Abstract
Screening for Chlamydia trachomatis was done for 280 endocervical swab samples by PCR specific for endogenous plasmid. Age dependency was seen in symptomatic patients, with a high chlamydial prevalence rate (28%) found in younger women. Genotyping by restriction fragment length polymorphism analysis of omp1 PCR-positive samples showed serovars D, E, and F to be the most prevalent.
Chlamydia trachomatis is a major cause of sexually transmitted disease (16). Serovars D to K are chiefly responsible for urogenital infections; of these serovars, E, F, and D account for up to 60 to 70% of these infections (1, 5, 14, 20). Epidemiological studies of C. trachomatis infections in sexual contacts have been few to date, which hampers the study of chlamydial transmission, its route of spread in a population, its virulence factors, and the associated risk factors of C. trachomatis infections (12, 19). Compared with immunotyping, the genotyping methods, particularly omp1 are more sensitive and precise in revealing C. trachomatis variants within serovars as well as in potential recombinants among serovars (13, 15, 18). The present study was undertaken to understand the occurrence of C. trachomatis serovars in the genital tracts of infected women, which will help in devising an effective screening program for routine diagnosis of chlamydial infections, understanding the immunopathogenesis, and developing an effective chlamydial control program.
Reference C. trachomatis standard serovars were kindly provided by T. Ossewaarde (National Institute of Public Health and Environment Protection, Bilthoven, The Netherlands). Cervical swabs (n = 280) were obtained from patients (17) attending the gynecology outpatient clinic for various gynecological reasons (abnormal vaginal discharge and pelvic pain) at Safdarjang Hospital, New Delhi, India. Chlamydial DNA was extracted from the clinical specimens by the alkali lysis method (2). The study protocol was approved by the committee responsible for the evaluation of work involving human subjects. Prior consent was obtained in all cases.
The clinical samples were first screened by a PCR specific for the human β-globin gene. The primers used for the human β-globin PCR were P1 (sense, 5′ ACA CAA CTG TGT TCA CTA GC) and P2 (antisense, 5′ GAA ACC CAA GAG TCT TCT CT). Samples positive in the β-globin PCR were used for C. trachomatis detection by using a plasmid PCR performed as described previously. The plasmid primers P3 (sense, 5′GAA CAA ATC GTA TCT CGG) and P4 (antisense, 5′GAA ACC AAC TCT ACG TCG) generated a fragment of 517 bp in the C. trachomatis-positive samples. The omp1 gene was amplified by primers selected from the published sequences of the omp1 gene of C. trachomatis (L2) (21). The PCR mixture contained 25 pmol of each of the forward and reverse primers, 200 μM concentrations of each of the deoxynucleoside triphosphates (dATP, dTTP, dGTP, and dCTP), 10× PCR buffer (containing 10 mM Tris HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin), and 0.1 U of Taq DNA polymerase (Gibco-BRL). PCR was performed in a thermocycler as follows: 5 min at 95°C; 40 cycles each consisting of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min; and a final elongation step of 10 min at 72°C. Restriction endonuclease digestion was performed with the restriction enzymes AluI, CfoI, HinfI, DdeI, and EcoRI (Promega, Madison, Wis.) on 5-μl amounts of the amplified 871-bp PCR products by following the manufacturer's instructions.
All samples shown positive by the β-globin PCR assay were further screened by a plasmid PCR specific for the C. trachomatis endogenous plasmid. A clear age dependency was observed, showing a high prevalence rate among younger women. The overall prevalence rates of C. trachomatis in cervical swabs as determined by plasmid PCR were 28.5% in group I (age, 18 to 25 years), 7.6% in group II (age, 25 to 35 years), and 3.2% in group III (age, 35 to 45 years). C. trachomatis-positive swabs detected by plasmid PCR were further examined by omp1 PCR for genotyping by restriction fragment length polymorphism. Purified PCR products were digested with AluI to differentiate serovars of C. trachomatis (Fig. 1). Serovars D and E showed fragments varying from 244 to 91 bp in size. Serovars B and D had a closely related pattern, except for the presence (serovar B) or absence (serovar D) of a 91-bp band in the digestion profile with AluI. The standard serovar F showed fragment sizes of 321 to 80 bp after digestion. Standard serovars L3 and I, which are in the C complex, showed fragment of sizes 486 to 92 bp after digestion with AluI.
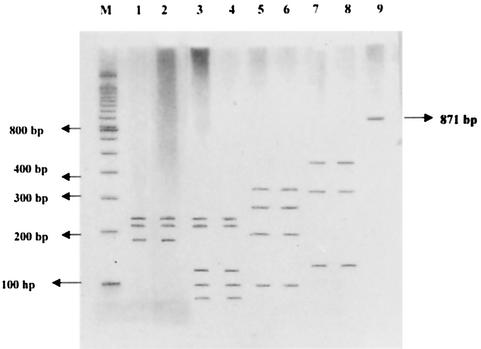
FIG. 1.
AluI restriction endonuclease digestion of PCR products of C. trachomatis-positive clinical samples visualized by silver staining on polyacrylamide gel electrophoresis. Lanes: M, molecular size marker; 1, standard serovar D; 2, clinical sample showing banding pattern matching that of standard serovar D; 3, standard serovar E; 4, clinical sample showing banding pattern matching that of standard serovar E; 5, standard serovar F; 6, clinical sample showing banding pattern matching that of standard serovar F; 7, standard serovar I; 8, clinical sample showing banding pattern matching that of standard serovar I; 9, uncut amplicon.
Serovars H, L3, and I were further distinguished by enzyme digestion with EcoRI and DdeI. With EcoRI, serovar I gave two bands, whereas with DdeI, it gave four bands varying between 323 to 80 bp in size. The clinical samples which were identified as having banding patterns matching that of serovar D after digestion with the AluI enzyme were further differentiated by digesting them with CfoI restriction enzyme. After analysis with the CfoI restriction enzyme, clinical samples were identified as serovar D, since they had an identical banding pattern as that of the standard serovar D. The restriction endonuclease sites for the various restriction enzymes are listed in Table 1.
TABLE 1.
Restriction endonuclease sites for different restriction enzymes
| Serovar | Size (bp) of restriction endonuclease site for restriction enzyme:
|
||||
|---|---|---|---|---|---|
| AluI | CfoI | DdeI | EcoRI | HinfI | |
| A | 486, 302, 138 | ||||
| B | 244, 225, 195, 91 | ||||
| Ba | 244, 204, 195, 91 | ||||
| D | 241, 225, 195 | 587, 408 | |||
| E | 241, 225, 132, 102, 95 | ||||
| F | 321, 256, 207, 101, 80 | ||||
| G | 321, 207, 195, 107, 96, 80 | ||||
| I | 458, 302, 138 | 323, 285, 183 | 110 | ||
| J | 435, 305, 138 | 540 | |||
| L2 | 227, 225, 132, 102, 96 | ||||
| L3 | 458, 305, 138 | 841, 131, 110 | |||
C. trachomatis serovar D (48%) was found to be the most prevalent, followed by serovars E (34%), F (12%), and I (6%) in urogenital samples.
The present study demonstrated that the prevalence of symptomatic C. trachomatis infection is high in cervical swabs, as determined by a sensitive PCR assay, and is age dependent. These data are in agreement with earlier published reports showing high C. trachomatis rates (6, 10). The clinical samples could be discriminated by restriction endonuclease digestion with AluI. However, for a few clinical samples, digestion with HinfI, DdeI, CfoI, and EcoRI was done, which further aided in discriminating among C. trachomatis serovars.
In our study, serovar D (48%) was found to be the most prevalent serovar, followed by serovars E (34%), F (12%), and I (6%) in the urogenital samples. A similar distribution of C. trachomatis serovars has been reported earlier with tissue culture isolates (11). This pattern of serovar prevalence was found to be in accordance with the distribution of C. trachomatis serovars in other regions of the world (4, 7, 8, 9). We were not able to find other serovars J, H, K, or G of C. trachomatis, which have been reported by few workers (1, 3, 12). We did not observe any restriction endonuclease pattern different from the prototype or known variant strains in the 50 clinical samples examined. But this would have not been surprising, as others have found only a limited number of variants by serologic methods. In conclusion, C. trachomatis genotyping in women younger than age 30 years revealed that serovars D and E were more frequently found in symptomatic patients. The procedure we have used for cervical swabs by PCR restriction fragment length polymorphism analysis should enable basic serovar identification and provide a springboard for studies of variation.
Acknowledgments
This work was partly supported by the Department of Biotechnology, New Delhi, India.
REFERENCES
- 1.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanparsain, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of C. trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp 1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acid Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrego, M. J., J. P. Gomes, J. F. Lefebvre, F. Eb, J. Orfila, and M. A. Catry. 1997. Genotyping of Portuguese Chlamydia trachomatis urogenital isolates. Genitourin. Med. 73:561-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frost, E. H., S. Deslandes, S. Veillexr, and D. Bourgauz-Ramoisy. 1991. Typing C. trachomatis by detection of RFLP in the gene encoding the MOMP. J. Infect. Dis. 163:1103-1107. [DOI] [PubMed] [Google Scholar]
- 5.Gaydos, C. A., L. Bobo, L. Welsh, E. W. Hook III, K. Viscidi, and T. C. Quinn. 1992. Genotyping of Chlamydia trachomatis by polymerase chain reaction and restriction endonuclease digestion. Sex. Transm. Dis. 19:303-308. [PubMed] [Google Scholar]
- 6.Hammerschlag, M. R., N. H. Golden, K. Oh, M. Gelling, M. Sturdevant, B. R. Brown, Z. Aras, S. Neuhoff, W. Dumornay, and P. M. Rublin. 1993. Single dose of azithromycin for the treatment of genital chlamydial infection in adolescents. J. Pediatr. 122:961-965. [DOI] [PubMed] [Google Scholar]
- 7.Kuo, C. C., S. P. Wang, K. K. Holmes, and J. T. Grayston. 1983. Immunotypes of Chlamydia trachomatis isolates in Seattle, Washington. Infect. Immun. 41:865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan, J., C. J. L. M. Meijer, A. R. van den Hoek, J. M. Ossewaarde, J. M. M. Walbommers, and A. J. C. van den Brule. 1995. Genotyping of C. trachomatis serovars derived from heterosexual partners and a detailed genomic analysis of serovar F. Genitourin. Med. 71:299-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan, J., I. Melgers, C. J. L. M. Meijer, J. M. M. Walboomers, R. Roosendaal, C. Burger, O. P. Bleker, and A. J. C. van den Burle. 1995. Prevalence and serovar distribution of asymptomatic cervical Chlamydiatrachomatis infections as determined by highly sensitive PCR. J. Clin. Microbiol. 33:3194-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittal, A., S. Kapur, and S. Gupta. 1993. Chlamydial cervicitis: role of culture, enzyme immunoassay and Giemsa cytology in diagnosis. Acta Pathol. Microbiol. Immun. Scand. 101:37-40. [DOI] [PubMed] [Google Scholar]
- 11.Mittal, A. 1998. Serovar distribution of C. trachomatis isolates collected from the cervix: use of the polymerase chain reaction and restriction endonuclease digestion. Br. J. Biomed. Sci. 55:179-183. [PubMed] [Google Scholar]
- 12.Moore, S. A., R. Moes, and I. Van Valkengoed. 1998. Genotyping of Chlamydia trachomatis in urine specimens will facilitate large epidemiological studies. J. Clin. Microbiol. 36:3077-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ossewaarde, J. M., M. Rieffe, M. Rozenberg-Arska, P. M. Ossenkoppele, R. P. Nawrocki, and A. M. van Loon. 1992. Development and clinical evaluation of a polymerase chain reaction test for detection of C. trachomatis. J. Clin. Microbiol. 30:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez, P., B. de Barbeyrac, K. Persson, B. Dutlith, and C. Sebear. 1993. Evaluation of molecular typing for epidemiology study of Chlamydiatrachomatis genital infections. J. Clin. Microbiol. 31:2238-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayada, C., E. Denamur, J. Orfila, and J. Elion. 1991. Rapid genotyping of the C. trachomatis major outer membrane protein by the PCR. FEMS Microbiol. Lett. 83:73-78. [DOI] [PubMed] [Google Scholar]
- 16.Schachter, J. 1999. Infection and epidemiology, p. 391-405. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C.
- 17.Singh, V., S. Rastogi, S. Garg, S. Kapur, A. Kumar, S. Salhan, and A. Mittal. 2001. Polymerase chain reaction for detection of endocervical C. trachomatis infection in women attending a gynecology outpatient department in India. Acta Cytol. 46:540-544. [DOI] [PubMed] [Google Scholar]
- 18.Stephens, R. S., E. A. Wagar, and G. K. Schoolnik. 1988. High resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 167:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viscidi, R. P., L. Bobo, E. W. Hook III, and T. C. Quinn. 1993. Transmission of C. trachomatis among sex partners assessed by polymerase chain reaction. J. Infect. Dis. 168:488-492. [DOI] [PubMed] [Google Scholar]
- 20.Wagenvoort, J. H. T., R. J. Suchland, and W. E. Stamm. 1988. Serovar distribution of urogenital C. trachomatis strains in the Netherlands. Genitourin. Med. 64:159-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang, C. L., I. Maclean, and R. C. Brunham. 1993. DNA sequence polymorphism of the C. trachomatis omp1 gene. J. Infect. Dis. 168:1225-1230. [DOI] [PubMed] [Google Scholar]