity of sows was significantly associated with the finding of Salmonella serotype Typhimurium in the nursery. However, that study included a larger number of herds, some with and some without Salmonella, and therefore its results cannot be directly compared with those of the present study, as weaners from negative herds were not at risk. Kjærsgaard et al. (14) found a significant association between the prevalence of exotic Salmonella serotypes in different batches of sows and that of their piglets after weaning. Funk et al. (11) detected fecal shedding of Salmonella in sows and piglets before weaning. Those authors concluded that the success of segregated early weaning as proposed by Dahl et al. (6) might be farm specific and that absolute exclusion of preweaning infection is unlikely to be achieved by contemporary early weaning alone. Funk et al. (11) furthermore found that even if sows were culture positive during gestation or lactation, the risk of their litters being culture positive did not significantly increase. Interestingly, these authors did not demonstrate a serotype-specific association between culture-positive sows and their piglets. However, this observation might be biased by the facts that many different serotypes were present and that only one isolate per pig was serotyped.
The rapid increase in Salmonella prevalence in the nursery might have been triggered by weaning stress. The most important stress factors are likely to be (i) a change in feed, (ii) the commingling of litters, and (iii) a piglet's being deprived of the antibodies found in sow's milk before the activation of its own immune response. The observations that no piglets were shedding Salmonella just before weaning but that 3 to 4 weeks later in the nursery between 5 and 50% of the piglets were culture positive strongly suggest that horizontal transmission occurred in the nursery. According to Berends et al. (5) and Funk et al. (11), residual infections in the nursing unit and in the transport vehicles seem to be the most plausible origins of infection of the piglets and can explain the sudden rise in prevalence in culture. Since no piglets were shedding Salmonella at weaning and residual infection was not measured in our study, a causal relationship between these factors and the sudden rise in prevalence in culture in the nursery could not be determined. Most likely, Salmonella shed by newly weaned piglets and residual infection act cooperatively, and stress is very likely to exacerbate the resulting infections.
Transmission patterns during the finisher period.
Overall, the present study shows that Salmonella shedding decreased during the fattening period (Fig. 1) but with considerable variation in onset and duration of shedding between and within cohorts (Fig. 3). Some pigs that were found to be shedding in the nursery apparently cleared themselves of infection, whereas others continued shedding in the finishing units. The data also show that despite the exposure, certain pigs were not infected in the nursery but started shedding in the finishing unit. This finding was reflected in the bacteriological curves for both cohorts in herd 1 (Fig. 2) around week 13. Data in Fig. 3 suggest that a shedding pig might infect at-risk penmates at any stage. Residual infection in the finisher units might also play a role, since some of the pigs started shedding after moving to those units. The results from herd 3 suggest that, irrespective of previous exposure or shedding, finisher pigs are still susceptible to a sudden substantial increase in exposure (e.g., through slurry overflow).
Association between serology and the onset and duration of shedding.
Experimental inoculations with Salmonella have shown that the onset of serological response and peak seroprevalence occur at approximately 7 and 30 days postinoculation, respectively (17). In our study there were somewhat longer time periods between peak prevalence in culture, the onset of a serological response, and peak seroprevalence (Fig. 1). The different course of the bacteriological and serological responses occurred because under natural conditions pigs are infected at different points in time, with variability in both exposure and host response (Fig. 3). The results from herds 1 and 2 indicate that even though Salmonella-infected herds might have a proportion of seroreactors sent for slaughter, most pigs have stopped shedding by the time of slaughter. Hence, unless they are carriers and resume shedding or are fecally cross-contaminated during transport or holding, these animals do not constitute a risk to human health with respect to Salmonella. This finding seems to be supported by the autoregressive pattern of the correlation structure for successive culture-positive events (see Table 2), for which self-cure is the most likely explanation. Progressive self-cure and carrier-state animals (19) minimize the amount of Salmonella shedding and consequently reduce the possibility of our detecting Salmonella by the culture technique. The corresponding correlation structure for successive seropositive events (Table 2) is not directly influenced by self-cure but might be due to continuous exposure and immunostimulation, to the half-life of serum antibodies, or to both.
In addition to the marked differences in prevalence in culture between cohorts (Fig. 2), differences in pattern of horizontal transmission were observed (Fig. 3). In some cohorts horizontal transmission seemed to occur frequently (e.g., cohorts 1A and 3A), which is represented in Fig. 3 by many overlapping bars. Other cohorts showed a more stable infection pattern, with a low incidence of shedding (e.g., cohorts 1B and 2B), which is represented in Fig. 3 by few overlapping bars.
In an attempt to describe the dynamics of Salmonella infection in pigs, we adopted a novel approach to estimating the times and patterns of shedding. To the best of our knowledge, such an approach has not previously been published. Using our definitions of shedding time and animals at risk, the results suggest that the dynamics of Salmonella infections are complex in nature. Infection can probably be explained as a random process where a previously uninfected pig has a certain probability of getting infected each time it is exposed. The definition of animals at risk was considered valid. It seems unlikely that an individual pig shedding on the first sampling occasion, not shedding on the second occasion, but shedding again on the third occasion would have been found to have a true-negative culture on the second occasion. Even though the sampling protocol allowed misclassification of intermittent shedders, because of the 3- to 4-week sampling interval, the low sensitivity of the culture technique seems a more reasonable explanation. It is not practically possible to distinguish between intermittent shedding and low-level shedding (i.e., close to detection level). Despite this, an animal with either intermittent or low-level shedding must be considered infected.
All individuals were seronegative on the sampling occasions prior to shedding Salmonella, with the exception of three animals from herd 3, which were seropositive long before Salmonella was detected in their feces. This result further indicates that the sensitivity of the bacteriological culture technique was acceptable but by no means perfect.
We estimated the average shedding time to be 18 or 26 days, depending on the approach used. These shedding times are in accordance with results of Salmonella experts, who estimated the shedding time to range between 17 and 38 days (18a). The fact that some pigs in herd 3 became reinfected following a substantial increase in exposure by a slurry overflow demonstrates that it is possible to penetrate the immunologic barrier in seropositive individuals.
Salmonella findings at slaughter.
Towards the end of the finishing period, cohorts 1A, 3A, and 3B showed a high seroprevalence and cohorts 3A and 3B also showed a high culture prevalence (Fig. 2 and Table 3). Interestingly, Salmonella was isolated most frequently at slaughter from these cohorts (Table 3). Although the data were limited in number, the results are in concordance with a large-scale study carried out by Sørensen et al. (18). Those authors demonstrated significant associations between serology, both at the individual level and at the herd level, and the occurrence of Salmonella in cecal lymph nodes, in cecal contents, and on carcasses. An approximately threefold increase in Salmonella prevalences between farm and slaughterhouse was found. This increase may be due to rapid cross-contamination during transport and lairage (1, 12). The fact that five of six pigs had culture-positive carcass swab samples and culture-negative lymph nodes and cecal contents suggests in-plant cross-contamination as a food safety risk.
Implications for assessment of preharvest prevalence.
The observed variations in Salmonella bacteriology and serology between cohorts and over time indicate that none of these methods on their own were reliable for point estimates of preharvest prevalence in subclinically infected herds. Repeated sampling in different cohorts of animals is required to correctly assess the infection dynamics in the particular herd under study or surveillance. Serological testing is inexpensive, has a higher sensitivity than bacteriology, and has proven its usefulness in the monitoring of Salmonella in finisher pigs (2). However, if additional sampling for bacteriological analysis is required in a herd, the temporal variability in Salmonella levels should be taken into consideration by making the necessary adjustments in the sampling strategy. In three of the six cohorts, the seroprevalence dropped to acceptable levels (<40%). Given that the seroprevalence on the last sampling occasion also applied to the two preceding months, only herd 3 would be classified as other than level 1. After we consulted the central database (the Zoonosis Register), from which all data from the surveillance program are available, herds 1 and 2 were classified as level 1 the month after the last sampling occasion. Herd 3 was classified as level 2 the month after the last sampling occasion, and yet another month later, it was classified as level 3.
Acknowledgments
We thank the participating farmers for their cooperation, Bjarne Nielsen and Leif Straarup Jørgensen for skillful technical assistance, and Henrik Wachmann for statistical advice during the data analysis.
REFERENCES
- 1.Alban, L., and K. Stärk. 2002. Simulating Salmonella prevalence from the growing pig to the slaughtered carcass: where should the effort be put to increase food safety?, p. 98-110. In Proceedings of the Society for Veterinary Epidemiology and Preventive Medicine, Cambridge, United Kingdom.
- 2.Alban, L., H. Stege, and J. Dahl. 2002. The new classification system for slaughter-pig herds in the Danish Salmonella surveillance-and-control program. Prev. Vet. Med. 53:133-146. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 2002. Annual report on zoonoses in Denmark 2001. Ministry of Food, Agriculture and Fisheries, Copenhagen, Denmark.
- 4.Baggesen, D. L., H. C. Wegener, F. Bager, H. Stege, and J. Christensen. 1996. Herd prevalence of Salmonella enterica infections in Danish slaughter pigs determined by microbiological testing. Prev. Vet. Med. 26:201-213. [Google Scholar]
- 5.Berends, B. R., H. A. P. Urlings, J. M. A. Snijders, and F. van Knapen. 1996. Identification and quantification of risk factors in animal management and transport regarding Salmonella spp. in pigs. Int. J. Food Microbiol. 30:37-53. [DOI] [PubMed] [Google Scholar]
- 6.Dahl, J., A. Wingstrand, and B. Nielsen. 1997. Elimination of Salmonella Typhimurium infection by the strategic movement of pigs. Vet. Rec. 140:679-681. [DOI] [PubMed] [Google Scholar]
- 7.Davies, P. R., F. G. E. M. Bovee, J. A. Funk, W. E. M. Morrow, F. T. Jones, and J. Deen. 1998. Isolation of Salmonella serotypes from feces of pigs raised in a multi-site production system. J. Am. Vet. Med. Assoc. 212:1925-1929. [PubMed] [Google Scholar]
- 8.Federal Register. 1996. Pathogen reduction: Hazard Analysis and Critical Control Point (HACCP) system, final rule. Fed. Regist. 61:38917-38928.
- 9.Fedorka-Cray, P. J., D. L. Harris, and S. C. Whipp. 1997. Using isolated weaning to raise Salmonella free swine. Vet. Med. 92:375-382.
- 10.Funk, J. A., P. R. Davies, and M. A. Nichols. 2000. The effect of fecal sample weight on detection of Salmonella enterica in swine feces. J. Vet. Diagn. Investig. 12:412-418. [DOI] [PubMed] [Google Scholar]
- 11.Funk, J. A., P. R. Davies, and M. A. Nichols. 2001. Longitudinal study of Salmonella enterica in growing pigs reared in multiple-site swine production systems. Vet. Microbiol. 83:45-60. [DOI] [PubMed] [Google Scholar]
- 12.Hurd, H. S., J. D. McKean, R. W. Griffith, I. V. Wesley, and M. H. Rostagno. 2002. Salmonella enterica infections in market swine with and without transport and holding. Appl. Environ. Microbiol. 68:2376-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansen, M., V. Møgelmose, J. Dahl, F. Udesen, and B. Nielsen. 1999. The influence of change in feeding and management on the prevalence of multi-resistant Salmonella Typhimurium DT-104 in Danish pig herds. Four case stories, p. 305-307. In P. B. Bahnson (ed.), Proceedings of the 3rd International Symposium on Epidemiology and Control of Salmonella in Pork. University of Illinois, Urbana-Champaign.
- 14.Kjærsgaard, H., L. Jørgensen, H. Wachmann, and J. Dahl. 2001. Effect on Salmonella prevalence by feeding sows meal or pelleted feed, p. 115-117. In P. J. van der Wolf (ed.), Proceedings of the 4th International Symposium on Epidemiology and Control of Salmonella and Other Food Borne Pathogens in Pork. Leipzig University, Leipzig, Germany.
- 15.Kranker, S., J. Dahl, and A. Wingstrand. 2001. Bacteriological and serological examination and risk factor analysis of Salmonella occurrence in sow herds, including risk factors for high Salmonella seroprevalence in receiver finisher herds. Berl. Muench. Tieraerztl. Wochenschr. 114:335-338. [PubMed] [Google Scholar]
- 16.Mousing, J., P. Thode Jensen, C. Halgaard, F. Bager, N. Feld, B. Nielsen, J. P. Nielsen, and S. Bech-Nielsen. 1997. Nation-wide Salmonella enterica surveillance and control in Danish slaughter swine herds. Prev. Vet. Med. 29:247-261. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, B., D. Baggesen, F. Bager, J. Haugegaard, and P. Lind. 1995. The serological response to Salmonella serovars Typhimurium and infantis in experimentally infected pigs. The time course followed with an indirect anti-LPS ELISA and bacteriological examination. Vet. Microbiol. 47:205-218. [DOI] [PubMed] [Google Scholar]
- 18.Sørensen, L., B. Nielsen, and J. Dahl. 2000. Association between Salmonella serology and bacteriological investigations of cecal-content, carcasses, throat and lymph nodes from Danish finishers: level 1-2-3 investigation). (In Danish.) Danish Bacon and Meat Council, Copenhagen, Denmark.
- 18a.Van der Gaag, M. A., and R. B. M. Huirne. 2002. Elicitation of expert knowledge on controlling Salmonella in the pork chain. J. Chain Network Sci. 2:135-146. [Google Scholar]
- 19.Wood, R. L., and R. Rose. 1992. Populations of Salmonella Typhimurium in internal organs of experimentally infected carrier swine. Am. J. Vet. Res. 53:653-658. [PubMed] [Google Scholar]