Abstract
Human herpesvirus 8 (HHV-8) can be classified into distinct subtypes on the basis of sequence polymorphisms in several open reading frames (ORFs). We analyzed the subtypes of HHV-8 in 59 human immunodeficiency virus-infected Japanese patients by using polymorphisms in ORF26 and found that over two-thirds of the HHV-8 isolates fell into major subtype A. We also found that single nucleotide polymorphisms (SNPs) at nucleotide positions 1032 (C-to-A substitution) and 1055 (G-to-T substitution) in HHV-8 ORF26 were correlated with increased susceptibility to Kaposi's sarcoma, compared to the results obtained with HHV-8 with wild-type nucleotides at these positions (P = 0.0106). This observation suggests that molecular heterogeneity of the HHV-8 genome affects the biological properties of HHV-8, resulting in different clinical phenotypes of HHV-8 infection. Since sensitive PCR of ORF26 allowed us to analyze the SNPs by using peripheral blood from HHV-8-infected patients, the ORF26 SNPs will be a potent tool for investigating the pathogenesis of HHV-8 infection.
Human herpesvirus 8 (HHV-8) or Kaposi's sarcoma (KS)-associated herpesvirus DNA is present in virtually all tumor samples from both classical and AIDS-associated forms of KS (1, 4, 5, 9, 16, 19). HHV-8 is thus thought to be involved in the pathogenesis of KS. However, some HHV-8-infected individuals fail to develop KS, suggesting that other factors, such as the virulence of HHV-8 or immune function and cytokine levels in infected hosts, may be required for the development of KS (3, 6, 17).
Previous studies revealed that HHV-8 has a highly variable open reading frame (ORF) K1 gene at the end of its genome and that this variability allows the ORF K1 gene to be a marker for HHV-8 subtyping and strain differentiation (5, 16). Major molecular subtypes A, B, C, and D have been described for specimens obtained from around the world (5, 16). Recently, the ORF26 gene of the HHV-8 genome was found to have single nucleotide polymorphisms (SNPs) that are linked to ORF K1 polymorphisms (16). The data suggested that the four major subtypes of HHV-8 with distinctive ethnic and geographic associations originated during the migration of modern humans from Africa in Paleolithic times (5, 16). It was speculated that the major evolutionary branches of HHV-8 occurred in Africa 100,000 years ago (subtype B), followed by the migration of ancestors of modern humans to southern Asia and Oceania 60,000 years ago (subtype D) and a second spread to Europe and northern Asia about 35,000 years ago (subtypes A and C) (5).
Given the limited nature of data regarding HHV-8 subtypes in Asia, we investigated the molecular characterization of HHV-8 derived from human immunodeficiency virus (HIV)-infected Japanese patients (7, 8, 12). We attempted to obtain DNA fragments of ORF K1 and ORF26 in HHV-8 genomes isolated from peripheral blood mononuclear cell (PBMC) DNA by PCR and found that ORF26 was an easier target for amplification. On the basis of our analysis of ORF26 SNPs, we describe here both the molecular epidemiology of HHV-8 in Japan and the relationship between ORF26 SNPs and clinical phenotypes of HHV-8 infection.
MATERIALS AND METHODS
Patients.
Fifty-nine HIV-infected patients who visited either The Research Hospital, The Institute of Medical Science, The University of Tokyo, or Komagome Metropolitan Hospital from April 1995 to June 2002 were selected for this study based on seropositivity for latency-associated nuclear antigen (LANA) (6) and clinical history of KS. All patients were HIV-infected men who had sex with men. Peripheral blood was collected from these patients with their informed consent. For some patients, previously resected KS tissues stored at −80°C were used as a source of DNA.
DNA isolation, PCR, and sequencing.
DNA was isolated from PBMCs by using a QIAamp DNA blood mini kit (Qiagen, Tokyo, Japan) and from KS tissues by using a QIAamp DNA mini kit (Qiagen) according to the manufacturer's instructions. The 671-bp ORF K1 coding region from nucleotide positions 193 to 864 was amplified from 100 ng of DNA by nested PCR with an outside primer pair (5′-GTTCTGCCAGGCATAGTC-3′ [21 to 38] and 5′-AATAAGTATCCGACCTCAT-3′ [1085 to 1067]) and an inside primer pair (5′-GAGTGATTTCAAAGAATTAC-3′ [193 to 212] and 5′-AGATACCACACATGGTT-3′ [840 to 864]) (5). Ex-Taq (TaKaRa, Tokyo, Japan) and a model 9700 GeneAmp PCR system (Applied Biosystems, Norwalk, Conn.) were used for PCR at 40 cycles of 94°C for 1 min, 59.7°C for 1 min, and 72°C for 2 min. Similarly, the 336-bp ORF26 coding region from nucleotide positions 890 to 1226 was amplified by nested PCR with an outside primer pair (5′-ATCTATCCAAGTGCACACTCG-3′ [826 to 846] and 5′-CTGGGAACCAAGGCTGATAGG-3′ [1291 to 1270]) and an inside primer pair (5′-GATGGATCCCTCTGACAACCT-3′ [890 to 910] and 5′-GGATCCGTGTTGTCTACG-3′ [1226 to 1204]) (16). The products of nested PCR were resolved on a 1.5% agarose gel, and the expected bands were excised and subjected to DNA sequencing (ABI PRISM 310 genetic analyzer; Applied Biosystems) by using both 5′ and 3′ inside primers.
Phylogenetic analysis.
Deduced amino acid sequences or nucleotide sequences were analyzed with a multiple-alignment system by using CLUSTAL W (DNA Data Bank of Japan, Mishima, Japan; http://www.ddbj.nig.ac.jp/Welcome-j.html) (18), and phylogenetic trees were produced. ORF K1 and ORF26 sequences (respective GenBank accession numbers) used in this analysis were BCBL-R, BCBL-B, 431KAP, and ASM72 (AF133038 to AF133041); TKS10 and ZKS3 (AF133042 and AF133044); BC1 (U75678); BCBL-1 (U86667); and those described by Poole et al. (BKS1, OKS3, OKS4, OKS7, RKS3, SKS6, BC3, TKS1, TKS6, SKS1, and SKS9) (16).
RESULTS
DNA was prepared from PBMCs from 59 HIV-infected patients who either had KS or were seropositive for LANA. All patients were men who had sex with men, and their ages and CD4 counts (means and standard deviations) in peripheral blood were 38.2 ± 10.2 years old (range, 20 to 61) and 231 ± 178 cells/μl (range, 6 to 856), respectively. When we initially tested the sensitivity of PCR for ORF K1 and ORF26, we were able to obtain the 671-bp target DNA fragment of ORF K1 from only 3 out of 29 samples (patients 3, 4, and 11), whereas all 29 samples were positive for PCR of the 336-bp ORF26 fragment (Table 1). For three patients whose KS tissues were available for analysis (patients 15, 16, and 17), DNA was extracted from the KS tissues and ORF K1 fragments were successfully obtained. HHV-8 subtypes determined on the basis of ORF K1 sequences according to the designations of Poole et al. (16) are shown in Table 1.
TABLE 1.
SNPs in HHV-8 ORF26 amplified from PBMC DNA
| Patient | SNP at the following nucleotide position in ORF26a:
|
ORF K1 subtypeb | KSc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 935 | 981 | 1032 | 1055 | 1086 | 1094 | 1103 | 1122 | 1132 | 1139 | |||
| BCBL-R | C | T | C | G | C | G | C | G | A | A | A1 | |
| 1 | — | — | A | T | — | — | — | — | — | — | NP | + |
| 2 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 3 | — | — | — | — | — | — | — | — | — | — | A4 | + |
| 4 | — | C | A | T | — | — | — | — | G | C | A5 | + |
| 5 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 6 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 7 | — | — | — | — | — | — | — | — | — | — | NP | + |
| 8 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 9 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 10 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 11 | — | — | — | — | — | — | — | — | — | — | A1 | + |
| 12 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 13 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 14 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 15 | — | — | — | — | — | — | — | — | — | — | A1′d | + |
| 16 | — | C | A | T | — | — | — | — | G | C | A4′d | + |
| 17 | A | C | — | — | T | — | — | — | — | C | C3′d | + |
| 18 | — | — | — | — | — | — | A | — | — | — | NP | + |
| 19 | — | — | — | — | — | — | A | — | — | — | + | |
| 20 | — | C | A | T | — | — | — | — | G | C | + | |
| 21 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 22 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 23 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 24 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 25 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 26 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 27 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 28 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 29 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 30 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 31 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 32 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 33 | — | — | — | — | — | — | A | — | — | — | NP | − |
| 34 | — | — | — | — | — | — | — | — | — | — | − | |
| 35 | — | — | — | — | — | — | — | — | — | — | − | |
| 36 | — | — | — | — | — | — | A | — | — | — | − | |
| 37 | — | — | — | — | — | — | — | — | — | — | − | |
| 38 | — | C | — | — | T | — | — | — | — | C | − | |
| 39 | — | — | — | — | — | — | A | — | — | — | − | |
| 40 | — | — | — | — | — | — | A | — | — | — | − | |
| 41 | — | — | — | — | — | — | — | — | — | — | − | |
| 42 | — | — | — | — | — | — | A | — | — | — | − | |
| 43 | — | — | — | — | — | — | A | — | — | — | − | |
| 44 | — | — | — | — | — | — | A | — | — | — | − | |
| 45 | — | — | — | — | — | — | A | — | — | — | − | |
| 46 | — | C | — | — | — | — | — | — | G | C | − | |
| 47 | — | — | — | — | — | — | A | — | — | — | − | |
| 48 | — | — | — | — | — | — | — | — | — | — | − | |
| 49 | — | — | — | — | — | — | A | — | — | — | − | |
| 50 | — | — | — | — | — | — | — | — | — | — | − | |
| 51 | — | — | — | — | — | — | A | — | — | — | − | |
| 52 | — | — | — | — | — | — | — | — | — | — | − | |
| 53 | — | C | — | — | — | — | — | — | — | C | − | |
| 54 | — | C | — | — | — | — | — | — | — | C | − | |
| 55 | — | — | — | — | — | — | — | — | — | — | − | |
| 56 | — | — | — | — | — | — | A | — | — | — | − | |
| 57 | — | — | — | — | — | — | A | — | — | — | − | |
| 58 | — | — | — | — | — | — | — | — | — | — | − | |
| 59 | — | — | — | — | — | — | A | — | — | — | − | |
Nucleotide positions of ORF26 SNPs reported by Poole et al. (16). Nucleotides in the first entry represent HHV-8 sequences derived from the BCBL-R cell line. —, no polymorphism.
ORF-K1 subtypes determined by phylogenetic analysis of ORF K1 sequences. NP, no PCR fragments were obtained by PCR of PBMC DNA.
Past or present history of KS. +, present; −, absent.
Data were obtained from KS tissue DNA. ORF K1 fragments could not be obtained from PBMC DNA for these patients.
Because of the low efficiency of the ORF K1 PCR, we decided to use HHV-8 ORF26 sequences for the molecular analysis of HHV-8. These sequences were found to be relatively conserved and were characterized by 10 SNPs (nucleotide positions 935, 981, 1032, 1055, 1086, 1094, 1103, 1122, 1132, and 1139) linked to ORF K1 polymorphisms (5). ORF26 fragments were successfully obtained from PBMC DNA from all 59 patients, and sequence analysis revealed that the patterns of the ORF26 SNPs could be classified into eight types (designated types 1 to 8; Tables 1 and 2). Since the nucleotides at positions 1094 and 1122 did not exhibit polymorphisms in the 59 samples, they were excluded from the list of SNPs. As shown in Table 2, ORF26 SNP type 1 was the most prevalent SNP pattern in our population (37 patients; 63%), and type 2 was the next most prevalent (13 patients; 22%). Although the numbers of patients with types 3 to 8 were small (one to three patients with each type), of note is that the frequency of KS patients with these types was high compared to the numbers with type 1 (29.7%) and type 2 (30.8%).
TABLE 2.
Eight SNPs types of ORF26 sequences obtained from 59 HHV-8-infected patients
| SNP type | SNP at the following nucleotide position (BCBL-R cell line nucleotide) in ORF26a:
|
Corresponding ORF K1 subtypesb | No. (%) of patientsc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 935(C) | 981(T) | 1032(C) | 1055(G) | 1086(C) | 1103(C) | 1132(A) | 1139(A) | Total | With KS | ||
| 1 | — | — | — | — | — | A | — | — | A3 | 37 (62.7) | 11 (29.7) |
| 2 | — | — | — | — | — | — | — | — | A1, A1′, A2, A4, C2, C3d | 13 (22.0) | 4 (30.8) |
| 3 | — | C | A | T | — | — | G | C | C3 | 3 (5.1) | 3 (100) |
| 4 | — | C | — | — | — | — | — | C | ND | 2 (3.4) | 0 (0) |
| 5 | A | C | — | — | T | — | — | C | C3′ | 1 (1.7) | 1 (100) |
| 6 | — | C | — | — | T | — | — | C | A5, B, C1, C2, C4, C5d | 1 (1.7) | 0 (0) |
| 7 | — | C | — | — | — | — | G | C | B | 1 (1.7) | 0 (0) |
| 8 | — | — | A | T | — | — | — | — | ND | 1 (1.7) | 1 (100) |
Nucleotides at positions 1094 and 1122 are not included because no SNPs were found at these positions in 59 samples. The nucleotides in bold type amino acid substitutions (T981C: Phe→Leu; C1032A: Leu→Ile; A1132G: Asp→Gly).
Based on the molecular data reported by Poole et al. (16). ND, no corresponding ORF26 SNPs.
Percentages represents the relative proportions of patients with each SNP type out of 59 patients (total) or the proportions of patients with each SNP out of patients with a past or present, history of KS.
ORF26 SNPs cannot distinguish these subtypes.
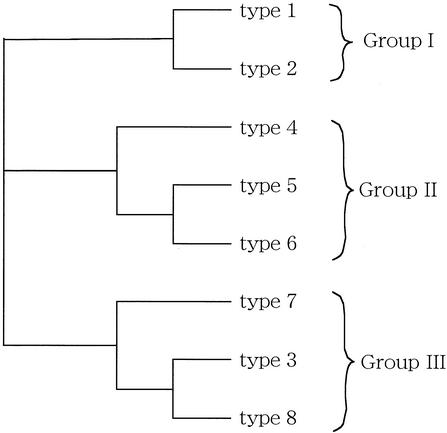
To study the relationship between the molecular diversity of HHV-8 and the clinical course of HHV-8 infection, the eight types of ORF26 SNPs were classified based on the similarities of the SNP patterns. The 8 nucleotides at each SNP position were regarded as 8-nucleotide oligomers and were aligned by using CLUSTAL W software. As shown in Fig. 1, the phylogenetic analysis of CLUSTAL W data showed that the eight types of ORF26 SNPs could be classified into three groups: group I, types 1 and 2; group II, types 4, 5, and 6; and group III, types 3, 7, and 8. The clinical parameters of the patients in these three groups were compared to each other, and a statistically significant difference was found between the numbers of patients with KS lesions in groups I and III (P = 0.0435) (Table 3). Although CD4 counts in group III seemed lower than those in groups I and II, the difference was not statistically significant.
FIG. 1.
Phylogenetic analysis of eight ORF26 SNP types. The sequences of eight types of ORF26 SNPs were aligned and analyzed by CLUSTAL W to produce a phylogenetic tree. The eight SNP types were divided into three groups: group I, types 1 and 2; group II, types 4, 5, and 6; and group III, types 3, 7, and 8.
TABLE 3.
Clinical parameters for three groups of ORF26 SNPsa
| ORF26 SNP group | Patients
|
CD4 counts
|
Mean log10 no. of HIV RNA copies/ml | No. (%) of patients with KSb | |||
|---|---|---|---|---|---|---|---|
| No. | Age (yr) | Total | KS+ | KS− | |||
| I | 50 | 37.8 ± 10.4 | 231 ± 189 | 132 ± 12 | 282 ± 196 | 4.75 | 15 (30.0) |
| II | 4 | 39.5 ± 10.7 | 283 ± 118 | 113 | 340 ± 91 | 4.73 | 1 (25.0) |
| III | 5 | 41.6 ± 7.5 | 177 ± 61 | 180 ± 70 | 168 | 4.98 | 4 (80.0) |
Data are reported as means and standard deviations unless otherwise indicated. KS+ and KS−, patients with and without KS, respectively.
The P value, as determined by Fisher's test, for a comparison of groups I and III was 0.0435. Other comparisons were not statistically significant.
To further analyze the relationship between ORF26 polymorphisms and KS susceptibility, we focused on all eight SNPs in ORF26. When KS frequencies were compared between patients with and without each SNP, the patients with either a C-to-A nucleotide substitution at position 1032 or a G-to-T substitution at position 1055 were significantly more susceptible to KS than those without these SNPs (P = 0.0106) (Table 4). These two SNPs were completely linked, and all patients (n = 4) who had a C-to-A SNP at nucleotide position 1032 had a G-to-T SNP at position 1055 and vice versa.
TABLE 4.
Relationship between ORF26 SNPs and KS susceptibilitya
| Position | Nucleotide | No. of patients who were:
|
Pb | |
|---|---|---|---|---|
| KS+ | KS− | |||
| 935 | C | 19 | 39 | 0.3389 |
| A | 1 | 0 | ||
| 981 | T | 16 | 35 | 0.2580 |
| C | 4 | 4 | ||
| 1032 | C | 16 | 39 | 0.0106 |
| A | 4 | 0 | ||
| 1055 | G | 16 | 39 | 0.0106 |
| T | 4 | 0 | ||
| 1086 | C | 19 | 38 | 0.5770 |
| T | 1 | 1 | ||
| 1103 | C | 9 | 13 | 0.2753 |
| A | 11 | 26 | ||
| 1132 | A | 17 | 38 | 0.1083 |
| G | 3 | 1 | ||
| 1139 | A | 16 | 35 | 0.2580 |
| C | 4 | 4 | ||
KS+ and KS−, KS positive and KS negative, respectively.
For comparisons of the nucleotides at the indicated positions, as determined by the Mann-Whitney U test.
DISCUSSION
Subtyping of viruses by using nucleotide polymorphism analysis is an important tool for investigating their geographic epidemiology, transmission routes, virulence, and differences in clinical manifestations. The 289-amino-acid ORF K1 membrane protein of HHV-8 displays unusually high levels of genetic variability, resulting in four major subtypes, A, B, C, and D, which differ by 15 to 30% at the amino acid level (5, 15). Although we initially tried to sequence the ORF K1 gene amplified from PBMC DNA, it was difficult to obtain the 671-bp DNA fragment unless KS tissues were used as the DNA source. In contrast, ORF26, which is characterized by relatively conserved nucleotide sequences, was an easier target for PCR than ORF K1; we were able to obtain by nested PCR the 336-bp ORF26 fragment from PBMCs from all 59 patients who were either seropositive for LANA or had previous or present histories of KS. The low efficiency of ORF K1 PCR was not due to mismatches of PCR primers to polymorphic ORF K1 sequences, because we sequenced approximately 60-bp fragments of primer binding sites and found no primer mismatch in eight out of nine samples which did not yield the 671-bp ORF K1 fragment (data not shown). Consequently, we speculate that the length of the target sequence, the secondary structures of the primers, and the target sequences themselves were not suitable for sensitive PCR of ORF K1. However, the high degree of ORF K1 sequence polymorphisms restricted the choice of PCR primer sites, and the other primers that we tried did not improve PCR sensitivity. Thus, we decided to use the ORF26 sequence for the molecular analysis of HHV-8 in this study, because ORF26 was shown to possess SNPs that were linked to HHV-8 subtypes, as determined by amino acid sequence analysis of ORF K1 (16).
The sequence polymorphisms of ORF26 in 59 Japanese samples revealed that the nucleotide patterns of eight SNP sites in ORF26 were classified into eight types. Type 1 was the most prevalent SNP (37 samples; 63%), and it corresponded to subtype A3, as determined on the basis of ORF K1 polymorphisms (16). The second most prevalent SNP was type 2, corresponding to subtype A1, A1′, A2, A4, C2, or C3 (13 samples; 22%). Similarly, type 3 corresponded to C3; type 5 corresponded to C3′; type 6 corresponded to A5, B, C1, C2, C4, or C5; and type 7 corresponded to B. Type 4 and 8 ORF26 SNPs did not appear in the data of Poole et al. (16) and could not be classified in the ORF K1-based subtyping system. Although subtypes A1, A1′, A2, A4, C2, and C3 of SNP type 2 were indistinguishable by ORF26 SNP analysis, ORF K1 sequences were available for 3 out of 13 type 2 samples; the phylogenetic analysis revealed them to be subtype A4 (patient 3), subtype A1 (patient 11), and subtype A1′ (patient 15) (Table 1). These results suggest that approximately two-thirds or more (at least 40 out of 59) of HHV-8 isolates in Japanese HIV-infected patients belong to major subtype A.
We found that HHV-8-infected patients with ORF26 SNP types 3, 7, and 8 (group III) developed KS more frequently than those with other SNP types (I versus III) (P = 0.435). When we analyzed each SNP and KS susceptibility, the C-to-A nucleotide substitution at position 1032 and the G-to-T substitution at position 1055 were found to be related to a higher level of susceptibility to KS (P = 0.0106). These SNPs were found in four patients who had ORF26 SNP type 3 (three patients) or type 8 (one patient) and were completely linked to each other. These observations suggest that ORF26 SNP group III is related to a high level of susceptibility to KS, because this group contains ORF26 SNPs at nucleotide positions 1032 and 1055 that are correlated with a high level of susceptibility to KS.
Although the mechanism by which ORF26 SNPs at nucleotide positions 1032 and 1055 affect KS susceptibility is not clear, speculation can be offered from knowledge about the molecular biology of HHV-8. ORF26 encodes one of the minor capsid antigens that are known to be immunogenic in humans and to serve as a specific serological marker for the presence of KS and body-cavity-based lymphoma (3, 7, 8, 10, 11). Since the C-to-A nucleotide substitution at position 1032 and not the G-to-T substitution at position 1055 results in an amino acid change from Leu to Ile, it is possible that the altered ORF26 product is less immunogenic and that the viruses escape host immune surveillance, resulting in a more virulent infection. To understand the relationship between ORF26 SNPs and KS susceptibility, we need to search sequence polymorphisms in the entire HHV-8 genome and study a larger number of patients.
Previous studies demonstrated that the molecular heterogeneity of HHV-8 may contribute to different biological properties and clinical phenotypes. Ma et al. showed that HHV-8 isolates derived from KS lesions had different ORF K1 sequences and more potent cytotoxic effects on cultured cells than HHV-8 isolates derived from body-cavity-based lymphomas (13). Boralevi et al. reported that HHV-8 isolates derived from 24 multiple myeloma patients consistently belonged to subtype C3, whereas HHV-8 isolates derived from KS patients were of subtype A (2). Similarly, Meng et al. showed that subtype A HHV-8 isolates resulted in significantly more frequent mucosal and/or visceral KS lesions and higher mean CD4 counts at the onset of KS in a French population (14). However, no reports have described the frequency of development of KS, and this report serves as the first evidence in support of a correlation between the molecular heterogeneity of HHV-8 and KS susceptibility. Past research has focused on patients with KS or body-cavity-based lymphoma, as opposed to HHV-8 carriers without KS, and has used tumor tissues as DNA sources. However, the latter source was expected because, as shown in this study, it is difficult to obtain ORF K1 sequences from samples other than KS tissues due to the low efficiency of ORF K1 PCR. In this regard, the study of ORF26 SNPs by using PBMC DNA is a feasible alternative for investigating overall HHV-8 subtype prevalence. Although ORF K1 polymorphisms allowed us to classify HHV-8 subtypes into more variants than ORF26 SNPs, the information obtained with ORF26 SNPs was sufficient to distinguish between patients who were and those who were not susceptible to KS. Further studies with ORF26 SNPs from a larger population of HHV-8-infected patients and from different geographic areas will help to lead to an understanding of the pathogenesis of HHV-8 infection.
Acknowledgments
This work was partly supported by grants for AIDS research from the Ministry of Health, Labor and Welfare of Japan; a grant-in-aid for scientific research from the Japan Society of the Promotion of Science (JSPS); and the Japan Human Sciences Foundation.
We thank David Chao for technical assistance.
REFERENCES
- 1.Beral, V., T. A. Peterman, R. C. Berkelman, and H. W. Jaffe. 1990. Kaposi's sarcoma among persons with AIDS: a sexually transmitted infection? Lancet 335:123-128. [DOI] [PubMed] [Google Scholar]
- 2.Boralevi, F., B. Masquelier, M. Denayrolles, M. Dupon, J. Pellegrin, J. Ragnaud, and H. J. A. Fleury. 1998. Study of human herpesvirus 8 (HHV-8) variants from Kaposi's sarcoma in France: is HHV-8 subtype a responsible for more aggressive tumors? J. Infect. Dis. 178:1546-1547. [DOI] [PubMed] [Google Scholar]
- 3.Boshoff, C., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus: a new DNA tumor virus. Annu. Rev. Med. 52:453-470. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y., E. Cesarman, M. S. Pession, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moor. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 5.Ciufo, A. M., D. J. Alcendor, X. Wan, J. Nicholas, P. J. Browning, P. L. Rady, S. K. Tyring, J. M. Orenstein, C. S. Rabkin, I.-J. Su, K. F. Powell, M. Croxson, K. E. Foreman, B. J. Nickoloff, S. Alkan, and G. S. Hayward. 1999. High-level variability in the open reading frame K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J. Virol. 73:4156-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujii, T., H. Taguchi, H. Katano, S. Mori, T. Nakamura, N. Nojiri, K. Nakajima, K. Tadokoro, T. Juji, and A. Iwamoto. 1999. Seroprevalence of human herpesvirus 8 in human immunodeficiency virus 1-positive and human immunodeficiency virus 1-negative populations in Japan. J. Med. Virol. 57:159-162. [DOI] [PubMed] [Google Scholar]
- 7.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. Moor. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 355:233-241. [DOI] [PubMed] [Google Scholar]
- 8.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. Moor. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 9.Harwood, A. R., D. Osoba, S. L. Hofstader, M. B. Goldstein, C. Cardella, M. J. Holecek, R. Kunynetz, and R. A. Giammarco. 1979. Kaposi's sarcoma in recipients of renal transplants. Am. J. Med. 67:759-765. [DOI] [PubMed] [Google Scholar]
- 10.Katano, H., T. Iwasaki, N. Baba, M. Terai, S. Mori, A. Iwamoto, T. Kurata, and T. Sata. 2000. Identification of antigenic proteins encoded by human herpesvirus 8 and seroprevalence in the general population and among patients with and without Kaposi's sarcoma. J. Virol. 74:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katano, H., T. Sata, T. Suda, T. Nakamura, N. Tachikawa, H. Nishizumi, S. Sakurada, Y. Hayashi, M. Koike, A. Iwamoto, T. Kurata, and S. Mori. 1999. Expression and antigenicity of human herpesvirus 8 encoded ORF59 protein in AIDS-associated Kaposi's sarcoma. J. Med. Virol. 59:346-355. [DOI] [PubMed] [Google Scholar]
- 12.Kedes, D. H., E. Oberskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 13.Ma, H. J., N. N. Sjak-Shie, R. A. Vescio, M. Kaminsky, A. Mikail, M. Pold, K. Parker, M. Beksac, D. Belson, T. J. Moss, C. H. Wu, J. Zhou, L. Zhang, G. Chen, J. W. Said, and J. R. Berenson. 2000. Human herpesvirus 8 open reading frame 26 and open reading frame 65 sequences from multiple myeloma patients: a shared pattern not found in Kaposi's sarcoma or primary effusion lymphoma. Clin. Cancer Res. 6:4226-4233. [PubMed] [Google Scholar]
- 14.Meng, Y. X., T. Sata, F. R. Stamey, A. Voevodin, H. Katano, H. Koizumi, M. Deleon, M. A. De Cristofano, R. Galimberti, and P. E. Pellett. 2001. Molecular characterization of strains of human herpesvirus 8 from Japan, Argentina and Kuwait. J. Gen. Virol. 82:499-506. [DOI] [PubMed] [Google Scholar]
- 15.Nicholas, J., J. C. Zong, D. J. Alcendor, D. M. Cifo, L. J. Poole, R. T. Sarisky, C. J. Chiou, X. Zhang, X. Wan, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 16.Poole, L. J., J. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spano, J. P., Y. Salhi, D. Costaglioa, W. Rozenbaum, and P. M. Girard. 2000. Factors predictive of disease progression and death in AIDS-related Kaposi's sarcoma. HIV Med 1:232-237. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, R. E. Sugget, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]