Abstract
Pulsed-field gel electrophoresis (PFGE) was used to investigate the diversity of Streptococcus suis isolates of various serotypes recovered from swine clinical samples in Spain. Capsular types 9 (64.9%) and 2 (14.8%) were the most frequently isolated serotypes followed by serotype 7 (5.9%) and serotype 8 (4.3%). The PFGE results of this study with 60 different pulsotypes indicate a great genetic diversity among the S. suis isolates, which is consistent with the broad distribution of S. suis in the swine population. Forty-five percent of the pulsotypes corresponded to single isolates, no pulsotype was common to all farms, and at least 3 different pulsotypes were isolated in 56% of herds in which more than 3 clinical isolates were analyzed. These results reveal a great diversity both between and within herds throughout the strains of S. suis studied, demonstrating that different strains of S. suis are associated with infection in pigs. Some pulsotypes were more frequently isolated and exhibited a wider distribution over herds than others, and were the unique or predominant strains in several herds, suggesting the existence of a prevalent or a few prevalent clones responsible for a large proportion of clinical cases. Overall, the great genetic heterogeneity of the clinical strains of S. suis, the isolation of different strains within the same herd, and the predominance of particular strains in some herds are evidence that infection by S. suis is a dynamic process and reinforce the idea that the epidemiology of S. suis infection is very complex.
Streptococcus suis is a well-recognized worldwide swine pathogen of emerging clinical significance in most countries with intensive swine industry. S. suis infections in pigs are associated with different clinical conditions such as meningitis, arthritis, endocarditis, and septicemia (13, 15). Moreover, clinically healthy pigs can carry S. suis in their nasal cavities, tonsils, and upper respiratory tracts, contributing to the dissemination and transmission of this pathogen. In addition to its significant sanitary and economic impact in the swine industry, S. suis is also a zoonotic agent, responsible for septicemia and meningitis in humans (24, 26).
The ability to identify specific strains of a bacterial pathogen is an essential tool for epidemiological investigations. Different typing techniques have been used to differentiate S. suis strains, but most of the epidemiological studies have been based on serotyping. Among the 34 different capsular types actually described, not all serotypes have similar clinical relevance or are equally important in the different countries. Serotype 2 is the most frequently isolated worldwide, including many European countries (2, 26, 30). Serotype 9 is most prevalent in Australia, Germany, Belgium, and The Netherlands (30), whereas in the United Kingdom, the most prevalent serotypes are 1 and 14 (1, 3, 12). However, serotyping is less discriminative than molecular typing methods (4). Ribotyping (18, 21, 22, 25) and randomly amplified DNA polymorphism (8) have been used for the typing of S. suis, mainly of serotype 2 isolates due to the special clinical relevance of this serotype. Pulsed-field gel electrophoresis (PFGE) is probably one of the most powerful molecular typing methods and has been successfully applied to different bacterial pathogens (4). Recently, PFGE has been used to study the genetic diversity of German and French S. suis swine isolates (2, 7). Infections by S. suis represent one of the most important sanitary and economic problems in the Spanish intensive swine industry (19). Nevertheless, molecular epidemiological studies of S. suis swine isolates have not been carried out in Spain. Thus, the purpose of the present work was the PFGE typing of S. suis clinical isolates to extend the knowledge about the epidemiology of infections caused by this microorganism and to explore the genetic diversity of the Spanish S. suis swine isolates.
MATERIALS AND METHODS
S. suis strains.
The study included 302 swine S. suis isolates (Table 1). Isolates were recovered in the course of routine diagnostic procedures from tissues of diseased pigs from 74 herds located in Spain between 1999 and 2002. Most pigs were 3 to 15 weeks old. As to the source of isolation, 185 isolates were isolated from the brains of pigs with meningitis, 58 isolates were isolated from the lungs of pigs with pneumonia, 12 isolates were isolated from lymph nodes, 40 isolates were isolated from the hearts of pigs with pericarditis, and 7 isolates were isolated from the joints of pigs with arthritis. Bacteria were isolated from clinical samples on Columbia blood agar plates (bioMérieux España, s.a.) after 48 h of incubation at 37°C.
TABLE 1.
Molecular characterization by PFGE of Spanish S. suis strains
| No. of strains | No. of herds | Serotype(s)a | Restriction pattern by digestion with:
|
Pulsotype | |
|---|---|---|---|---|---|
| ApaI | SmaI | ||||
| 18 | 10 | 2 | 1 | 4 | I |
| 4 | 3 | 2 | 1 | 9 | II |
| 2 | 1 | 1-14 | 2 | 4 | III |
| 1 | 1 | 1-14 | 2 | 16 | IV |
| 2 | 1 | 9 | 2 | 31 | V |
| 1 | 1 | 2 | 3 | 17 | VI |
| 1 | 1 | 1 | 4 | 18 | VII |
| 1 | 1 | 19 | 4 | 36 | VIII |
| 2 | 2 | 2 | 5 | 33 | IX |
| 2 | 2 | 2 | 6 | 35 | X |
| 3 | 3 | 2 | 7 | 3 | XI |
| 1 | 1 | 9 | 8 | 9 | XII |
| 4 | 1 | 9 | 8 | 20 | XIII |
| 1 | 1 | 7 | 9 | 9 | XIV |
| 4 | 4 | 8 | 10 | 13 | XV |
| 1 | 1 | 16 | 10 | 27 | XVI |
| 1 | 1 | 3 | 11 | 22 | XVII |
| 1 | 1 | 4 | 12 | 19 | XVIII |
| 1 | 1 | 8 | 13 | 29 | XIX |
| 1 | 1 | 3 | 14 | 5 | XX |
| 4 | 1 | 9 | 15 | 5 | XXI |
| 6 | 3 | 9 | 15 | 32 | XXII |
| 1 | 1 | 9 | 16 | 5 | XXIII |
| 140 | 46 | 9 | 17 | 5 | XXIV |
| 1 | 1 | 9 | 17 | 23 | XXV |
| 12 | 8 | 9 | 17 | 32 | XXVI |
| 5 | 3 | 9 | 18 | 26 | XXVII |
| 3 | 1 | 8 | 18 | 31 | XXVIII |
| 2 | 1 | 3 | 18 | 34 | XXIX |
| 1 | 1 | 9 | 19 | 24 | XXX |
| 1 | 1 | 9 | 20 | 5 | XXXI |
| 4 | 1 | 9 | 21 | 32 | XXXII |
| 5 | 2 | 3 | 22 | 32 | XXXIII |
| 1 | 1 | 3 | 23 | 26 | XXXIV |
| 3 | 2 | 9 | 24 | 25 | XXXV |
| 3 | 1 | 9 | 25 | 29 | XXXVI |
| 1 | 1 | 9 | 26 | 25 | XXXVII |
| 1 | 1 | 9 | 27 | 29 | XXXVIII |
| 3 | 2 | 9 | 28 | 7 | XXXIX |
| 1 | 1 | 9 | 28 | 9 | XL |
| 2 | 1 | 9 | 28 | 10 | XLI |
| 1 | 1 | 9 | 29 | 28 | XLII |
| 5 | 3 | 1-14 | 30 | 4 | XLIII |
| 3 | 2 | 2 | 31 | 1 | XLIV |
| 7 | 3 | 7 | 32 | 12 | XLV |
| 1 | 1 | ND | 32 | 18 | XLVI |
| 2 | 2 | ND | 33 | 16 | XLVII |
| 1 | 1 | NT | 34 | 6 | XLVIII |
| 4 | 1 | 2 | 35 | 8 | XLIX |
| 3 | 1 | 2 | 36 | 1 | L |
| 4 | 1 | 2 | 36 | 4 | LI |
| 3 | 1 | 7 | 36 | 21 | LII |
| 1 | 1 | 7 | 37 | 2 | LIII |
| 1 | 1 | 7 | 37 | 5 | LIV |
| 4 | 1 | 8 | 37 | 14 | LV |
| 1 | 1 | 8 | 37 | 30 | LVI |
| 5 | 1 | 7 | 38 | 14 | LVII |
| 1 | 1 | 9 | 39 | 11 | LVIII |
| 1 | 1 | 9 | 39 | 15 | LIX |
| 2 | 1 | 9 | 39 | 18 | LX |
NT, not typeable by serotyping; ND, not determined.
Biochemical identification and serotyping.
Identification of S. suis strains was carried out by using the commercial identification system Rapid ID32 Strept (bioMérieux España, s.a.) according to the manufacturer's instructions. Serotyping was performed by an agglutination test based on polysaccharide capsular antigens 1 to 34 as described previously (30).
PFGE.
Streptococcal cells were grown aerobically in brain heart infusion broth (Difco) at 37°C to reach an absorbance (A610) of approximately 0.6. The cells were harvested by centrifugation (3,500 × g, 10 min, 4°C), washed twice with buffer (10 mM Tris-HCl, 10 mM EDTA, 10 mM EGTA, 1 M NaCl [pH 8]), and resuspended in 1 ml of cold Tris-EDTA buffer supplemented with 1 mg of lysozyme ml−1. Agarose plugs were made from a 1:1 mixture of 2% low-melting-point agarose and the cell suspension. Plugs were incubated in buffer (6 mM Tris-HCl, 100 mM EDTA, 1 M NaCl, 0.5% [wt/vol] Brij 58, 0.2% [wt/vol] sodium deoxycholate, 0.5% [vol/vol] lauroyl sarcosine) supplemented with 1 mg of lysozyme ml−1 for 4 h at 37°C. Cells were then treated overnight at 56°C with the same volume of solution (0.25 M EDTA, 20 mM NaCl, and 1% lauroyl sarcosine [pH 9]) containing 500 μg of proteinase K ml−1 and washed three times with Tris-EDTA buffer for 1 h at 4°C. ApaI (Promega Co.) and SmaI (MBI Fermentas) enzymes were used for restriction endonuclease digestion according to the manufacturer's instructions. The fragments were resolved by PFGE within electrophoresis grade agarose (1%; Boehringer Mannheim) by using a CHEF-DR III system (Bio-Rad). The following parameters were used: running time, 21 h; temperature, 14°C; voltage gradient, 200 V; included angle, 120°C; ApaI initial pulse time, 0.1 s; ApaI final pulse time, 25 s; SmaI initial pulse time, 1 s; and SmaI final pulse time, 30 s. The gels were stained with ethidium bromide (0.5 μg/ml) for 15 min, destained in distilled water, and photographed under UV light. A lambda ladder PFGE marker (Boehringer Mannheim) was used for molecular weight and size determinations.
PFGE pattern analysis.
Similarities between restriction endonuclease digestion profiles were expressed by Jaccard similarity index, with the numerical taxonomy program TAXAN (Information Resources Group, Maryland Biotechnology Institute, University of Maryland, College Park). A similarity matrix was computed and transformed into an agglomerative cluster by using the unweighted pair group method with arithmetic averages (UPGMA) (23).
Statistical analysis.
Statistical significance analysis was performed to analyze the relationship between S. suis PFGE patterns, capsular types, and clinical sources. The chi-square test was used with the SPSS, version 11.0, software. Differences were considered significant when probabilities were lower than 0.05.
RESULTS
Biochemical identification and serotyping.
All S. suis isolates were recovered in pure culture and were properly identified with the commercial Rapid ID32 identification system. Except for three S. suis strains in which the serotype was not determined, all but one of the 299 S. suis isolates (99.7%) could be serotyped by using the 34 capsular types. The results of serotyping for S. suis isolates are shown in Table 1. Capsular types 9 (67.4%) and 2 (14.8%) were the most frequently isolated S. suis serotypes, followed by serotype 7 (6.0%) and serotype 8 (4.4%). The rest of the serotypes were isolated in proportions lower than 3%.
The majority of the strains of serotype 9 (67.6%) and serotype 7 (61.1%) were isolated from pigs with meningitis. On the other hand, most strains isolated from pigs with pneumonia belonged to serotype 3 (80.0%). The association between these serotypes and clinical sources was statistically significant (P < 0.05). Most of the serotype 2 strains (40.9%) were isolated from pigs with meningitis, but no significant association was observed (P > 0.05). The same result was observed with the other serotypes.
PFGE analysis.
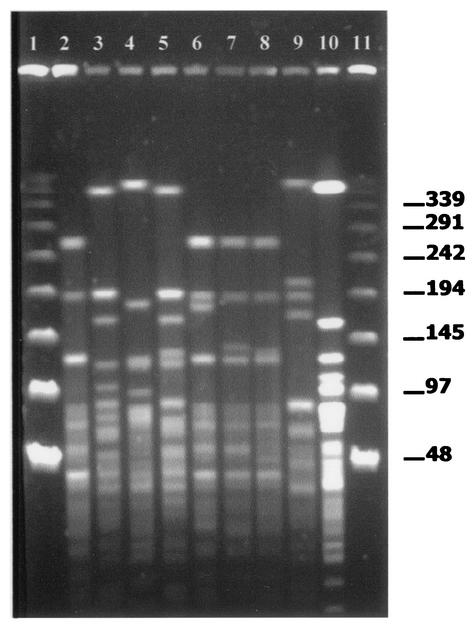
All S. suis isolates displayed ApaI and SmaI restriction endonuclease digestion profiles. The restriction patterns of the same isolate generated for each of the two restriction enzymes were found to be stable and reproducible in at least two different trials (data not shown). The ApaI enzyme generated 12 to 17 major fragments over a size range from ca. 48.5 to 440 kb (Fig. 1), whereas SmaI-digested DNA generated restriction endonuclease digestion profiles with 9 to 15 major fragments over a size range from ca. 28.5 to 582 kb (data not shown). Visual comparison of macrorestriction patterns generated with the SmaI enzyme revealed 36 different DNA fragment profiles, whereas the ApaI enzyme generated 39 different DNA fragment profiles (Table 1). The SmaI and ApaI restriction patterns revealed the great genetic diversity of S. suis. Although the ApaI enzyme was slightly more discriminative than SmaI, some S. suis isolates with identical ApaI profiles displayed different SmaI profiles (Table 1).
FIG. 1.
Representative PFGE fingerprint patterns of S. suis strains after ApaI digestion. Lanes 1 and 11, bacteriophage lambda ladder PFGE marker (Boehringer Mannheim); lane 2, S. suis isolate B194-31-01 (pattern 15); lane 3, S. suis isolate L215-71-01 (pattern 38); lane 4, S. suis isolate L83-1-99 (pattern 1); lane 5, S. suis isolate L160-45-00 (pattern 36); lane 6, S. suis isolate B125-13-99 (pattern 21); lane 7, S. suis isolate L282-4-01 (pattern 22); lane 8, S. suis isolate B78-72-99 (pattern 17); lane 9, S. suis isolate L47-66-99 (pattern 10); lane 10, S. suis isolate B19-19-99 (pattern 30).
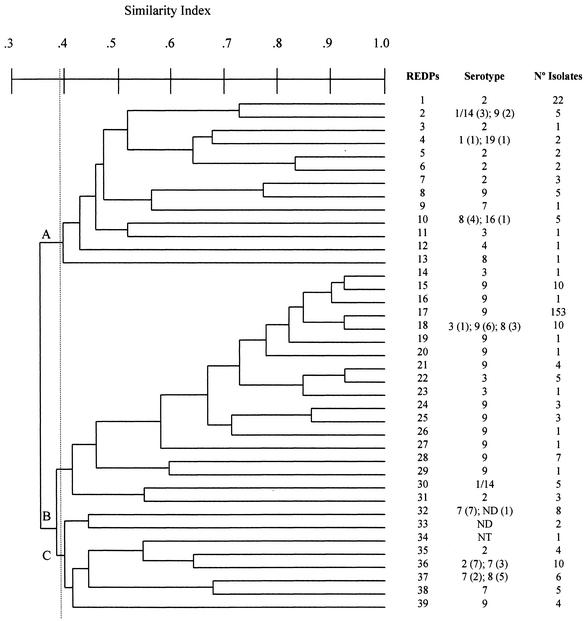
The genetic relationship between the S. suis isolates on the basis of the ApaI-digested DNA restriction patterns is presented in the dendrogram shown in Fig. 2. Cluster analysis by UPGMA revealed three major groups, A, B, and C, with a similarity of at least 38% (Fig. 2), with the groups containing 51 (16.9%), 210 (69.5%), and 41 (13.2%) isolates, respectively. There was a statistically significant association (P < 0.05) between serotype 2 and PFGE group A (68.2% of serotype 2 strains were included in this cluster), serotype 9 and PFGE group B (95.5% of serotype 9 strains were included in group B), and serotype 7 and PFGE group C (94.4% of serotype 7 strains were included in group C). Clustering was not observed for the other serotypes.
FIG. 2.
Dendrogram showing the genetic relationship between 302 S. suis isolates based on UPGMA cluster analysis of the 39 PFGE patterns obtained after macrorestriction with the ApaI enzyme. REDP, restriction endonuclease digestion profile.
After composite profiling, a total of 60 pulsotypes (I to LX) were obtained from the 302 S. suis isolates analyzed (Table 1). A high degree of genetic diversity was observed within the most frequently recovered S. suis serotypes. Twenty-four different pulsotypes were observed for serotype 9, 10 pulsotypes were observed for serotype 2, 6 pulsotypes were observed for serotype 7, and 5 pulsotypes were observed for serotype 8. A marked heterogeneity was observed between and within herds by PFGE in S. suis isolates. Forty-five percent of pulsotypes corresponded to single isolates, and 43 pulsotypes (71.7%) were unique to individual herds. No pulsotype was found to be common to all herds, but some pulsotypes exhibited a higher frequency of isolation. Thus, pulsotype XXIV was the most frequent, with 140 isolates (46.3%), followed by pulsotype I (18 isolates; 5.9%) and pulsotype XXVI (12 isolates; 3.9%). Pulsotypes XXIV and I were widely distributed among herds, being isolated from 46 (62.1%) and 10 (13.5%) herds, respectively. These pulsotypes were repeatedly isolated from different animals of the same herd for several months or years. Nevertheless, at least 3 different pulsotypes were identified in 19 of the 34 herds (55.9%) in which more than 3 isolates were isolated.
DISCUSSION
Most of the molecular characterization studies of S. suis have been focused on the comparison between isolates from diseased or healthy pigs or between human and swine isolates (2, 7, 8). In the present study, PFGE was used for the genetic characterization of a total of 302 S. suis clinical isolates of various serotypes isolated from 74 herds. Special attention was paid to the variation and diversity of clinical S. suis isolates between and within herds. PFGE was chosen due to its higher discrimination power (4). The PFGE results of this study with 60 different pulsotypes indicate a great genetic heterogeneity among the S. suis isolates studied, which correlates well with the wide distribution of S. suis in the swine population. Similar genetic diversity has been observed in other widespread microorganisms (29).
S. suis can be differentiated on the basis of distinct polysaccharide capsular antigens. In this study, the results of serotyping were in agreement with those of other studies (1, 2, 5, 15, 20, 26, 30) in that capsular types 2 and 9 are the most prevalent in many European countries, including Spain. However, the frequency of serotype 2 isolation was about 50% lower than usual in Spain while serotype 9 was isolated in a much higher proportion (15, 20, 26, 30). These results may indicate an emergence in the distribution of S. suis serotype 9 causing infections in Spanish pigs. A great diversity was observed by PFGE within the most frequently isolated capsular types in this study (9, 2, and 7) with 24, 10, and 6 different pulsotypes, respectively. Thus, PFGE was proven to be a more discriminatory and accurate technique than capsular serotyping for differentiating S. suis isolates from the swine population and therefore more suitable for epidemiological studies (2, 7). As expected, the discriminatory power of this technique improved when both enzymes were combined (29). The great diversity observed in this study agrees with previous reports (2, 7, 18, 22, 24, 25) and could explain the differences in virulence observed not only between serotypes but also between S. suis strains of the same serotype (24).
The facts that nearly 50% of the pulsotypes corresponded to single isolates, no strain was common to all farms, and the isolation of at least 3 different pulsotypes in most herds in which more than 3 isolates were analyzed indicate a great genetic heterogeneity of the S. suis isolates between and within herds and demonstrate that many different strains of S. suis can be responsible for infections in pigs. Pathogen diversity represents a challenge for the development of broadly protective vaccines (10). The diversity of S. suis strains observed by PFGE and the absence of heterologous protection (14) would explain the limited efficacy provided by commercially available vaccines used for the control of S. suis infections (13, 16, 24). In these circumstances, the use of autovaccines based on the results of molecular epidemiological studies could be a more rational approach than generic vaccines for an adequate control of S. suis infections.
Despite the significant number of pulsotypes found and the diversity of S. suis strains between herds, some strains of S. suis were more frequently isolated than others (Table 1). This fact is particularly evident with pulsotype XXIV, which represented 46.3% of the swine isolates, and it may be related to a higher pathogenic potential of this strain or to a wider environmental distribution. The isolation of this strain from 62.1% of the herds would be consistent with the idea of a wider distribution in the swine population studied, which is likely related to the existence of carrier pigs (24) involved in the spread of infection by vertical (3) and/or horizontal transmission (6, 11). In this sense, the demonstration of identical strains of S. suis in different pigs within the same herd suggests a direct transmission from pig to pig. Pulsotype XXIV was the unique or predominant strain in several herds, being repeatedly isolated from different animals during several months or years. These results are consistent with the idea that in a closed population there is a tendency for only one strain to be established and be responsible for all or the majority of clinical cases (17, 27) and confirm the ability of certain strains of S. suis to persist during prolonged periods of time in the same herd (27). All these results indicate that, together with the diversity of S. suis strains found in many herds, the existence of a prevalent or of a few prevalent clones responsible for a great proportion of clinical cases is also possible, as has been observed with other pathogens (9, 28, 29).
In summary, the great genetic heterogeneity of the clinical strains of S. suis, the isolation of different strains within the same herd, and the persistence of some strains during prolonged periods of time in the same herd are evidence that infection by S. suis is a dynamic process in which animals are usually exposed to multiple S. suis strains and corroborate the idea of the complexity of the epidemiology of S. suis infections.
Acknowledgments
We thank A. Casamayor for technical assistance.
REFERENCES
- 1.Aarestrup, F. M., S. E. Jorsal, and N. E. Jensen. 1998. Serological characterization and antimicrobial susceptibility of Streptococcus suis isolates from diagnostic samples in Denmark during 1995 and 1996. Vet. Microbiol. 60:59-66. [DOI] [PubMed] [Google Scholar]
- 2.Allgaier, A., R. Goethe, H. J. Wisselink, H. E. Smith, and P. Valentin-Weigand. 2001. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J. Clin. Microbiol. 39:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amass, S. F., P. SanMiguel, and L. K. Clark. 1997. Demonstration of vertical transmission of Streptococcus suis in swine by genomic fingerprinting. J. Clin. Microbiol. 35:1595-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbeit, R. D. 1995. Laboratory procedures for the epidemiological analysis of microorganisms, p. 190-208. In P. R. Murray, E. J. O. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 5.Berthelot-Hérault, F., H. Morvan, A. M. Kéribin, M. Gottschalk, and M. Kobisch. 2000. Production of muraminidase-released protein (MRP), extracellular factor (EF) and suilysin by field isolates of Streptococcus suis capsular types 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet. Res. 31:473-479. [DOI] [PubMed] [Google Scholar]
- 6.Berthelot-Hérault, F., M. Gottschalk, A. Labbé, R. Cariolet, and M. Kobisch. 2001. Experimental airborne transmission of Streptococcus suis capsular type 2 in pigs. Vet. Microbiol. 82:69-80. [DOI] [PubMed] [Google Scholar]
- 7.Berthelot-Hérault, F., C. Marois, M. Gottschalk, and M. Kobisch. 2002. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatellier, S., M. Gottschalk, R. Higgins, R. Brousseau, and J. Harel. 1999. Relatedness of Streptococcus suis serotype 2 isolates from different geographic origins as evaluated by molecular fingerprinting and phenotyping. J. Clin. Microbiol. 37:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, C. J. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, P., S. Self, M. Rao, A. Naficy, and J. Clemens. 2001. Sieve analysis: methods for assessing from vaccine trial data how vaccine efficacy varies with genotypic and phenotypic pathogen variation. J. Clin. Epidemiol. 54:68-85. [DOI] [PubMed] [Google Scholar]
- 11.Gottschalk, M., and M. Segura. 2000. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76:259-272. [DOI] [PubMed] [Google Scholar]
- 12.Heath, P. J., B. W. Hunt, J. P. Duff, and J. D. Wilkinson. 1996. Streptococcus suis serotype 14 as a cause of pig disease in the UK. Vet. Rec. 139:450-451. [PubMed] [Google Scholar]
- 13.Higgins, R., and M. Gottschalk. 1999. Streptococcal disease, p. 563-570. In B. E. Straw, S. D'Állaire, W. L. Mengeling and D. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames.
- 14.Kebede, M., M. M. Chengappa, and J. G. Stuart. 1990. Isolation and characterization of temperature-sensitive mutants of Streptococcus suis: efficacy trial of the mutant vaccine in mice. Vet. Microbiol. 22:249-257. [DOI] [PubMed] [Google Scholar]
- 15.Luque, I., C. Tarradas, A. Arenas, A. Maldonado, R. Astorga, and A. Perea. 1998. Streptococcus suis serotypes associated with different disease conditions in pigs. Vet. Rec. 142:726-727. [DOI] [PubMed] [Google Scholar]
- 16.MacInnes, J. I., and R. Desrosiers. 1999. Agents of the “suis-ide diseases” of swine: Actinobacillus suis, Haemophilus parasuis, and Streptococcus suis. Can. J. Vet. Res. 63:83-89. [PMC free article] [PubMed] [Google Scholar]
- 17.Mogollon, J. D., C. Pijoan, M. P. Murtaugh, J. E. Collins, and P. P. Cleary. 1991. Identification of epidemic strains of Streptococcus suis by genomic fingerprinting. J. Clin. Microbiol. 29:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okwumabua, O., J. Staats, and M. M. Chengappa. 1995. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping). J. Clin. Microbiol. 33:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perea, A. 2000. Estreptococias porcinas: importancia y estrategias de control. Anaporc 197:5-24. [Google Scholar]
- 20.Prieto, C., J. Peña, P. Suárez, M. Imaz, and J. M. Castro. 1993. Isolation and distribution of Streptococcus suis capsular types from diseased pigs in Spain. J. Vet. Med. B 40:544-548. [DOI] [PubMed] [Google Scholar]
- 21.Rasmussen, S. R., F. M. Aarestrup, N. E. Jensen, and S. E. Jorsal. 1999. Association of Streptococcus suis serotype 2 ribotype profiles with clinical disease and antimicrobial resistance. J. Clin. Microbiol. 37:404-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, H. E., M. Rijnsburger, N. Stockhofe-Zurwieden, H. J. Wisselink, and M. A. Smits. 1997. Virulent strains of Streptococcus suis serotype 2 and highly virulent strains of Streptococcus suis serotype 1 can be recognized by a unique ribotype profile. J. Clin. Microbiol. 35:1049-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sneath, P. H., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 24.Staats, J. J., I. Feder, O. Okwumabua, and M. M. Chengappa. 1997. Streptococcus suis: past and present. Vet. Res. Commun. 21:381-407. [DOI] [PubMed] [Google Scholar]
- 25.Staats, J. J., B. L. Plattner, J. Nietfeld, S. Dritz, and M. M. Chengappa. 1998. Use of ribotyping and hemolysin activity to identify highly virulent Streptococcus suis type 2 isolates. J. Clin. Microbiol. 36:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarradas, C., I. Luque, D. De Andrés, Y. E. Abdel-Aziz Shahein, P. Pons, F. González, C. Borge, and A. Perea. 2001. Epidemiological relationship of human and swine Streptococcus suis isolates. J. Vet. Med. B 48:347-355. [DOI] [PubMed] [Google Scholar]
- 27.Torremorell, M., and C. Pijoan. 1998. Prolonged persistence of an epidemic Streptococcus suis strain in a closed pig population. Vet. Rec. 143:394-395. [DOI] [PubMed] [Google Scholar]
- 28.Vela, A. I., J. Vázquez, A. Gibello, M. M. Blanco, M. A. Moreno, P. Liébana, C. Albendea, B. Alcalá, A. Méndez, L. Domínguez, and J. F. Fernández-Garayzábal. 2000. Phenotypic and genetic characterization of Lactococcus garvieae isolated from lactococcosis outbreaks and comparison with isolates of other countries and sources. J. Clin. Microbiol. 38:3791-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vela, A. I., J. F. Fernández-Garayzábal, J. A. Vázquez, M. V. Latre, M. M. Blanco, M. A. Moreno, L. De La Fuente, J. Marco, C. Franco, A. Cepeda, A. A. Rodríguez, G. Suárez, and L. Domínguez. 2001. Molecular typing by pulsed-field gel electrophoresis of Spanish animal and human Listeria monocytogenes isolates. Appl. Environ. Microbiol. 67:5840-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisselink, H. J., H. E. Smith, N. Stockhofe-Zurwieden, K. Peperkamp, and U. Vecht. 2000. Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet. Microbiol. 74:237-248. [DOI] [PubMed] [Google Scholar]