Abstract
Four spirochete strains were isolated from papillomatous digital dermatitis (PDD) lesions in Iowa dairy cattle and compared with two previously described spirochete strains isolated from dairy cattle in California. These six strains shared an identical 16S ribosomal DNA sequence that was 98% similar to Treponema phagedenis and 99% similar to the uncultivated PDD spirochete sequence DDLK-4. The whole-cell protein profiles resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of these six strains were similar. However, these strains showed differences in the antigenic diversity of lipopolysaccharide (LPS). Genetic diversity was also detected by pulsed-field gel electrophoresis of genomic DNA digests, revealing differences among five of the six strains. Serum immunoglobulin G antibodies from dairy cattle with active PDD lesions reacted with the LPS of all but one PDD spirochete strain. Likewise, peripheral blood mononuclear cells from cattle with active PDD lesions produced blastogenic responses to one of the two California isolates. Both antibody and lymphocyte blastogenic responses were reduced in convalescent dairy cattle, suggesting the immune response to these spirochetes has short duration. These results demonstrate genetic and antigenic diversity among T. phagedenis-like treponemes and provide further evidence for the involvement of these spirochetes in the pathogenesis of PDD.
Papillomatous digital dermatitis (PDD) is a leading cause of lameness in dairy cattle (23). This disease state is referred to by a variety of names including digital dermatitis, interdigital papillomatosis, hairy heelwart, and hairy footwart. PDD begins as a mild, superficial dermatitis which progresses to an erosive lesion. The tissue becomes granulated and can form a hyperkeratotic papillomatous lesion with long hair-like projections (3, 29, 30). These lesions usually form on the plantar surface, proximal to the bulb of the heel, or occasionally, within the interdigital cleft, and are thought to arise due to constant exposure to fecal slurry (2, 28).
The first report of PDD was in 1974 in Europe (8), and the disease is now found with increasing frequency in most geographical regions of the world. In the United States, the disease was identified in 43% of 1,182 U.S. dairy herds surveyed in 1996 and 78% of these herds reported their first cases in 1993 or later. These data confirm a rapid spread of the disease (46), which may in part be due to changes in herd management (31, 32, 46).
The etiology of PDD remains equivocal. Application of topical or parenteral antibiotics results in rapid resolution of the lesions, suggesting that PDD is caused by a bacterial infection. A mixed population of gram-negative bacteria, including anaerobes, microaerophilic organisms, and spirochetes have been demonstrated in or isolated from PDD lesions (4, 26, 29). Spirochetes are prevalent in PDD lesions and may be important in pathogenesis. Spirochetes are found deep within the epidermis, invading the stratum spinosum and dermal papillae (2, 4, 22, 29).
At least five different phylogenetic groups of Treponema (9) were detected in PDD lesions by analysis of PCR-amplified rRNA, and three of these groups (DDKL-3, DDKL-4, and DDKL-13) closely resemble cultivatable human spirochetes Treponema denticola, Treponema phagedenis, and Treponema vincentii/Treponema medium, respectively. Because these bacteria are strict anaerobes with fastidious nutritional requirements, few have been isolated in pure culture (44). To date, four different genetic groups of Treponema have been isolated from PDD lesions: Walker and colleagues first isolated T. phagedenis-like, T. denticola-like, and T. vincentii/T. medium-like spirochetes (35, 44; R. L. Walker, D. H. Read, S. J. Sawyer, and K. J. Loretz, Abstr. 79th Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 17, 1998). Demirkan et al. (12) isolated T. denticola-like spirochetes, and Schrank et al. (33) isolated and characterized the newly proposed species Treponema brennaborense. While PDD can be reproduced experimentally by inoculating the feet of fecal slurry-exposed cattle with lesion material, bacteria isolated from PDD lesions have not fulfilled Koch's postulates in experimental infections (27).
In the present study, we report on the genetic and immunologic characterization of four Treponema spirochete strains isolated from PDD lesions in Iowa dairy cattle and compare them to two strains previously isolated from PDD lesions in California dairy cattle (44). Our findings show that all six bovine spirochetes are T. phagedenis-like and exhibit genetic and antigenic diversity. In addition, we report that dairy cattle with erosive PDD lesions develop an immune response primarily against the lipopolysaccharide (LPS) of these spirochetes.
MATERIALS AND METHODS
Bacterial strains and media.
Spirochete strains 9-3301 and 2-1498 were originally isolated from dairy cattle in California with PDD lesions (44). Brachyspira hyodysenteriae strain B204 was provided by Thad Stanton, Enteric Diseases and Food Safety Research Group, National Animal Disease Center, Ames, Iowa. Prereduced, anaerobic oral Treponema isolation (OTI) broth and agar were prepared as described under 100% N2 (34). Most experiments used OTI medium supplemented with 10% heat-treated newborn calf serum (OTIS). Media with antibiotics (OTISER) contained enrofloxacin (5 μg/ml) and rifampin (25 μg/ml). In some experiments, 100 μg of polymyxin B/ml was added to the OTISER agar.
Blood sampling.
Sera for immunoblotting and blood in 10% anticoagulant citrate-dextrose for collection of peripheral blood mononuclear cells (PBMC) were obtained from 13 Jersey cows at an Iowa dairy farm with a history of PDD. PDD lesions were classified as erosive or papillomatous following clinical examination of the foot by a veterinarian. The samples were obtained from cows with erosive lesions (n = 4), papillomatous lesions (n = 4), and cows that had been previously treated with topical antibiotics for PDD and had recovered (n = 5). Sera were also obtained from 3 of the cows with erosive lesions and 2 of the cows with papillomatous lesions 42 days after topical antimicrobial treatment, by which time 4 of the 5 cows had recovered. Negative-control samples were obtained from 6 healthy cows from a second herd with no history of PDD.
Collection of PDD biopsy samples.
Biopsy samples were obtained from a second group of 10 Jersey cows with erosive PDD lesions on a subsequent visit to the farm. The cattle were restrained on a tilt table, and the feet were washed with water and scrubbed with a soft brush to remove excess fecal material. Lidocaine hydrochloride (5 to 10 ml) was injected subcutaneously in a ring block around the lesion and a 1- by 0.5- by 0.5-cm full-skin-thickness wedge biopsy sample was obtained with a sterile scalpel and forceps from the margin of the lesion. The biopsy samples were gently rinsed in sterile, distilled water, placed in semisolid anaerobic transport medium (Anaerobe Systems, San Jose, Calif.), and kept on ice for a maximum of 4 h before transport back to the laboratory.
Isolation of spirochetes.
Lesion material was prepared under anaerobic conditions for culture as described previously (44), with modifications. Lesion material and cultures were examined by phase-contrast microscopy to identify morphotypes and confirm the presence of spirochetes throughout the isolation procedure. Tissue was cut into 1-mm2 cubes with a sterile scalpel blade, the cubes were macerated with a sterile swab in 400 μl of OTI broth, and 200 μl of this suspension was used to inoculate 7 ml of OTISER broth to enrich for spirochetes. After an initial incubation at 37°C for 24 h, 100-μl aliquots were spread onto OTISER agar plates and incubated at 37°C for up to 14 days. Agar plugs containing bacterial colonies were removed from the OTISER agar plates with a Pasteur pipette, confirmed to be composed of spirochetes by phase-contrast microscopy, and cultivated in OTIS broth without antibiotics. Growth in OTIS broth was measured by equating optical density at 620 nm (OD620) in a Bausch and Lomb spectrophotometer with bacterial counts obtained by using a Petroff-Hausser counting chamber. The purity of the spirochete cultures was determined by subculturing them onto OTIS agar and Trypticase soy blood agar plates incubated aerobically and anaerobically for up to 5 days. Spirochete cultures were stored at −70°C in 50 to 100× their original concentrations in fresh OTIS containing 20% glycerol as recommended previously (36).
Electron microscopy.
Spirochetes grown to an OD620 of 0.8 were harvested by centrifugation at 2,000 × g for 5 min, washed twice in TBS (10 mM Tris, 150 mM Tris-buffered saline heavy and light chain [pH 7.4]), and suspended in distilled water. Samples were negatively stained with an equal volume of 2.5% phosphotungstic acid (pH 7) and examined at 80 kV under a Phillips model 410 transmission electron microscope as described previously (40).
16S rDNA sequencing and analysis.
Genomic DNA was isolated from each PDD spirochete in the exponential phase of growth (37). 16S ribosomal DNA (rDNA) was amplified from genomic DNA by PCR with universal bacterial primers 410 (5′-GAG TTT GAT C[A/C]T GGC TCA G-5′) and 408 (5′-GGT TAC CTT GTT ACG ACT T-5′), corresponding to positions 9 to 27 and 1492 to 1510 of the Escherichia coli 16S rRNA gene, respectively (5). Amplicons were generated by using a three step program of (i) 98°C for 2 min; (ii) 30 cycles of 95°C for 1 min, 42°C for 1 min, and 72°C for 8 min; and (iii) 72°C for 8 min. PCR products were purified in a Microcon 100 ultrafiltration cartridge (Millipore, Bedford, Mass.), and the sequence was determined by dideoxynucleotide termination reactions resolved on Applied Biosystems 377 Prims DNA sequencers at the Iowa State University DNA facility. Sequence data were analyzed and assembled with Vector NTI, version 5.5 (North Bethesda, Md.). The sequence data were submitted to GenBank.
Pulsed-field gel electrophoresis.
PDD spirochetes were grown to an OD620 of 0.8 in 100 ml of OTIS broth and encapsulated in agarose beads, and genomic DNA was prepared as described previously (47). Encapsulated DNA was digested at 37°C with approximately 10 U of restriction endonuclease for 4 to 6 h, then separated in 1% agarose gels by clamped homogenous electric field electrophoresis as described previously (47). Restriction fragments were visualized by UV illumination after staining with ethidium bromide.
SDS-PAGE.
Each spirochete strain was grown to an OD620 of 0.8 in 7 ml of OTIS, harvested by centrifugation (8,000 × g, 10 min, 4°C), washed twice, and resuspended in TBS. Protein concentrations were determined by the modified Lowry assay (20). Samples (100 μg of protein) were precipitated in 10 volumes of ice-cold acetone, dissolved in 1× sample buffer, boiled for 5 min, and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using standard techniques (18). Proteins were visualized by staining with Coomassie blue. For immunoblot analysis, each lane contained either 5 to 10 μg of cellular protein or 5 to 10 μg of cellular protein digested with 200 μg of proteinase K/ml at 56°C for 2 h.
Immunoblot analysis.
Standard methods were used for immunologic detection of antigens separated by SDS-PAGE (39). Immunodetection was achieved with the Enhanced Chemiluminescence System (Amersham Pharmacia Biotech, Arlington Heights, Ill.) following the manufacturer's recommendations. All antibody solutions were made in 5% skim milk and 0.01% Tween 20 in TBS. A 1:1,000 dilution of pooled sera from cows with erosive lesions was used as the primary antibody, and a 1:5,000 dilution of horseradish peroxidase labeled goat anti-bovine immunoglobulin G H&L was used as the secondary antibody. Slot blot analysis was done with a miniblotter (Immunetics, Cambridge, Mass.) probed with 1:250 dilutions of sera obtained from cows with no lesions, cows with erosive or papillomatous lesions, and convalescent cows 42 days following treatment.
Lymphocyte blastogenesis assay.
PBMC were isolated from peripheral blood by discontinuous density gradient separation (1.077 g of Ficoll-Hypaque [Sigma, St. Louis, Mo.]/ml). Wells of 96-well round-bottom microtiter plates were seeded with 2 × 105 PBMC in a total volume of 200 μl/well. The medium was RPMI 1640 (Fisher Scientific, Pittsburgh, Pa.) supplemented with 100 U of penicillin/ml, 0.1 mg of streptomycin/ml, 5 × 10−5 M 2-mercaptoethanol (Sigma Chemical Co.), and 10% fetal bovine serum (Atlanta Biologics, Atlanta, Ga.). Treatments included media alone (no stimulation), media plus antigen, or mitogen stimulation. Antigens tested included whole-cell sonicates of PDD strains 2-1498 and 9-3301 (both at 2 μg/ml and 10 μg/ml on a dry matter basis), B. hyodysenteriae strain B204 (5 μg/ml), and Bacteroides vulgatus ATCC strain 8482 (5 μg/ml). Mitogen stimulation was with 5 μg of concanavalin A (Sigma Chemical Co.)/ml. Plates were then incubated at 37°C in a 5% CO2 humidified atmosphere for 5 days. On day 4, 0.5 mCi of methyl-[3H]thymidine in 10 μl of medium was added to each well, and plates were incubated overnight for an additional 18 to 20 h. Well contents were harvested onto glass fiber filters with a PHD Cell Harvester (Cambridge Technology, Cambridge, Mass.), and incorporated radioactivity was measured by liquid scintillation counting. Treatments were run in triplicate, and stimulation indices (SI) were calculated by dividing counts per minute of stimulated wells by counts per minute from nonstimulated wells. Data are presented as SI ± standard errors of the mean, and an SI of ≥2 was considered to be significant.
Nucleotide sequence accession number.
Sequence data were submitted to GenBank with the accession numbers AF546873, AF546874, AF546875, AF546876, AF546877, and AF546878.
RESULTS
Isolation of spirochetes from PDD lesions.
Highly motile spirochetes were observed in all 10 biopsy suspensions. In addition, we also detected motile rods, long, filamentous, clumping rods, short nonmotile rods, and cocci in chains. The bacterial population, after an initial incubation in OTISER broth for 24 h at 37°C, was enriched for motile spirochetes, but some of the other bacterial morphotypes were also still present. Samples of the OTISER broth were spread onto OTISER agar. Colonies containing short, gram-negative rods and gram-positive cocci appeared on the agar plates within 48 h of incubation. Extended incubation of these plates was required to isolate spirochetes. Colonies of spirochetes appeared by 9 days incubation as small (1- to 2-mm diameter), white, opaque, firmly adherent colonies that grew down into the agar. Examination under the phase-contrast microscope confirmed that these colonies consisted of spirochetes. Following subculture and repeated cycles of growth in OTISER broth and plates, spirochetes were obtained in pure culture from 4 of the 10 biopsy samples (strains 1A, 3A, 4A, and 5B).
Characterization of PDD spirochete strains.
Transmission electron microscopy of spirochete strains 1A, 3A, 4A, and 5B showed that these bacteria are 10 to 15 μm in length and 0.35 to 0.40 μm wide and have 7 to 9 periplasmic flagella inserted subterminally at each end, giving a total of 14 to 18 flagella per cell. These bacteria resemble the PDD spirochete strains (2-1498 and 9-3301) isolated by Walker et al. (44) from dairy cattle in California. Analyses of 16S rDNA sequences (1,455 bp) were obtained for strains 1A, 3A, 4A, and 5B. These strains had identical 16S rDNA sequences that were 99% identical to that obtained previously for strain 2-1498 (GenBank accession number L78126). These 16S rDNA sequences also share 99% identity to the uncultivated PDD spirochete DDLK-4 (GenBank accession number Y08894) described by Choi et al. (9) and 98% identity to T. phagedenis (GenBank accession number L78126). The sequences were only 92% identical to the uncultivated spirochete sequence DDLK-3 (GenBank accession number Y08893) and 91% identical to T. brennaborense (GenBank accession number Y1658.1).
Genetic diversity among T. phagedenis-like spirochetes.
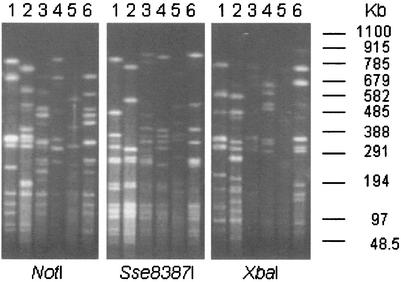
Pulsed-field agarose gel electrophoresis of large restriction fragments is a useful method for the comparison of bacterial strains. Genomic DNA from PDD spirochetes was digested with a variety of rare cutting enzymes. Digests obtained with three restriction endonucleases (NotI, Sse8387I, and XbaI) generated well-resolved fragments from all six PDD isolates. With the exception of the patterns of isolates 9-3301 and 3A, which were closely related by this method, the digestion patterns of all the other isolates were unique (Fig. 1). In contrast, the SDS-PAGE profile of proteins from these PDD spirochete strains is quite similar (Fig. 2).
FIG. 1.
Pulsed-field gel electrophoresis analysis of spirochete DNA. Genomic DNA from each strain was isolated in agarose beads and digested with the restriction endonucleases indicated on the figure. Lanes 1, 9-3301; lanes 2, 2-1498; lanes 3, 4A; lanes 4, 5B; lanes 5, 1A; lanes 6, 3A. The ethidium bromide-stained gel is shown. The migration of size markers (lambda phage concatemers and S. cerevisiae chromosomes) is shown on the right.
FIG. 2.
SDS-PAGE profiles of whole-cell protein extracts of six bovine T. phagedenis-like spirochete strains isolated from PDD lesions in dairy cattle (100 μg of protein per lane). Lane 1, 2-1498; lane 2, 9-3301; lane 3, 1A; lane 4, 3A; lane 5, 4A; lane 6, 5B. The relative mobility of molecular mass markers (in kilodaltons) is shown on the left.
Dairy cattle with PDD develop a humoral immune response to T. phagedenis-like spirochetes.
Western blotting showed that sera from cows with erosive PDD lesions consistently reacted with many bands from each PDD spirochete strain except 2-1498. The reactivity detected was specific for the T. phagedenis-like bacteria, as there was no detectable reaction with antigens from another spirochete, B. hyodysenteriae strain B204 (Fig. 3A). Sera from healthy cattle with no history of PDD did not react with any of the spirochete strains (data not shown). Immunoblot analysis of whole-cell extracts pretreated with proteinase K showed that the immunoreactivity was directed primarily against spirochete LPS. Immunoreactive bands had a regular, ladder-like pattern that resembled O-antigen side chains of smooth gram-negative LPS (Fig. 3B). In the corresponding SDS-PAGE gel of proteinase K-treated whole-cell protein extract, the protein bands were entirely digested except for a proteinase K-resistant band of 34 kDa, confirming that the immunoreactive material was not proteinaceous in nature (data not shown). Differences in the pattern and intensity of the immunoreactive bands in the proteinase K-resistant material were apparent among the five strains. The immunoreactive bands of strain 9-3301 had a regular, ladder-like structure of repeating O-antigen subunits. The four strains isolated from Iowa dairy cattle also possessed immunoreactive, ladder-like O antigens. However, additional broad bands of approximately 40 to 55 kDa and 80 to 90 kDa were also apparent. The antigenic structures of strains 1A and 5B were very similar. Strains 3A and 4A were also similar, except that strain 4A possessed an 80-kDa band that was intensely reactive and 3A did not possess an antigenic band in this region.
FIG. 3.
Immunoblot analysis of whole-cell (A) and proteinase K-treated (B) extracts of bovine T. phagedenis-like spirochetes and B. hyodysenteriae strain B204 probed with pooled sera from three Iowa dairy cows with erosive PDD lesions (5 μg of protein per lane). Lane 1, 2-1498; lane 2, 9-3301; lane 3, 1A; lane 4, 3A; lane 5, 4A; lane 6, 5B; lane 7, B204. The relative mobility of molecular mass markers (in kilodaltons) is shown on the left.
As strain 4A exhibited the most intense humoral response, it was used as the antigen for the comparison of the reactivities of cattle in the active and convalescent stages of PDD. Sera from 2 of 3 cows with erosive PDD lesions and 1 of the 2 cows with papillomatous PDD lesions reacted strongly with strain 4A whole-cell extracts pretreated with proteinase K. Following recovery, sera from these cows did not react as strongly, indicating that the antibody response had waned considerably during convalescence (Fig. 4).
FIG. 4.
Immunoblot analysis of proteinase K-treated whole-cell extracts of bovine T. phagedenis-like spirochete strain 4A probed with sera from cows in various stages of PDD infection. Lanes 1 to 2, healthy cows with no history of PDD; lane 3, cow 1, erosive lesion; lane 4, cow 1, recovered; lane 5, cow 2, erosive lesion; lane 6, cow 2, recovered; lane 7, cow 3, erosive lesion; lane 8, cow 3, erosive lesion 42 days after the first serum sample; lane 9, cow 4, papillomatous lesion; lane 10, cow 4, recovered; lane 11, cow 5, papillomatous lesion; lane 12, cow 5, recovered. The relative mobility of molecular mass markers (in kilodaltons) is shown on the left.
Cattle with PDD develop a cellular response to T. phagedenis-like spirochetes.
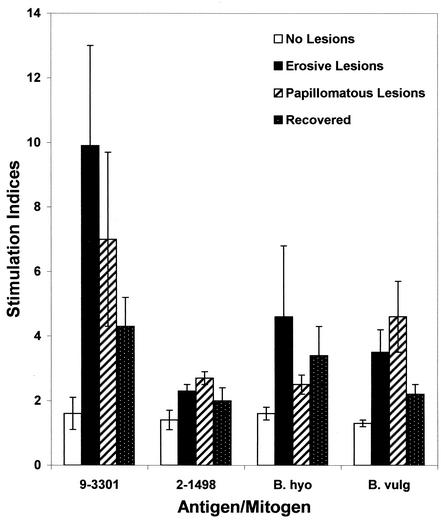
Results of the lymphocyte blastogenesis assay indicated that cattle with erosive (SI = 9.9 ± 3.1) and papillomatous (SI = 7.0 ± 2.7) PDD lesions produced a specific, cellular immune response to PDD spirochete strain 9-3301 in comparison to cattle with no prior history of the disease (SI = 1.6 ± 0.5). Cattle with erosive PDD lesions also produced significant cellular responses to B. hyodysenteriae (SI = 4.6 ± 2.2) and Bacteroides vulgaris (SI = 3.5 ± 0.7). However, PDD strain 2-1498 was relatively inert (SI = 2.3 ± 0.3) (Fig. 5). The cell-mediated immune response of recovered cows to strain 9-3301 was reduced in comparison to cows with active lesions (SI = 4.3 ± 0.9), a finding that was confirmed when lymphocyte blastogenic responses were compared for the same animals in the active and convalescent stages of the disease (Fig. 6).
FIG. 5.
Mean blastogenic responses of lymphocytes from Iowa cattle to California bovine T. phagedenis-like spirochete antigens. PBMC from cattle with erosive or papillomatous lesions, convalescent cattle, or cattle with no history of PDD were cultured with and without antigen. Antigens included California PDD spirochete strains 9-3301 and 2-1498 (10 μg of each/ml), B. hyodysenteriae (B. hyo) strain B204 (5 μg/ml), and B. vulgatus (B. vulg) ATCC strain 8482 (5 μg/ml). SI of ≥2 were considered significant. The SI results for concanavalin A stimulation were 85.6, 67.3, 54.5, and 14.7, respectively.
FIG. 6.
Waning of spirochete-specific lymphocyte blastogenic responses following topical antimicrobial treatment of cattle with PDD. On day 0, cows 1 to 3 were treated for erosive lesions and cows 4 and 5 were treated for papillomatous lesions. When reexamined on day 42, all of the cows had recovered except for cow 3. Decreased spirochete-specific lymphocyte blastogenic responses to bovine T. phagedenis-like spirochete strain 9-3301 were associated with recovery in 3 of 4 cows.
DISCUSSION
Despite much indirect evidence linking spirochetes with the etiology of PDD (2, 11, 14, 28, 29, 44), the pathogenic capability of these organisms still remains to be established. A significant hindrance to progress is the difficulty many researchers have experienced in isolating spirochetes from PDD lesions. PDD spirochetes are strict anaerobes that do not survive even short exposures to oxygen, they have fastidious substrate requirements, and they are easily overgrown by more-rapidly growing organisms that are present in the lesions. In addition, PDD spirochetes comprise a number of distinct species and the relative contribution of each species to lesion development (either alone or in combination) is unknown. At least five Treponema phylotypes have been demonstrated in PDD lesions by DNA-based techniques (9).
Moter et al. (22) used fluorescence in situ hybridization to determine the spatial distribution of these phylogenetic groups in PDD lesions. T. denticola-like (DDLK-3) spirochetes were only observed in the surface debris and superficial layers, indicating that they may not be key pathogens but merely secondary invaders. In contrast, T. phagedenis-like (DDLK-4) spirochetes and spirochetes that reacted with human oral treponeme probes TRE-I (T. vincentii/T. medium-like) and TRE-IV (21) were present deep within the epidermis next to the dermal papillae. A long, stretched bacterial morphotype that did not react with any of the spirochete probes but reacted with the universal bacterial probe was located even deeper within the dermis. Based on both 16S rDNA and 16S-23S intergenic spacer region sequence analysis, spirochetes previously isolated from California dairy herds fall within three phylogenetic groups: T. denticola-like, T. phagedenis-like, and T. vincentii/T. medium-like, although 16S-23S intergenic spacer region sequence analysis was more discriminatory for the T. denticola-like spirochetes (35, 44; Walker et al., Abstr. 79th Annu. Meet. Conf. Res. Workers Anim. Dis.). However, in the United Kingdom and Germany, only T. denticola-like spirochetes (12) and T. brennaborense, respectively, have successfully been isolated from PDD lesions (33). Spirochetes that are closely related to T. vincentii/T. medium and T. denticola have also been isolated from sheep with severe virulent foot rot, but the potential for cross-species transmission of these organisms between sheep and cattle is unknown (10, 13, 24).
By adopting stringent anaerobic culture techniques and utilizing the selective medium recommended by Walker et al. (44), we successfully isolated Treponema spirochetes from PDD lesions in dairy cattle in Iowa. However, less than optimal recovery was achieved due to the same problems with overgrowth of other organisms experienced by Walker et al. (44). Spirochetes were observed microscopically in all 10 biopsy samples but could only be isolated in pure culture from 4 of the biopsy samples. The spirochetes isolated from Iowa dairy cattle had a similar ultrastructure to 7 of the 8 California PDD spirochetes isolated by Walker et al. (44), shared an identical 16S rDNA sequence with two of these strains, and were closely related to the T. phagedenis-like phylotype identified by Choi et al. (9). Furthermore, the results of 16S rDNA sequencing for the Iowa strains were in agreement with 16S-23S intergenic spacer region sequence analysis. This technique also placed the four Iowa strains together with 2-1498 and 9-3301 in the T. phagedenis-like phylotype (35). The isolation procedure used may favor the selection of this group of spirochetes in preference to other PDD phylogenetic groups, or it is also possible that T. phagedenis-like spirochetes are the predominant phylotype in PDD lesions in the dairy cattle sampled during these studies.
The 16S rDNA sequence obtained for the six PDD spirochete strains was 98% identical to the corresponding T. phagedenis sequence (37), a nonpathogenic treponeme that shares some cross-reactive antigens with Treponema pallidum (7). A sequence identity of 98% is on the borderline for assigning these spirochetes within the T. phagedenis species. Furthermore, comparison of cloned 16S-23S intergenic spacer regions from strain 2-1498 and the type strain of T. phagedenis showed that they were 99.2% identical (35). However, further techniques such as DNA-DNA relative reassociation and complete phenotypic analysis will be required to determine the true taxonomic position of this phylotype. Preliminary results showed that bovine T. phagedenis-like spirochetes produced acetate, formate, butyrate, and proprionate as fermentation end products when grown in OTIS broth (D. J. Trott, unpublished data). In comparison, T. phagedenis produces acetate, propionate, and butyrate but not formate (7).
The assignment of these spirochetes to T. phagedenis would certainly raise questions concerning their origin and pathogenic significance. The type strain of T. phagedenis, the Reiter treponeme, was originally isolated from a human syphilitic sore. Interestingly, this strain was initially pathogenic in rabbits, but then it lost its virulence and is now believed to be a nonpathogenic commensal of the normal genital flora of humans and other primates (7, 45). In contrast, bovine T. phagedenis-like spirochetes have only been demonstrated in PDD lesions and not in healthy bovine feet. Bovine T. phagedenis-like spirochetes migrate deep within the epidermis and penetrate the stratum spinosum (22). In contrast, bovine T. denticola-like spirochetes were only demonstrated to invade the superficial layer and associated debris of PDD lesions (22), even though they possess a T. denticola-like hemolysin and were initially thought to have a major role in PDD by analogy with T. denticola pathogenesis in periodontal disease (11). T. denticola possesses a number of other virulence factors that damage the host, including a chymotrypsin-like protease and a pore-forming major surface protein (16). It remains to be determined if the bovine T. phagedenis-like spirochetes possess similar virulence determinants that may cause the typical lesions associated with PDD, such as necrosis of the epidermis.
The six bovine T. phagedenis-like spirochetes displayed both genetic and antigenic diversity. It is not unusual for certain species of spirochete, such as Brachyspira pilosicoli, to demonstrate marked within-species genetic and antigenic variability while having highly conserved 16S rDNA sequences (19, 38, 41). Based on pulsed-field gel electrophoresis patterns obtained with three different restriction endonucleases, the four spirochete isolates obtained from a single Iowa dairy farm each represented a distinct strain. In a previous study, 7 PDD spirochetes isolated from California dairy cattle were separated into 7 distinct restriction endonuclease patterns, although these isolates were obtained from four different farms (44). In combination, these results suggest that bovine T. phagedenis-like spirochetes are genetically heterogeneous. Interestingly, California strain 9-3301 and Iowa strain 3A were closely related by pulsed-field gel electrophoresis, suggesting that they belong to the same clonal group. The examination of further isolates would help determine the within- and between-herd diversity of these spirochetes. It also may be possible that an individual animal is colonized with more than one strain. However, in the present study, only one isolate from each biopsy sample was characterized.
Immunoblotting and blastogenic response assays confirmed that Iowa dairy cattle with erosive lesions produce both humoral and cellular responses to bovine T. phagedenis-like spirochetes. This supported previous studies that demonstrated humoral responses to T. phagedenis-like and also T. denticola-like spirochetes in cattle with PDD lesions (14, 43). Interestingly, in these previous studies, a serum immunoglobulin G response was demonstrated to strain 2-1498 and immunoblotting showed that the predominant antigens were proteins of 34, 41, and 55 kDa, although some cows also produced antibody to LPS-like material (14, 43). In the present study, Iowa dairy cattle with PDD lesions produced a strong humoral response to the LPS of five of the six spirochetes. Interestingly, strain 2-1498 was consistently unreactive with serum antibodies from Iowa dairy cattle and did not induce a significant cellular immune response, suggesting that it was antigenically distinct from the other T. phagedenis-like isolates. It is possible that a less dilute serum sample may have detected the protein antigens identified in this strain in a previous study (14). However, it is more likely that strain 2-1498 may represent a distinct serological type that was not present in the Iowa dairy herd. Serological specificity to LPS is involved to some degree in protective immunity in other spirochete diseases such as leptospirosis and swine dysentery (6, 17, 25, 42).
The significant blastogenesis SI observed for B. hyodysenteriae suggest that cell-mediated immunity may be directed against bacterial components that are common between PDD treponemes and other species of spirochete. Antisera from cattle with PDD have previously been shown to react with Borrelia burgdorferi, either because the cattle were exposed to this spirochete or because of immunologic cross-reactivity with PDD spirochetes (1, 14). Immunologic cross-reactivity may be due to flagellar proteins, which tend to have conserved antigenic regions shared between spirochete genera. The lymphocyte blastogenic response to B. vulgatus suggests that gram-negative anaerobes also have some involvement in the pathogenesis of PDD.
On the basis of pulsed-field gel electrophoresis, strain 9-3301 was genetically related to strain 3A, and Iowa dairy cattle with PDD produced both humoral and cellular immune responses to this strain. The four Iowa PDD strains and strain 9-3301 were all reactive with pooled sera from cattle with active lesions, although each strain had a different antigenic LPS profile. Strain 4A possessed a strongly antigenic, proteinase K-resistant antigen of approximately 80 kDa. In future studies, it will be interesting to determine if strain 4A also produces a stronger lymphocyte blastogenic response than those of the other strains.
Histologically, PDD spirochetes advance along the horny columns of the epidermis, and large numbers of inflammatory cells (predominantly neutrophils and monocytes) infiltrate these sites. In chronic PDD infections, antibody-secreting plasma cells become a significant part of the inflammatory cell population (15). Our results suggest that T. phagedenis-like LPS may be an important target for the inflammatory response in PDD lesions; however, confirmation will require analysis of the inflammatory reactivity of purified LPS in the lymphocyte blastogenesis assay. We have demonstrated that cattle in the active stages of infection produce significant cellular and serum antibody responses to bovine T. phagedenis-like spirochetes. However, this immune response appears to be of short duration and rapidly diminishes, suggesting that reinfection of convalescent animals is possible. If the immune response to the other PDD spirochetes is also of short duration, control of PDD via the use of Treponema bacterins would be difficult to achieve or may require multiple doses to sustain protective immunity, particularly if these bacterins only induce a serum antibody response in the host.
Acknowledgments
The excellent technical assistance of Sam Humphrey and Tonia McNunn and the advice of Thad Stanton are greatly appreciated.
This work was supported in part by funds provided by the Iowa Livestock Health Advisory Council.
REFERENCES
- 1.Blowey, R. W., S. D. Carter, A. G. White, and A. Barnes. 1994. Borrelia burgdorferi infections in UK cattle: a possible association with digital dermatitis. Vet. Rec. 135:577-578. [PubMed] [Google Scholar]
- 2.Blowey, R. W., S. H. Done, and W. Cooley. 1994. Observations on the pathogenesis of digital dermatitis in cattle. Vet. Rec. 135:112-115. [DOI] [PubMed] [Google Scholar]
- 3.Blowey, R. W., and M. W. Sharp. 1988. Digital dermatitis in dairy cattle. Vet. Rec. 122:505-508. [DOI] [PubMed] [Google Scholar]
- 4.Blowey, R. W., M. W. Sharp, and S. H. Done. 1992. Digital dermatitis. Vet. Rec. 131:39. [DOI] [PubMed] [Google Scholar]
- 5.Both, B., G. Krupp, and E. Stackebrandt. 1991. Direct sequencing of double-stranded polymerase chain reaction-amplified 16S rDNA. Anal. Biochem. 199:216-218. [DOI] [PubMed] [Google Scholar]
- 6.Bulach, D. M., T. Kalambaheti, A. de la Pena Moctezuma, and B. Adler. 2000. Lipopolysaccharide biosynthesis in Leptospira. J. Mol. Microbiol. Biotechnol. 2:375-380. [PubMed] [Google Scholar]
- 7.Canole Parola, E. 1984. The spirochetes, p. 38-70. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkens, Baltimore, Md.
- 8.Cheli, R., and C. M. Mortellaro. 1974. La dermatite digitale del bovino, p. 208-213. In P. Gallarati (ed.), Proceedings of the 8th International Conference on Diseases of Cattle. Piacenza, Milan, Italy.
- 9.Choi, B. K., H. Natterman, S. Grund, W. Haider, and U. B. Göbel. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47:175-181. [DOI] [PubMed] [Google Scholar]
- 10.Collighan, R. J., R. D. Naylor, P. K. Martin, B. A. Cooley, N. Buller, and M. J. Woodward. 2000. A spirochete isolated from a case of severe ovine foot disease is closely related to a treponeme isolated from human periodontitis and bovine digital dermatitis. Vet. Microbiol. 74:249-257. [DOI] [PubMed] [Google Scholar]
- 11.Collighan, R. J., and M. J. Woodward. 1997. Spirochaetes and other bacterial species associated with bovine digital dermatitis. FEMS Microbiol. Lett. 156:37-41. [DOI] [PubMed] [Google Scholar]
- 12.Demirkan, I., S. D. Carter, C. A. Hart, and M. J. Woodward. 1999. Isolation and cultivation of a spirochaete from bovine digital dermatitis. Vet. Rec. 145:497-498. [DOI] [PubMed] [Google Scholar]
- 13.Demirkan, I., S. D. Carter, C. Winstanley, K. D. Bruce, N. M. McNair, M. Woodside, and C. A. Hart. 2001. Isolation and characterisation of a novel spirochaete from severe virulent ovine footrot. J. Med. Microbiol. 50:1061-1068. [DOI] [PubMed] [Google Scholar]
- 14.Demirkan, I., R. L. Walker, R. D. Murray, R. W. Blowey, and S. W. Carter. 1999. Serological evidence of spirochaetal infections associated with digital dermatitis in dairy cattle. Vet. J. 157:69-77. [DOI] [PubMed] [Google Scholar]
- 15.Döpfer, D., A. Koopmans, F. A. Meijer, I. Szakall, H. Schukken, W. Klee, R. B. Bosma, J. L. Cornelisse, A. J. A. M. van Asten, and A. A. H. M. ter Huurne. 1997. Histological and bacterial evaluation of digital dermatitis in cattle, with special reference to spirochaetes and Campylobacter faelis. Vet. Rec. 140:620-623. [DOI] [PubMed] [Google Scholar]
- 16.Fenno, J. C., G. W. K. Wong, P. M. Hannam, and B. C. McBride. 1998. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol. Lett. 163:209-215. [DOI] [PubMed] [Google Scholar]
- 17.Joens, L. A., S. C. Whipp, R. D. Glock, and M. E. Nuessen. 1983. Serotype-specific protection against Treponema hyodysenteriae infection in ligated colonic loops of pigs recovered from swine dysentery. Infect. Immun. 39:460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lee, B. J., and D. J. Hampson. 1999. Lipo-oligosaccharide profiles of Serpulina pilosicoli strains and their serological cross-reactivities. J. Med. Microbiol. 48:411-415. [DOI] [PubMed] [Google Scholar]
- 20.Markwell, M., S. Haas, L. Bieber, and N. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 21.Moter, A., C. Hoenig, B. K. Choi, B. Riep, and U. B. Göbel. 1998. Molecular epidemiology of oral treponemes associated with periodontal disease. J. Clin. Microbiol. 36:1399-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moter, A., G. Leist, R. Rudolph, K. Schrank, B. K. Choi, M. Wagner, and U. B. Göbel. 1998. Flourescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459-2467. [DOI] [PubMed] [Google Scholar]
- 23.Murray, R. D., D. Y. Downham, M. J. Clarkson, W. B. Faull, J. W. Hughes, F. J. Manson, J. B. Merritt, W. B. Russell, J. E. Sutherst, and W. R. Ward. 1996. Epidemiology of lameness in dairy cattle: description and analysis of foot lesions. Vet. Rec. 138:586-591. [DOI] [PubMed] [Google Scholar]
- 24.Naylor, R. D., P. K. Martin, J. R. Jones, and M. C. Burnell. 1998. Isolation of spirochaetes from an incident of severe virulent ovine footrot. Vet. Rec. 143:690-691. [PubMed] [Google Scholar]
- 25.Nuessen, M. E., and L. A. Joens. 1982. Serotype-specific opsonization of Treponema hyodysenteriae. Infect. Immun. 38:1029-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohya, T., H. Yamaguchi, Y. Nii, and H. Ito. 1999. Isolation of Campylobacter sputorum from lesions of papillomatous digital dermatitis in dairy cattle. Vet. Rec. 145:316-318. [DOI] [PubMed] [Google Scholar]
- 27.Read, D. H., and R. L. Walker. 1996. Experimental transmission of papillomatous digital dermatitis (footwarts) in cattle. Vet. Pathol. 33:607. [Google Scholar]
- 28.Read, D. H., and R. L. Walker. 1998. Papillomatous digital dermatitis (footwarts) in California dairy cattle: clinical and gross pathologic findings. J. Vet. Diagn. Investig. 10:67-76. [DOI] [PubMed] [Google Scholar]
- 29.Read, D. H., R. L. Walker, A. E. Castro, J. P. Sundberg, and M. C. Thurmond. 1992. An invasive spirochaete associated with interdigital papillomatosis of dairy cattle. Vet. Rec. 130:59-60. [DOI] [PubMed] [Google Scholar]
- 30.Rebhun, W. C., R. M. Payne, J. M. King, M. Wolf, and S. N. Begg. 1980. Interdigital papillomatosis in dairy cattle. J. Am. Vet. Med. Assoc. 5:437-440. [PubMed] [Google Scholar]
- 31.Rodriguez-Lainz, A., D. W. Hird, T. E. Carpenter, D. H. Read, and A. R. Lainz. 1996. Case-control study of papillomatous digital dermatitis in southern Californian dairy farms. Prev. Vet. Med. 28:117-131. [Google Scholar]
- 32.Rodriguez-Lainz, A., P. Melendez-Retamal, D. W. Hird, D. H. Read, and R. L. Walker. 1999. Farm- and host-level risk factors for papillomatous digital dermatitis in Chilean dairy cattle. Prev. Vet. Med. 42:87-97. [DOI] [PubMed] [Google Scholar]
- 33.Schrank, K., B. K. Choi, S. Grund, A. Moter, K. Heuner, H. Nattermann, and U. B. Göbel. 1999. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int. J. Syst. Bacteriol. 49:43-50. [DOI] [PubMed] [Google Scholar]
- 34.Smibert, R. M. 1991. Anaerobic spirochetes, p. 572-578. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 35.Stamm, L. V., H. L. Bergen, and R. L. Walker. 2002. Molecular typing of papillomatous digital dermatitis-associated Treponema isolates based on analysis of 16S-23S ribosomal DNA intergenic spacer regions. J. Clin. Microbiol. 40:3463-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanton, T. B. 2002. The genus Brachyspira. In M. Dworkin (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 37.Stanton, T. B., N. S. Jensen, T. S. Casey, L. A. Tordoff, F. E. Dewhirst, and B. J. Paster. 1991. Reclassification of Treponema hyodysenteriae and Treponema innocens in a new genus, Serpula gen. nov., as Serpula hyodysenteriae comb. nov. and Serpula innocens comb. nov. Int. J. Syst. Bacteriol. 41:50-58. [DOI] [PubMed] [Google Scholar]
- 38.Stanton, T. B., D. J. Trott, J. I. Lee, A. J. McLaren, D. J. Hampson, B. J. Paster, and N. S. Jensen. 1996. Differentiation of intestinal spirochaetes by multilocus enzyme electrophoresis and 16S rRNA sequence comparisons. FEMS Microbiol. Lett. 136:181-186. [DOI] [PubMed] [Google Scholar]
- 39.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trott, D. J., T. B. Stanton, N. S. Jensen, G. E. Duhamel, J. L. Johnson, and D. J. Hampson. 1996. Serpulina pilosicoli sp. nov.: the agent of porcine intestinal spirochetosis. Int. J. Syst. Bacteriol. 46:206-215. [DOI] [PubMed] [Google Scholar]
- 41.Trott, D. J., A. S. J. Mikosza, B. G. Combs, S. L. Oxberry, and D. J. Hampson. 1998. Population structure and molecular epidemiology of Serpulina pilosicoli in villages in the Eastern Highlands of Papua New Guinea. Int. J. Syst. Bacteriol. 48:659-668. [DOI] [PubMed] [Google Scholar]
- 42.Vinh, T., S. Faine, C. J. Handley, and B. Adler. 1994. Immunochemical studies of opsonic epitopes of the lipopolysaqccharide of Leptospira interrogans serovar hardjo. FEMS Immunol. Med. Microbiol. 8:99-107. [DOI] [PubMed] [Google Scholar]
- 43.Walker, R. L., D. H. Read, K. J. Loretz, D. W. Hird, and S. L. Berry. 1997. Humoral response of dairy cattle to spirochetes isolated from papillomatous digital dermatitis lesions. Am. J. Vet. Res. 58:744-748. [PubMed] [Google Scholar]
- 44.Walker, R. L., D. H. Read, K. J. Loretz, and R. W. Nordhausen. 1995. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol. 47:343-355. [DOI] [PubMed] [Google Scholar]
- 45.Wallace, A. L., and A. Harris. 1967. Reiter treponeme: a review of the literature. Bull. W. H. O. 36(Suppl. 2):5-103. [PMC free article] [PubMed] [Google Scholar]
- 46.Wells, S. J., L. P. Garber, and B. A. Wagner. 1999. Papillomatous digital dermatitis and associated risk factors in US dairy herds. Prev. Vet. Med. 38:11-24. [DOI] [PubMed] [Google Scholar]
- 47.Zuerner, R. L., and T. B. Stanton. 1994. Physical and genetic map of the Serpulina hyodysenteriae B78 chromosome. J. Bacteriol. 176:1087-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]