Abstract
Forty-eight Mycobacterium tuberculosis strains were obtained from patients living in metropolitan Manila, Republic of the Philippines. Three molecular typing methods, IS6110 restriction fragment length polymorphism, spoligotyping, and DNA sequencing of the oxyR, gyrA, and katG loci, established that these strains have restricted diversity and are members of a related genetic group of organisms. Comparison of the DNA fingerprint patterns with those in international databases confirmed the uniqueness of this group of isolates, which we designate the Manila family of M. tuberculosis.
The advent of IS6110-based restriction fragment length polymorphism (RFLP) typing of Mycobacterium tuberculosis has led to fingerprinting studies throughout the world (13, 16). These studies have provided insight on the transmission of tuberculosis within and among different geographical areas of the world (10, 16). One finding of IS6110 DNA fingerprinting is that the population structure of M. tuberculosis differs in regions in that some geographical areas are associated with families or groups of related isolates, such as the Beijing, Haarlem, and African genotype families (2, 5, 7, 8, 15; L. Qian, H. Traore, D. van Soolingen, J. D. A. van Embden, Z. Huang, F. Portaels, and J. T. Douglas, Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995, abstr. U-4, p. 117, 1995.). Additional DNA typing technologies, such as spoligotyping (6), analysis of the pseudogene oxyR (12), polymorphic GC-rich sequence RFLP (9), and typing of variable numbers of tandem repeats (1) confirmed these family groupings of M. tuberculosis.
Since we noticed that tuberculosis isolates from Filipinos, which account for 50% of the M. tuberculosis isolates from foreign-borne patients in Hawaii (State of Hawaii Department of Health data, www.hawaiistate/health/tuberculosis, 2001), had related spoligotyping patterns, we decided to directly investigate isolates from Manila, Republic of the Phillipines (J. T. Douglas, L. Qian, J. C. Montoya, S. Sreevatsan, J. M. Musser, and J. D. A. van Embden, Abstr. 97th Gen. Meet. Am. Soc. Microbiol., abstr. U-166, p. 572, 1997). Manila is the largest city in the Philippines (about 10 million people); it has a high population density and socioeconomic problems, and tuberculosis is highly endemic. Manila is also geographically isolated within an island nation, which could be a fertile ground for clonal expansion of M. tuberculosis. Because little is known about the population structure of tuberculosis in the Philippines, we subjected a random sample of M. tuberculosis isolates originating in Manila to various molecular typing techniques to search for genetic markers that might be useful in studying M. tuberculosis isolates recovered from immigrant populations within Hawaii.
The Tuberculosis Study Group at the Research Institute for Tropical Medicine and the Philippine General Hospital provided 48 M. tuberculosis isolates from separate individuals in the metropolitan Manila area. Initially 18 isolates were collected in December 1995, followed by 30 isolates 8 months later. Of these, 35 came with some demographic data. These isolates were from 18 males and 17 females. The mean age for 22 patients was 40.3 years. The patients were broken down into two age groups; the first group consisted of 15 patients (15 to 50 years old) with a mean age of 30.7 years, and the second group consisted of 7 patients (51 to 66 years old) with a mean age of 59.4 years. These M. tuberculosis isolates, obtained from 16 healthcare centers around Manila, were heat killed at 80°C in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) for 1 h and transported to the University of Hawaii in Honolulu, where DNA was extracted and stored for further use.
RFLP analysis was conducted using IS6110 as a probe according to a standardized method previously described (13, 16). Briefly, this DNA fingerprinting technique entails the growth of M. tuberculosis, extraction of DNA, PvuII restriction endonuclease digestion, Southern blotting, and probing for the IS6110 element. Each Southern blot contained DNA from M. tuberculosis reference strain Mt14323 as an external standard.
Spoligotyping (spacer oligotyping) is a DNA fingerprinting method that exploits the DNA polymorphism at a unique chromosomal locus, the direct repeat (DR) region, in M. tuberculosis complex bacteria. Spoligotyping relies on determining the presence or absence of spacers interspersing the DRs by amplifying the DR locus by PCR with one biotin-labeled primer, followed by hybridization of the PCR product with 43 oligonucleotides that are immobilized on a membrane. These oligonucleotides are designed from known spacer sequences of M. tuberculosis strain H37Rv and Mycobacterium bovis BCG strain P3. Detection of hybridization signals is done by using the enhanced chemiluminescence direct nucleic acid labeling and detection system (Amersham Life Science). Details of the method were described by Kamerbeek et al. (6). Sequence data for the oxyR gene were generated by automated DNA sequencing with an Applied Biosystems model 377 sequencer, and polymorphisms in gyrA codon 95 and katG codon 463 were identified by PCR-RFLP (11, 12). Sequence data were assembled and edited electronically with the EDITSEQ, ALIGN, and MEGALIGN software programs (DNASTAR, Madison, Wis.).
Comparison of the IS6110 RFLP and spoligotyping patterns was performed with the GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium) at the National Institute of Public Health and the Environment (RIVM; Bilthoven, The Netherlands) (4). The international database of IS6110 RFLP patterns consisted of 5,906 patterns of M. tuberculosis complex strains originating from 30 different countries, covering Europe, Asia, Africa, and North and South America (excluding the United States and Canada). The international database of spoligotyping patterns consisted of 3,575 patterns originating from 55 countries around the world. This database included 19 spoligotyping patterns from M. tuberculosis strains isolated from Philippine immigrants living in Hawaii. These databases are maintained at RIVM. Comparisons were made by using the Dice coefficient for calculation of the similarities, allowing 1.0% position tolerance and 1.0% optimization.
A further combined comparison of RFLP and spoligotyping patterns for each isolate was accomplished by using the BioNumerics software (version 2.0; Applied Maths). For this purpose patterns were exported from GelCompar into BioNumerics. The combined dendrogram (see Fig. 1) was prepared by using the Dice coefficient for similarity calculations and the unweighted pair group method using arithmetic averages for clustering.
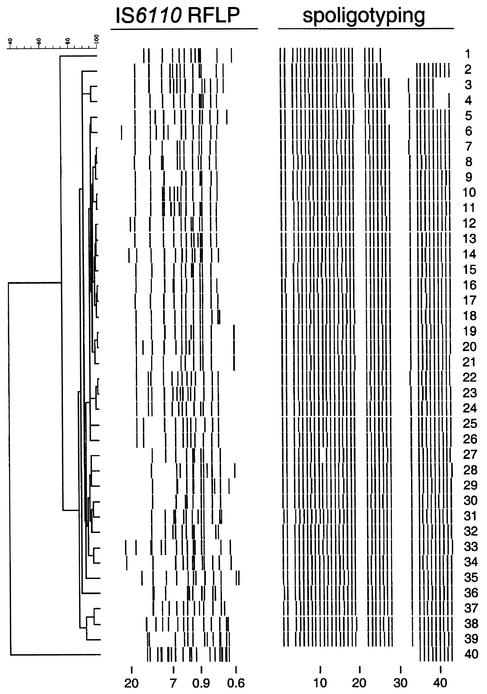
FIG. 1.
IS6110 RFLP and spoligotype patterns of 40 M. tuberculosis isolates originating from the Philippines. Shown is a combined dendrogram based on the similarities of the patterns on the basis of both typing methods. Strains 5 to 39 represent the Manila family, described in this study. Note that the spoligotype patterns of these strains are all identical, except those of strains 5 and 27, which lack one spacer. Strain 40 exhibits fingerprints characteristic of the Beijing genotype of M. tuberculosis. Numbers at the bottom indicate the sizes of the PvuII restriction fragments in kilobase pairs (left) and the spacer oligonucleotides (right).
IS6110 RFLP data.
The IS6110 RFLP patterns of the M. tuberculosis strains from the Philippines exhibited a high degree of similarity, yet none of them were identical (Fig. 1). Thirty-eight (95%) of the 40 isolates for which RFLP patterns were available showed similarities of 80% or greater among the patterns. We designated these 38 isolates the Manila family. Of the two remaining RFLP patterns one (Fig. 1, lane 1) exhibited more than 75% similarity to those of the Manila family and the other (lane 40) was very distinct from the other patterns, exhibiting only 40% similarity. Comparison with the international database proved that the latter strain had the Beijing genotype of M. tuberculosis (2, 8, 15; Qian et al., Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995). The uniqueness of the Manila family was established by comparing the 38 IS6110 RFLP patterns with an international database. Only 12 out of the 5,906 IS6110 DNA fingerprints present in the database showed more than 90% similarity to the fingerprints from Manila (Table 1). These strains were all isolated in The Netherlands. Of these 12 isolates, 6 could be associated with the ethnic origins of the patients. Four immigrants were from the Philippines, one was from Indonesia, and one was from Somalia. Extending the search to include patterns that had more than 80% of the IS6110-containing DNA restriction fragments in common with the Manila isolates identified 76 isolates originating from The Netherlands (58 isolates, including the 12 previously mentioned), Denmark (12 isolates), Thailand (5 isolates), and Iran (1 isolate) (Table 1). Thus, it was clear that the high frequency of occurrence, restricted geographical range, and similarities of IS6110 RFLP patterns of these isolates from Manila helped establish this unique family of M. tuberculosis. In addition, the high association with Filipinos, where ethnic data were available, helped establish these isolates as a unique family of M. tuberculosis.
TABLE 1.
Frequencies of the Manila family IS6110 RFLP and spoligotype patterns found in the RIVM worldwide databases
| Category | No. of patterns matched (%) | No. of countries with matching pattern | No. of countries in database | Total no. of patterns |
|---|---|---|---|---|
| IS6110 with 90% similaritya | 12 (0.2) | 1 | 30 | 5,906 |
| IS6110 with 80% similaritya | 76 (1.3) | 4 | 30 | 5,906 |
| Spoligotyping | 31 (0.9) | 3 | 55 | 3,575 |
Comparisons were made by using the Dice coefficient for calculation of the similarities, allowing 1.0% position tolerance and 1.0% optimization.
Spoligotyping data.
The spoligotyping of 41 of the 48 M. tuberculosis isolates from metropolitan Manila yielded characteristic identical patterns lacking hybridization to eight spacers (3, 20, 21, 29 to 32, and 34); the remaining 35 spacers showed a positive hybridization signal. Forty of these isolates are shown in Fig. 1, along with their matching IS6110 RFLP patterns. Five isolates exhibited spoligotyping patterns that were highly similar to the pattern observed for the 41 isolates. Two strains (Fig. 1, lanes 5 and 27) lacked hybridization to a single spacer oligonucleotide and by our convention would also be classified as Manila family members. Thus spoligotyping classified 43 or 90% of the isolates as Manila family. Additional deletions occurred in three strains in lanes 2, 3, and 4 of Fig. 1. These lacked three or four additional spacers to the right of spacer 24. One isolate, in lane 1 of Fig. 1, lacked 13 additional spacers to the right of spacer 24. The IS6110 RFLP pattern of this isolate had a similarity of 75% to those of the Manila family, as established by Bionumerics software analysis. The remaining isolate exhibited a different spoligotype pattern, consisting of the last nine spacers (Fig. 1, lane 40). Eight isolates did not have RFLP data available but did have spoligotyping data. Two of these isolates lacked eight spacers (pattern not shown), and the remaining six isolates were typical Manila family members.
All of the spoligotype patterns illustrated in Fig. 1 and two additional patterns not shown represented nine different spoligotypes and were compared with 3,575 patterns of M. tuberculosis complex strains in the international database (Table 1). The spoligotypes represented a very restricted cluster of 41 of 48 isolates that matched with 17 of 19 patterns from isolates from Philippine patients living in Hawaii, 7 from The Netherlands, and 4 from Mexico. In addition, three matches with spoligotype patterns lacking one spacer in comparison with the major Manila spoligotype were found in the database. The spoligotype patterns of one isolate from Hawaii and one from The Netherlands matched the pattern of lane 27 in Fig. 1. The spoligotype pattern of another isolate from Hawaii matched the pattern of lane 5 in Fig. 1. These findings are included in Table 1, for a total of 31 isolates matching the Manila family out of the 3,575 isolates in the RIVM international database.
The nine-spacer spoligotype pattern (Fig. 1, lane 40) matched the pattern representing a large family of strains, mainly originating from China and other northern Asian countries, and was recognized to represent the Beijing genotype (2, 7, 8, 15). The remaining four spoligotypes were unique.
oxyR, gryA, and katG polymorphism data.
An additional test was used to evaluate this related group of strains: oxyR gene polymorphism. Polymorphism at position 37 of this pseudogene results in the C→T substitution. This polymorphism had been recognized in isolates from Filipino immigrants in Texas previously. All of the isolates from Manila contained this polymorphism, except the isolate that belonged to the Beijing genotype family. In addition, all isolates had the combination of gryA codon 95 ACC (Thr) and katG codon 463 CTG (Leu). This combination has been designated evolutionary group 1 of M. tuberculosis (11, 12).
Summary.
IS6110 RFLP, spoligotyping and gene polymorphisms revealed a closely related group of M. tuberculosis isolates found in high frequency in Manila. These isolates showed a consistent polymorphism in the pseudogene oxyR, nearly identical and characteristic spoligotype patterns, and highly similar IS6110 RFLP patterns. The uniqueness of this closely related group of strains was demonstrated after comparison with large international databases of IS6110 RFLP and spoligotype patterns, and we have therefore designated this group of strains the Manila family. This finding contributes to the collection of information on the worldwide population structure of M. tuberculosis, particularly in the Pacific regions.
The isolates used in this study had been collected at random at different time periods and locations around Manila and were from patients of diverse ages, ranging between 15 and 68 years, and the sexes were equally represented. Ninety percent (43 of 48) of the isolates in this study were of the homogeneous Manila family, and a single isolate of the Beijing genotype family was observed. To investigate the spread of the Manila family and possibly other genotypes throughout the Philippines requires more sampling from other areas.
The M. tuberculosis Beijing family, which is frequently observed in Asia, is associated with resistance to antituberculosis drugs in some areas (2). It will be interesting to investigate the association between the Manila family and drug resistance, especially in the light of the study of Grimaldo et al., who performed a hospital-based study in the city of Makati in the Philippines. They found 17.2% of the M. tuberculosis isolates to be multidrug resistant, resistance to ciprofloxacin in 26.8%, and resistance to ofloxacin in 35.3% (3). Furthermore, virulence factors and other characteristics of the Manila family will be worthwhile investigating.
Acknowledgments
This work was supported by grants from the American Lung Association of Hawaii, the Leahi Trust, the Hawaiian Community Foundation, and the European Union (grant QLK2-CT-2000-00630).
We thank Petra de Haas at RIVM for her support and the Tuberculosis Study Group at the Philippine General Hospital for their extraordinary effort in providing M. tuberculosis isolates. Also we acknowledge, the cooperation of the State of Hawaii Department of Health and Xiaotan Zheng of Diagnostic Laboratory Services for providing cultures in Hawaii.
REFERENCES
- 1.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 2.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimaldo, E. R., T. E. Tupasi, A. B. Rivera, M. I. Quelapio, R. C. Cardano, J. O. Derilo, and V. A. Belen. 2001. Increased resistance to ciprofloxacin and ofloxacin in multidrug-resistant Mycobacterium tuberculosis isolates from patients seen at a tertiary hospital in the Philippines. Int. J. Tuberc. Lung Dis. 5:546-550. [PubMed] [Google Scholar]
- 4.Heersma, H. F., K. Kremer, and J. D. van Embden. 1998. Computer analysis of IS6110 RFLP patterns of Mycobacterium tuberculosis. Methods Mol. Biol. 101:395-422. [DOI] [PubMed] [Google Scholar]
- 5.Hermans, P. W., F. Messadi, H. Guebrexabher, D. van Soolingen, P. E. de Haas, H. Heersma, H. de Neeling, A. Ayoub, F. Portaels, and D. Frommel. 1995. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 171:1504-1513. [DOI] [PubMed] [Google Scholar]
- 6.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. Van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, B. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian, L., J. D. A. van Embden, A. G. M. van der Zanden, E. F. Weltevereden, H. Duanmu, and J. T. Douglas. 1999. Retrospective analysis of the Beijing family of Mycobacterium tuberculosis in preserved lung tissues. J. Clin. Microbiol. 37:471-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross, B. C., K. Raios, K. Jackson, and B. Dwyer. 1992. Molecular cloning of a highly repeated DNA element from Mycobacterium tuberculosis and its use as an epidemiological tool. J. Clin. Microbiol. 30:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 11.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sreevatsan, S., X. Pan, Y. Zhang, V. Deretic, and J. M. Musser. 1997. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob. Agents Chemother. 41:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 15.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Soolingen, D., P. E. W. de Haas, and K. Kremer. 2001. Restriction fragment length polymorphism typing of Mycobacteria, p. 165-203. In T. Parisch and N. G. Stoker (ed.), Mycobacterium tuberculosis protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]