Abstract
A 2-month interruption of only nonnucleoside reverse transcriptase inhibitors (NNRTIs) for patients carrying mutations associated with resistance to NNRTIs was followed by no change in either viral load or CD4 cell counts. These data suggest that these compounds have lost all of their in vivo antiviral activity in such cases.
In addition to the nucleoside reverse transcriptase inhibitors (NRTIs) and protease inhibitors (PIs), nonnucleoside reverse transcriptase inhibitors (NNRTIs), especially nevirapine and efavirenz, have gained a definitive place in the treatment of human immunodeficiency virus type 1 (HIV-1) infection. Studies have shown that a triple therapy including one NNRTI and two NRTIs had a similar effect to therapy involving one PI and two NRTIs. However, rapid emergence of resistance mutations both in vitro (12, 13) and in vivo (4, 8, 9, 14, 15) is a well-known characteristic of this drug class.
Prospective controlled studies have demonstrated that genotype resistance testing is useful for adapting and choosing new compounds in case of failure of previous drugs (10, 11). Several algorithms have been constructed to provide rules to interpret the presence or the absence of mutations associated with resistance to NRTIs, NNRTIs, and PIs.
Usually, when some NNRTI resistance mutations are present, the interpretation of most of the algorithms is to avoid the use of the drug of this class, ruling out any residual effect of the NNRTIs in such cases. The high levels of in vitro resistance observed for the most common NNRTI resistance mutations and their frequent cross-resistance support this attitude (3, 14).
The aim of this study was to measure in vivo the magnitude of the residual effect of NNRTIs in the plasma of patients harboring viruses with mutations associated with NNRTI resistance.
There are different ways to measure the effect of one compound in the context of an antiretroviral combination. One way is to add the drug to the current treatment and observe the magnitude of the decrease in viral load (M. D. Miller, N. A. Margot, and B. Lu, Abstr. 9th Conf. Retrovir. Opportunistic Infect., abstr 43, 2002). Another way is to stop administration of a drug for a relatively short period and then measure whether there is an increase in viral load. Thus, we have measured the residual effect of NNRTIs in patients who harbored viruses with NNRTI resistance mutations and who had stopped taking only the compounds of this class.
This study was conducted with 11 patients on an NNRTI regimen involving efavirenz (n = 5) or nevirapine (n = 6) plus two to four NRTIs and who demonstrated a stable HIV-1 RNA level (variation, <0.5 log10) on the same antiretroviral combination for at least 6 months before the interruption of treatment. The subjects were selected consecutively by our therapeutic community, including virologists and clinicians who had decided to stop NNRTI treatment if mutations associated with resistance to NNRTIs were present at day −30. In fact, in all patients, viruses in plasma harbored NNRTI and NRTI resistance mutations (Table 1). These results agree with previous studies that have shown the quasisystematic presence of NNRTI resistance mutations associated with failure of NNRTI regimens (4, 5, 7).
TABLE 1.
Treatments, reverse transcriptase inhibitor resistance-associated mutations before stopping NNRTI treatment, viral load, and CD4 cell count changes
| Patient | Treatmenta | Resistance-associated mutation(s)
|
Result forb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | Mean day 30/day 0
|
Day 15
|
Day 30
|
Day 60
|
||||||
| Viral load | CD4 count/mm3 (%) | Viral load | CD4 count/mm3 (%) | Viral load | CD4 count/mm3 (%) | Viral load | CD4 count/mm3 (%) | ||||
| 1 | d4T, 3TC, NVP | M41L, M184V, T215Y | Y188L | 2.93 | 343 (25) | 3.38 | 287 (25) | 3.34 | 285 (25) | 3.25 | 347 (23) |
| 2 | ZDV, 3TC, ABC, ddI EFV | M41L, D67N, M184V, T215Y, K219N | K103N, Y181C, G190S | 2.75 | 335 (13) | 2.30 | 264 (15) | 2.30 | 324 (17) | 3.03 | 245 (16) |
| 3 | d4T, ddI, ABC, NVP | A62V, D67N, K70R, V75I, F116Y, Q151M, K219Q | Y181C, G190A | 3.98 | 295 (22) | ND | ND | 4.33 | 277 (16) | 4.13 | 223 (18) |
| 4 | d4T, 3TC, EFV | M41L, D67N, K70R, M184V, T215Y, K219Q | L100I, K103N | 3.29 | 217 (15) | 3.28 | 280 (16) | 3.15 | 249 (12) | 3.03 | 285 (14) |
| 5 | d4T, 3TC, NVP | M41L, M184V, T215Y | K103N, V108I | 3.59 | 294 (17) | 3.78 | 280 (16) | 3.99 | 313 (19) | 4.45 | 283 (16) |
| 6 | ABC, ddI, 3TC, EFV | M41L, L74V, M184V, L210W, T215Y | A98G, Y188L | 3.34 | 242 (11) | 3.35 | 198 (12) | 3.14 | 222 (11) | 3.16 | 236 (10) |
| 7 | ABC, 3TC, NVP | M41L, D67N, M184V, L210W, T215Y | A98G, K103N | 3.55 | 378 (31) | 3.46 | 425 (32) | 3.47 | 428 (30) | 3.54 | 482 (31) |
| 8 | d4T, ddI, EFV | D67N, K70R, T215F, K219Q | K103N, G190A | 3.13 | 380 (22) | 3.42 | 256 (22) | 2.92 | 266 (21) | 3.35 | 381 (21) |
| 9 | d4T, 3TC, NVP | A62V, M184V | K103N | 3.01 | 609 (31) | 2.30 | 667 (24) | 2.30 | 624 (27) | 2.68 | 797 (28) |
| 10 | ZDV, 3TC, EFV | M41L, M184V, T215F | L100I, K103N | 3.41 | 304 (21) | 3.73 | 388 (23) | 3.74 | 290 (20) | 3.87 | 330 (19) |
| 11 | ddI, 3TC, NVP | M41L, L74V, M184V, L210W, T215Y | Y181C | 2.99 | 490 (20) | 3.32 | 485 (20) | 2.30 | 658 (20) | 3.12 | 438 (20) |
d4T, stavudine; 3TC, lamivudine; NVP, nevirapine; ZDV, zidovudine; ABC, abacavir; ddI, didanosine; EFV, efavirenz.
Viral load is measured as log10 copies of RNA per milliliter. ND, not done.
Viral load (Roche Amplicor HIV-1 Monitor assay 1.5), CD4 cell counts (flow cytometric analysis [Coulter]), and genotype (automated population-based full-sequence analysis [ABI System]) were determined at day 0 (the time of NNRTI interruption) and then at days 15, 30, and 60. Trough NNRTI concentrations in plasma were measured at days 0 and 15 as previously described (2).
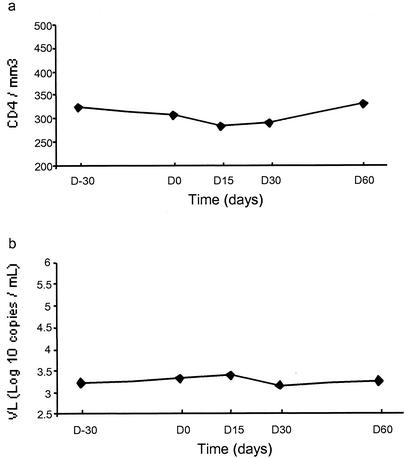
For 10 of the 11 patients, no genotypic change was detected between days −30 and 60. Only one patient (no. 5) lost the NNRTI and PI resistance mutations between days 30 and 60. For all patients taken together, neither viral load nor CD4 cell counts showed a significant change between day 0 and day 60 according to nonparametric Wilcoxon tests (Fig. 1). The trough NNRTI concentrations in plasma were in the range of therapeutic values at day 0 and were undetectable at day 15. No clinical event and no biological abnormality were observed during the follow-up.
FIG. 1.
Medians of the CD4 count (a) and HIV-1 viral load (b) during the follow-up study (2 months).
This study has demonstrated that, in patients with viruses harboring mutations causing resistance to NNRTIs, these compounds have lost all their in vivo antiviral effect. Thus, the interruption of the NNRTIs in these patients was not followed by any change of CD4 count and viral load during 2 months. This is in accordance with the large increases in 50% inhibitory concentration (IC50) and IC90 measured by phenotypic assays for viruses with NNRTI resistance mutations.
However, despite the loss of their antiviral potency, we cannot exclude the possibility that these compounds could have a positive effect on maintaining NNRTI mutations in the genetic background of the viruses. It is widely appreciated that many PI and some NRTI mutations impair HIV-1 replication to various degrees, whereas the effects of NNRTI mutations are largely uncharacterized. It is often assumed that NNRTI resistance is not associated with impaired viral replication, but in vitro studies have shown that not all NNRTI resistance mutations preserve HIV-1 replication capacity (1; W. Huang, T. Wrin, A. Gamarnick, J. Beauchaine, J. M. Whitcomb, and C. J. Petropoulos, Abstr. 11th Int. HIV Drug Resistance Workshop, abstr. 72, 2002). According to these authors, mutations such as V106A, G190C/S/E/Q, P225H, M230L, and P236L can reduce replication capacity to various degrees. However, these mutations are not the most frequently selected or are usually associated with primary substitution in clinical context (4-7). In fact, depending on the NNRTI compound used, primary mutations such as K103N, Y181C/I, Y188C, and G190A are the most common substitutions selected in NNRTI regimen failure (6). The fact that these mutations have little or no effect on HIV-1 replication capacity in vivo, according to Huang et al., can explain their high frequency.
In our study, as expected, most of the patients at baseline harbored viruses with NNRTI resistance mutations that are not associated with impaired virus replication (Table 1). Although the follow-up was relatively short (60 days), this is in accordance with the observation that all but one patient did not harbor any reversion of NNRTI mutations during the study. Interestingly, the one patient (no. 5) who has presented a reversion to the wild type from NNRTI mutations is also the one whose viral load increased significantly, with a change above 0.5 log10.
In conclusion, this study has demonstrated that antiviral activity is totally lost when viruses harboring NNRTI resistance mutations are selected. However, the discussion remains open about the reduction in replication capacity associated with some specific NNRTI resistance mutations that are less frequently selected in clinical practice. Studies are necessary to evaluate the real impact of these particular mutations on HIV-1 replication capacity in vivo.
Acknowledgments
This work was supported by Sidaction and ANRS.
REFERENCES
- 1.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aymard, G., M. Legrand, N. Trichereau, and B. Diquet. 2000. Determination of twelve antiretroviral agents in human plasma sample using reversed-phase high-performance liquid chromatography. J. Chromatogr. B Biomed. Appl. 744:227-240. [DOI] [PubMed] [Google Scholar]
- 3.Bacheler, L., S. Jeffrey, G. Hanna, R. D'Aquila, L. Wallace, K. Logue, B. Cordova, K. Hertogs, B. Larder, R. Buckery, D. Baker, K. Gallagher, H. Scarnati, R. Tritch, and C. Rizzo. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casado, J. L., K. Hertogs, L. Ruiz, F. Dronda, A. Van Cauwenberge, A. Arno, I. Garcia-Arata, S. Bloor, A. Bonjoch, J. Blazquez, B. Clotet, and B. Larder. 2000. Non-nucleoside reverse transcriptase inhibitor resistance among patients failing a nevirapine plus protease inhibitor-containing regimen. AIDS 14:F1-F7. [DOI] [PubMed] [Google Scholar]
- 6.Deeks, S. G. 2001. International perspectives on antiretroviral resistance. Nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S25-S33. [DOI] [PubMed] [Google Scholar]
- 7.Delaugerre, C., R. Rohban, A. Simon, M. Mouroux, C. Tricot, R. Agher, J. M. Huraux, C. Katlama, and V. Calvez. 2001. Resistance profile and cross-resistance of HIV-1 among patients failing a non-nucleoside reverse transcriptase inhibitor-containing regimen. J. Med. Virol. 65:445-448. [PubMed] [Google Scholar]
- 8.Havlir, D., S. H. Cheeseman, M. McLaughlin, R. Murphy, A. Erice, S. A. Spector, T. C. Greenough, J. L. Sullivan, D. Hall, M. Myers et al. 1995. High-dose nevirapine: safety, pharmacokinetics, and antiviral effect in patients with human immunodeficiency virus infection. J. Infect. Dis. 171:537-545. [DOI] [PubMed] [Google Scholar]
- 9.Havlir, D., M. M. McLaughlin, and D. D. Richman. 1995. A pilot study to evaluate the development of resistance to nevirapine in asymptomatic human immunodeficiency virus-infected patients with CD4 cell counts of >500/mm3: AIDS Clinical Trials Group Protocol 208. J. Infect. Dis. 172:1379-1383. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch, M. S., and D. D. Richman. 2000. The role of genotypic resistance testing in selecting therapy for HIV. JAMA 284:1649-1650. [PubMed] [Google Scholar]
- 12.Mellors, J. W., G. E. Dutschman, G. J. Im, E. Tramontano, S. R. Winkler, and Y. C. Cheng. 1992. In vitro selection and molecular characterization of human immunodeficiency virus-1 resistant to non-nucleoside inhibitors of reverse transcriptase. Mol. Pharmacol. 41:446-451. [PubMed] [Google Scholar]
- 13.Richman, D., C. K. Shih, I. Lowy, J. Rose, P. Prodanovich, S. Goff, and J. Griffin. 1991. Human immunodeficiency virus type 1 mutants resistant to nonnucleoside inhibitors of reverse transcriptase arise in tissue culture. Proc. Natl. Acad. Sci. USA 88:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, C.-K. Shih, M. Myers, and J. Griffin. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staszewski, S., J. Morales-Ramirez, K. T. Tashima, A. Rachlis, D. Skiest, J. Stanford, R. Stryker, P. Johnson, D. F. Labriola, D. Farina, D. J. Manion, and N. M. Ruiz. 1999. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. N. Engl. J. Med. 341:1865-1873. [DOI] [PubMed] [Google Scholar]