Abstract
The diagnosis of bloodstream infection with coagulase-negative staphylococci is frequently based on the isolation of the same organism from more than one blood culture. Phenotypic variation is a common characteristic of pathogenic strains of Staphylococcus epidermidis which may affect species identification by the microbiology laboratory. We describe a patient with a new onset of nephritis and gram-positive bacteremia. Gram-positive cocci grew in multiple blood cultures and were identified by the Vitek 2 system as Kocuria varians, Staphylococcus hyicus, and S. epidermidis. Bacterial isolates grew on blood agar and Congo red agar plates as two distinct morphotypes and exhibited phenotypic variation. Neither morphotype could be identified by the API-Staph assay. Cellular fatty acid analysis identified one of the morphotypes as S. epidermidis but could not identify the other morphotype. All isolates were found to be identical by pulsed-field gel electrophoresis, and both colonial morphotypes were identified as S. epidermidis by 16S ribosomal gene sequencing. Phenotypic variation of S. epidermidis may affect identification to the species level by phenotype-based identification systems. Caution should be exercised when differentiating between true infection and contamination based on strain identification.
Coagulase-negative staphylococci (CoNS) are a frequent cause of nosocomial infection and bacteremia, especially in patients with indwelling medical devices. CoNS are, however, also a part of the normal microflora of skin and mucous membranes, and they often contaminate clinical specimens. Differentiation of true infection from contamination may, therefore, pose a challenge (9). When two or more blood cultures drawn from the same patient grow CoNS, identification of isolates as belonging to different species is considered evidence of contamination rather than true bloodstream infection (7, 8).
Phenotypic variation is a recognized trait of pathogenic strains of Staphylococcus epidermidis (12). The degree with which phenotypic variation of CoNS may affect species identification is unclear. We present a case of ventriculoatrial (VA) shunt infection caused by a phenotypically variable strain of S. epidermidis which presented as polymicrobial gram-positive bacteremia with widely differing bacterial isolates, suggesting contamination.
CASE REPORT
A 47-year-old woman was admitted for progressive peripheral edema. She had been diagnosed with hydrocephalus resulting from aqueductal stenosis 10 years earlier for which a ventriculoperitoneal shunt was inserted. The shunt was subsequently replaced several times due to recurring dysfunction. Three years prior to the current admission a VA shunt was inserted and appeared to be functioning well.
On examination, the patient appeared dyspneic but was alert and well oriented. Her body temperature was 36.2°C, her pulse was 68 beats/min, and her blood pressure was 140/80 mm Hg. There was no evidence of meningeal irritation. The neurological examination was unremarkable. Bipedal pitting edema (severity, +2) was present. Laboratory studies were notable for normocytic anemia (hemoglobin, 8.1 g/dl; hematocrit, 24%; mean corpuscular volume, 85 fl), hypoalbuminemia (serum albumin, 3.2 g/dl), and hypercholesterolemia (serum cholesterol, 269 mg/dl). Urinalysis showed dysmorphic red blood cells and granular casts. Urine protein excretion was 2,865 mg/24 h. Calculated creatinine clearance was 52 ml/min. There was marked hypocomplementemia (C-3, 0.63 g/liter [normal, 0.9 to 1.8 g/liter]; C-4, 0.06 g/liter [normal, 0.1 to 0.4 g/liter]). Blood cultures from the date of admission grew gram-positive cocci. Transesophageal echocardiography showed the VA catheter tip within the right atrium, with no evidence of intracardiac thrombus, dilatation, or dysfunction.
Blood cultures were repeated, and treatment with intravenous vancomycin was initiated. Subsequently, additional blood cultures grew gram-positive cocci. Isolates were identified by the Vitek 2 system (bioMerieux, St. Louis, Mo.) on different dates as Kocuria varians, Staphylococcus hyicus, S. epidermidis, and CoNS (Table 1). Cerebrospinal fluid drawn from the shunt contained no leukocytes; the culture was negative. Creatinine clearance deteriorated to 25 ml/min.
TABLE 1.
Identification by Vitek 2 of clinical isolates cultured from patients' blood
| Day of hospitalization | Identification (Vitek 2) | No. of positive bottles/total no. of bottles | Antimicrobial susceptibility pattern |
|---|---|---|---|
| 1 | K. varians | 2/2 | |
| 5 | K. varians | 3/3 | Aa |
| 7 | K. varians | 1/1 | |
| 8 | S. hyicus | 4/5 | A |
| S. epidermidis | 1/5 | ||
| 9 | K. varians | 1/2 | |
| CoNS | 1/2 | A | |
| 10 | K. varians | 2/2 | |
| 11 | CoNS | 1/2 | |
| 12 | CoNS | 1/2 | |
| 14 | S. hyicus | 1/2 | A |
| 15 | CoNS | 1/2 | |
| 19 | No growth | 0/4 | |
| 42 | No growth | 0/2 | |
| 43 | No growth | 0/2 |
Antimicrobial susceptibility pattern A indicates susceptibility to oxacillin, vancomycin, rifampin, clindamycin, trimethoprim-sulfamethoxazole, and chloramphenicol.
A presumptive diagnosis of VA shunt infection and shunt nephritis was made. On day 18 of hospitalization, the patient underwent exteriorization of the shunt, which was removed and replaced by a new ventriculoperitoneal shunt 3 weeks later. Culture of the VA shunt was negative. Treatment with vancomycin was continued for 2 weeks after this shunt replacement, and repeated blood cultures were sterile. During the following month, creatinine clearance increased to 55 ml/min, urinary protein excretion declined to 142 mg/24 h, and the peripheral edema resolved.
MATERIALS AND METHODS
Colonial morphology of clinical isolates.
Thirteen isolates from the patient's blood were analyzed. Colonial morphology was studied on Trypticase soy agar plates containing 5% sheep blood (Hy-Labs, Rehovot, Israel) and on homemade Congo red agar (CRA) plates. CRA was prepared as described by Deighton et al. (6), except that Congo red (0.01%) was incorporated into Mueller-Hinton agar plates (Difco Laboratories, Detroit, Mich.). To maximize differences in colonial morphologies, the plates were incubated for 48 h at 35°C with CO2 (5%) followed by a further 2 days of incubation at room temperature. Distinct colonial morphotypes identified by these methods were selected for further biochemical and molecular analysis.
Species identification by biochemical analysis.
Isolates were identified by the Vitek 2 automated system and the API-Staph commercial kit (API systems SA, La Balme Les Grottes, France). Susceptibility of isolates to antibiotics was determined by the Vitek 2 system.
Cellular fatty acid analysis.
Strains were grown aerobically on Mueller-Hinton agar (Hy-Labs) for 24 to 48 h at 35°C. Well-grown cells were analyzed for their fatty acid methyl ester (FAME) composition using the services of Accugenix (Newark, Del). The FAME profiles obtained from each clinical isolate were compared to the Accugenix database. Similarity indexes obtained from the FAME analysis of each clinical isolate indicated the relative match of the isolate with a specified species from the database. A similarity index of >0.5 indicates an excellent match, an index in the range of 0.3 to 0.5 suggests that the isolate is a strain of the species, and an index of <0.3 suggests a species that does not appear in the database.
Determination of genetic relatedness of isolates.
Pulsed-field gel electrophoresis (PFGE) analysis was performed on all available clinical isolates. DNA preparation and cleavage were performed as described previously (2). Chromosomal restriction fragments were separated electrophoretically in a 1.0% agarose gel (SeaKem LE agarose; FMC BioProducts, Rockland, Maine) with the CHEF-DR III apparatus (Bio-Rad, Hercules, Calif.). Electrophoresis conditions were 6 V/cm for 22 h at 14°C in 0.5× TBE buffer (50 mM Tris-HCl, 50 mM boric acid, 1 mM EDTA), with pulse times ranging from 5 to 40 s. Lambda Ladder (New England BioLabs, Beverly, Mass.) was used as a molecular weight marker. The DNA was visualized by staining with ethidium bromide (0.5 μg/ml) in 0.5× TBE buffer, and the gel was photographed using a Bio-Rad Gel Doc 2000 system. Patterns were analyzed visually and by using diversity fingerprinting software (Bio-Rad).
16S ribosomal DNA sequencing.
DNA was prepared by using the Wizard genomic DNA purification kit according to the manufacturer's instructions (Promega, Madison, Wis.). After extraction, the DNA solution was stored at 4°C. The nearly complete sequence of the 16S rRNA gene was amplified by PCR with the conserved primers 8F (5′-AGAGTTTGATYMTGGCTCAG-3′) and 1942R (5′-ACCTTGTTACGACTT-3′). DNA amplification was carried out using Vent DNA polymerase (New England BioLabs) and genomic DNA template. The PCR conditions were as follows: 5 min of initial denaturation at 95°C; 30 cycles of 60 s at 95, 56, and 72°C; and a final elongation step of 10 min at 72°C. The PCR product was purified by using the Promega Wizard PCR Preps kit (Promega) according to the manufacturer's instructions. Automated DNA sequencing was performed by the dideoxynucleotide chain termination method using an Applied Biosystems (Foster City, Calif.) model 373A sequencer. The obtained sequences were compared to sequences available in the GenBank database by using BLASTN software (http://www.ncbi.nlm.nih.gov/BLAST/) (1) and then compared with each other.
RESULTS
Colonial morphology and phase variation.
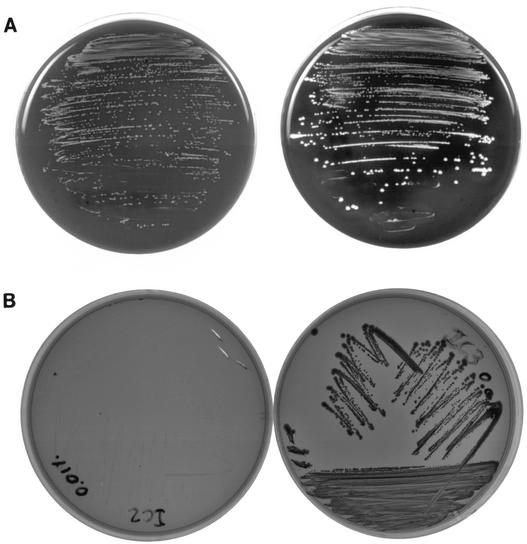
The bacterial isolates grew on Trypticase soy agar plates as two morphologically distinct colonies, i.e., large white beta-hemolytic colonies and pinpoint white nonhemolytic colonies (Fig. 1A). A subculture of the large colonies yielded a mixture of both types, indicating a high rate of phenotypic variation. A subculture of the small colonies yielded mostly small colonies with occasional reversion to the large colony phenotype, indicating a low rate of phenotypic variation.
FIG. 1.
(A) Appearance of two colonial morphotypes on blood agar. Predominantly large, beta-hemolytic colonies (at right) and predominantly small nonhemolytic colonies (at left) are shown. (B) Appearance of colonial morphotypes on Congo red agar. Large dark red colonies (at right) and small transparent colonies (at left) are shown.
We used growth on CRA, as previously described (6), in order to better differentiate the two colonial morphotypes. The large colonies stained dark red with Congo red, whereas the small colonies were transparent on CRA (Fig. 1B).
Species identification by biochemical analysis.
Both morphotypes were reported as K. varians by the Vitek 2 system and scored as a being a “very good identification” (45 tests for the diagnosis of K. varians and 2 tests against it).
Identification of the large colony strain with the API-Staph system yielded an “unacceptable profile,” with the most closely matching taxa being Staphylococcus xylosus and Aerococcus viridans (no identification likelihood given). Analysis of the small colony strain yielded a “doubtful profile,” with the most closely matching taxa being Staphylococcus hominis (46.4%), Staphylococcus capitis (18.9%), and Staphylococcus warneri (16.9%) (Table 2). The profiles of the two morphotypes differed from one another by 1 of 26 tests (acid production from mannitol).
TABLE 2.
Comparative phenotypic and genotypic analysis of the two colonial morphotypes
| Analysis method | Result for morphotype
|
|
|---|---|---|
| Large colony | Small colony | |
| Hemolysis on blood agar | Beta-hemolytic | Nonhemolytic |
| Congo red staining | Dark red | Transparent |
| Vitek 2 | K. varians (“very good ID”) | Kocuria varians (“very good ID”) |
| API-Staph | Profile: 067330320 (“unacceptable profile”) | Profile: 067130300 (“doubtful profile”) |
| Fatty acid profile | No good match | S. epidermidis (similarity index = 0.467) |
| 16S ribosomal gene sequencing | S. epidermidis (99% identity) | S. epidermidis (99% identity) |
Identification by cellular fatty-acid analysis.
The fatty acid profile of the large colony strain did not match any of the microbial profiles in the database. Staphylococcus aureus was the most closely related taxon (a similarity index of 0.205). The small colony strain was identified by fatty acid analysis as S. epidermidis, with a similarity index of 0.467 (Table 2).
Genetic relatedness of isolates.
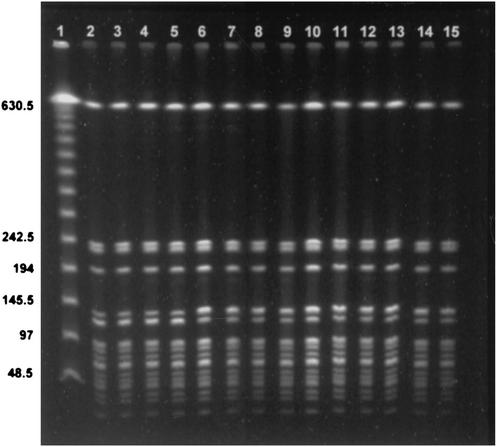
PFGE was performed on 13 isolates from hospital days 1, 5, 8, 9, 10, 11, 12, 14, and 15. There was complete identity of all isolates (Fig. 2).
FIG. 2.
DNA fingerprinting analysis of the clinical isolates by PFGE. Lane 1, lambda concatemers as a molecular size marker; lanes 2 to 14, each lane contains a different clinical isolate.
Identification by 16S ribosomal gene sequencing.
16S ribosomal gene sequencing yielded a 1,449-bp sequence for the large colony strain and a 1,448-bp sequence for the small colony strain. Both sequences showed 99% identity with the S. epidermidis strain KL-096 16S ribosomal gene sequence (GenBank accession number AY030342 [10]), and were 99% homologous with each other (Table 2).
DISCUSSION
Phenotypic variation is a well-recognized feature of S. epidermidis, with the variability being evident in colonial morphology, production of exopolysaccharide, adhesion to plastic surfaces, and antimicrobial resistance patterns (5). Furthermore, phenotypic variation may contribute to bacterial virulence: pathogenic strains of S. epidermidis isolated from blood exhibit phenotypic variation, whereas saprophytic strains from skin do not (12). It has been suggested that phenotypic variation enhances bacterial survival and growth under changing environmental conditions (12).
The case presented herein illustrates a pitfall in diagnosing S. epidermidis infection of indwelling foreign devices: a highly phenotypically variable strain of S. epidermidis was identified by the Vitek 2 system as three widely different bacterial species, and both API-Staph and FAME analysis could not correctly identify the isolates. Two of the isolates from the patients' blood were identified as K. varians and S. hyicus, which have only anecdotally been associated with infection in humans, further supporting a diagnosis of pseudo-bacteremia (3). The diagnosis of true shunt infection was based on the typical clinical presentation of a VA shunt infection, on the PFGE demonstrating that all isolates were identical, and on the favorable response to removal of the infected shunt and institution of appropriate antibiotic therapy.
Deighton et al. (6) described a phenotypically variable S. epidermidis isolated from a resected valve from a patient with endocarditis. The two colonial morphotypes were identified as S. hominis and Staphylococcus simulans by the Vitek system, and both were identified as S. hominis by the API-Staph system. As had been reported by those authors, we also noted the appearance of two morphotypes: one was a small nonhemolytic colony which grew transparent on CRA and exhibited a low rate of phenotypic variation, and the other was a large hemolytic colony which grew dark red on CRA and was highly phenotypically variable. However, in the case we now present, the isolates were identified by Vitek 2 as belonging to different genera (i.e., Kocuria and Staphylococcus). Furthermore, FAME analysis, which is highly dependable in characterizing coagulase-negative staphylococci (4, 11), failed to identify the large colony strain. These features probably indicate particularly extensive phenotypic variation.
Phenotypic variation of clinical isolates of S. epidermidis depends upon several factors. It increases in frequency with prolonged incubation of cultures: it is observed on CRA in 14% of isolates when cultures are incubated overnight but this figure reaches 68% of isolates with longer incubation (6). Variation is also dependent upon the medium used. Moreover, under stress conditions, such as high salt concentration, phenotypic variation becomes more frequent (6). Finally, clinical isolates cultured from blood are more likely to be phenotypically variable than saprophytic strains cultured from skin or mucosal surfaces (12).
Since phenotypic variation is common in S. epidermidis isolates, misidentification by commercial systems may not be uncommon. Failure to correctly identify S. epidermidis might lead to the erroneous diagnosis of polymicrobial bacteremia, which is commonly interpreted as signifying contamination rather than true bloodstream infection. Most troubling, such errors are likely to be particularly prevalent with highly virulent strains of S. epidermidis which are known to be phenotypically variable. Recognition of this potential problem should prompt the use of genomic identification tools, such as PFGE and 16S ribosomal DNA sequencing, when polymicrobial bacteremia with CoNS or related bacteria is encountered and infection is clinically suspected.
Acknowledgments
Esther Eshkol is thanked for editorial assistance.
REFERENCES
- 1.Altschul, S. F., A. A. Madden, J. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basaglia, G., E. Carretto, D. Barbarini, L. Moras, S. Scalone, P. Marone, and P. De Paoli. 2002. Catheter-related bacteremia due to Kocuria kristinae in a patient with ovarian cancer. J. Clin. Microbiol. 40:311-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnbaum, D., L. Herwaldt, D. E. Low, M. Noble, M. Pfaller, R. Sherertz, and A. W. Chow. 1994. Efficacy of microbial identification system for epidemiologic typing of coagulase-negative staphylococci. J. Clin. Microbiol. 32:2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, G. D., L. M. Baddour, B. M. Madison, J. T. Parisi, S. N. Abraham, D. L. Hasty, J. H. Lowrence, J. A. Josephs, and W. A. Simpson. 1990. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to β-lactam antibiotics, and virulence. J. Infect. Dis. 161:1153-1169. [DOI] [PubMed] [Google Scholar]
- 6.Deighton, M., S. Pearson, J. Capstick, D. Spelman, and R. Borland. 1992. Phenotypic variation of Staphylococcus epidermidis isolated from a patient with native valve endocarditis. J. Clin. Microbiol. 30:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khatib, R., K. M. Riederer, J. A. Clark, S. Khatib, L. E. Briski, and F. M. Wilson. 1995. Coagulase-negative staphylococci in multiple blood cultures: strain relatedness and determinants of same-strain bacteremia. J. Clin. Microbiol. 33:816-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, S. D., L. C. McDonald, W. R. Jarvis, S. K. McAllister, R. Jerris, L. A. Carson, and J. M. Miller. 2000. Determining the significance of coagulase-negative staphylococci isolated from blood cultures at a community hospital: a role for species and strain identification. Infect. Control Hosp. Epidemiol. 21:213-217. [DOI] [PubMed] [Google Scholar]
- 9.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 10.Venkateswaran, K., M. Satomi, S. Chung, R. Kern, R. Koukol, C. Basic, and D. White. 2001. Molecular microbial diversity of a spacecraft assembly facility. Syst. Appl. Microbiol. 24:311-320. [DOI] [PubMed] [Google Scholar]
- 11.Welch, D. F. 1991. Applications of cellular fatty acid analysis. Clin. Microbiol. Rev. 4:422-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]