Abstract
The level of in vitro detection of viral genomes in mixes with two different hepatitis C virus (HCV) subtypes was investigated by artificially mixing previously measured subtype-specific HCV RNA genomes. The RNAs in these mixtures were reverse transcribed and then PCR amplified by using two sets of primers corresponding to the 5′ untranslated region and digested with endonucleases to analyze the restriction fragment length polymorphism patterns. This approach facilitated detection of a wider range of type-specific HCV genomes than originally described, beyond equimolar concentrations of contributing HCV subtypes. Moreover, by using computerized image analysis, this study also demonstrated that the true contribution of each virus type—and consequently of mixed infections—may be underestimated when only visual observation is carried out. These results may be useful for comparing data obtained from this and other currently used methodologies.
At present, there are at least six different clades (1 to 6) of hepatitis C virus (HCV), which include 11 types and numerous subtypes (16, 17). This hierarchical organization seems to have crucial significance for the epidemiology, diagnosis, clinical course, and pathogenesis of HCV-related disease (21).
Molecular genotyping methods are widely used for research purposes. A frequently used noncommercial strategy, restriction fragment length polymorphism (RFLP), employs endonuclease digestion of amplicons from the 5′ untranslated region (UTR) after conventional reverse transcription-nested PCR (RT-nested PCR) (2, 3, 10, 14).
This study was designed to evaluate the ability of RFLP to detect HCV dual infections when two of the three HCV types most prevalent around the world (1, 2, and 3) are combined. Taking into account the known limitation on HCV (sub)type assignment based on 5′ UTR analysis, in the present work we included HCV isolates from different viral (sub)types determined by analysis of genomic coding regions. Although previous reports have compared RFLP to other methodologies as appropriate techniques for HCV typing (3), the aim of this study was to evaluate whether the coexistence of two different HCV (sub)types influenced the ability to detect one or both (sub)types based upon molar concentrations and, if this were the case, whether one of the (sub)types was preferentially detected.
Serum samples.
Sera were obtained from 14 patients serologically proven to be chronically infected with HCV, each exhibiting infection with a single virus type, 1, 2, 3, or 4, as documented by cDNA sequencing of viral genomic coding regions (NS5B or core) followed by phylogenetic analysis. To minimize the risk of potential mixed infections in the samples, the selection criteria for these 14 patients were their lack of parenteral risk of infection and the absence of any known exposure to a potential infectious source as indicated by their answers to an exhaustive questionnaire. Therefore, such strains were ascribed to sporadic (community-acquired) cases of HCV infections. The samples from these patients were selected and grouped according to their viral loads, which were very similar irrespective of the titration method used. A summary of the experimental procedures performed for sample characterization is given in Table 1.
TABLE 1.
Serum sample characterization and primers used in RNA mixing assays
| Serum sample | Viral load detected by:
|
Virus type determined by:
|
Primers used for RT-nested PCR (Set[s]) | ||||
|---|---|---|---|---|---|---|---|
| Limiting dilution method (105 PCR U/ml) | Roche Monitor version 2.0 (log10 no. of genomic copies/ml) | Sequences analysis
|
5′ UTR RFLP | INNO-LiPA | |||
| NS5B | Core | ||||||
| 1 | 8 | 5.91 | 1a | 1 | 1a | 1 and 2 | |
| 2 | 8 | 5.84 | 1b | 1 | 1b | 1 and 2 | |
| 3 | 0.8 | 4.76 | 2c | 2 | 2a or 2c | 1 and 2 | |
| 4 | 8 | 6.19 | 2c | 2 | 2a or 2c | 1 and 2 | |
| 5 | 0.8 | 5.08 | 3a | 3 | 3a | 1 and 2 | |
| 6 | 8 | 5.86 | 1b | 1 | 1a | 1 and 2 | |
| 7 | 8 | 5.87 | 1b | 1 | 1b | 1 and 2 | |
| 8 | 8 | 5.93 | 2c | 2 | 2a or 2c | 1 and 2 | |
| 9 | 8 | 5.93 | 2c | 2 | 2b or 2c | 1 and 2 | |
| 10 | 0.04 | 3.88 | 3a | 3 | 3a | 1 and 2 | |
| 11 | 8 | 5.80 | 1b | 1 | 1b | 2 | |
| 12 | 8 | 5.59 | 2c | 2 | 2a or 2c | 2 | |
| 13 | 0.8 | 5.07 | 3a | 3 | 3a | 2 | |
| 14 | 0.8 | 5.15 | 4a | 3 | 3a | 2 | |
RT-nested PCR for detection of HCV RNAs in serum samples.
Details of blood processing as well as the RNA extraction procedure have been described elsewhere (15). Two sets of primers were used throughout these studies: set 1 (13, 15) and set 2 (2), both of them corresponding to the 5′ UTR. Outer antisense primers from both sets overlap at their 3′ ends, except the last nucleotide from set 1.
Moloney murine leukemia virus (MMLV) was used for reverse transcription with the respective outer antisense primer (from both set 1 and set 2 primers) in an adaptation of a protocol previously described (1). HCV RNAs were previously heated at 95°C for 5 min, immediately kept on ice for 5 min, and added to the reaction mix. Reverse transcriptase was inactivated by heating the mixture at 95°C for 1 h.
First-round and nested PCRs were performed with Taq polymerase (Promega, Madison, Wis.). To validate the results, one negative control (sterilized bidistilled water plus tRNA) was included for the extraction and subsequent steps. Another negative control was added from the reverse transcription. Positive controls for each subtype assayed were also used from the extraction step. Precautions to avoid PCR contamination were strictly followed, as previously reported (15).
Viremia levels in samples used for in vitro RNA mixing assays.
The viral load of each sample was determined by two different methodologies: (i) an in-house limiting dilution method and (ii) a commercially available PCR-based assay (HCV Roche Monitor, version 2.0).
For in-house titration, viral RNAs were sequentially diluted on a fivefold scale. Each sample was analyzed in duplicate by 5′ UTR RT-nested PCR. Viral titer for each serum sample was calculated as the reciprocal of the maximum RNA dilution ratio at which the sample showed a positive result when analyzed by 5′ UTR RT-nested PCR. This procedure has been reported to be more accurate than other assays for quantitative purposes (4, 11).
The HCV Roche Monitor assay was used by strictly following the manufacturer's instructions. Numbers of genomic copies were determined twice by using two aliquots of each analyzed serum sample.
HCV genotyping.
Three different approaches were performed for HCV typing in all samples, including direct sequencing of PCR products from genomic coding regions (core or NS5B) (Table 1).
First, 15 μl of 5′ UTR RT-nested PCR amplicons from all studied samples (obtained with both set 1 and set 2 primers) was cleaved with 10 U of the following enzymes used for combined digestions: HaeIII/RsaI, ScrFI/HinfI, and BstNI/HinfI. We followed the method described by Davidson et al. (2), modifying it slightly when using set 1 primers (13). Thereafter, according to the genotype, a fresh aliquot of each amplicon was digested with either BstUI (for type 1 subtyping) or ScrFI (for subtyping of types 2 and 3).
After incubation at 37°C (or at 60°C for both BstNI and BstUI) for 4 h, each aliquot was electrophoresed in a 3% agarose gel stained with ethidium bromide.
Secondly, subsequently studied samples were also typed by using a reverse hybridization line probe assay (INNO-LiPa HCV II; Innogenetics, Ghent, Belgium) which is based on the 5′ UTR of the HCV genome (19). (This kit is now distributed, sold, and marketed by Bayer Diagnostics NAD under the VERSANT trade name.) This method was used because it has been reported to be able to detect minor HCV genotypes (if present) in 1 to 2% of the viral population (14).
Final HCV subtype assignment was based on the results of respective phylogenetic analyses of partial sequences derived from either the NS5B region (15) or the 5′ UTR-core region. Both HCV type 1 and type 2 isolates were properly characterized by using NS5B amplicons. In contrast, since no detectable NS5B amplicons were observed with either type 3 or type 4 isolates, fragments encompassing the 5′ UTR-core region (510-bp amplicons) were chosen for RT-nested PCR amplification for subsequent phylogenetic analysis of these samples. These products were obtained by using both outer and inner sense oligonucleotides from set 2 primers (5′ UTR) (2) and antisense primers 186 and 339 (core region) (13).
Mixing studies.
All RNA samples were diluted to obtain uniform titers. Therefore, initial viral input RNA derived from each isolate was calibrated by adding the same amount of PCR units (and similar numbers of copies per milliliter) of each selected HCV RNA type. After the dissimilar proportions of type-specific RNAs were mixed, the 5′ UTR RT-nested PCR amplicons obtained were digested by using the appropriate set of endonucleases. Both type 1 and type 2 cDNA products were differentiated from each other by using BstNI/HinfI. For those mixtures involving either types 1 and 3 or types 2 and 3, ScrFI was used throughout.
Visual analysis of each gel was blindly performed by three researchers working independently. Results were confirmed and further extended when National Institutes of Health Image (version 1.62) software was applied.
Computerized RNA secondary structure analysis.
Partial HCV sequences comprising the first 400 nucleotides from the 5′ end and belonging to different subtypes (GenBank accession numbers m62321 [subtype 1a], d90208 [1b], d00944 [2a], d10988 [2b], d50409 [2c], d17763 [3a], and y11604 [4a]) were selected from this database. The secondary structures of these sequences were analyzed at 37°C by using both RNAdraw (9) and mFold (22) programs and at 70°C by applying the latter program.
Statistical analysis.
Quantitative variables were expressed as means ± standard deviations (SD). The statistical analysis of ratios of HCV type-specific RT-nested PCR products from mixing assays (measured in pixels and calculated as relative percentages) was performed with the Student t test after means and SD of results from each set of experiments were determined. For this purpose, the Tadpole III program (Biosoft, Cambridge, United Kingdom) was used throughout this study. P values of <0.05 were considered statistically significant.
HCV isolate characterization.
Initial typing was performed with both 5′ UTR RFLP and INNO-LiPA assays (Table 1), thus allowing us to perform RNA mixing experiments focused on detecting such 5′ UTRs. With these methodologies, two samples were assigned to subtype 1a, three were assigned to 1b, three to 2a or 2c, two to 2b or 2c, and four to 3a. These samples were from three batches, as shown in Table 2.
TABLE 2.
Combinations of serum samples used for mixing assays
| Sample no. (subtype) | Type of mix for batcha
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A
|
B
|
C
|
||||||||||||
| 1 (1a) | 2 (1b) | 3 (2c) | 4 (2c) | 5 (3a) | 6 (1b) | 7 (1b) | 8 (2c) | 9 (2c) | 10 (3a) | 11 (1b) | 12 (2c) | 13 (3a) | 14 (4a) | |
| 1 (1a) | X | X | X | X | ||||||||||
| 2 (1b) | X | X | X | X | ||||||||||
| 3 (2c) | X | X | X | X | ||||||||||
| 4 (2c) | X | X | X | X | ||||||||||
| 5 (3a) | X | X | X | X | ||||||||||
| 6 (1b) | X | X | X | X | ||||||||||
| 7 (1b) | X | X | X | X | v | v | ||||||||
| 8 (2c) | X | X | X | X | v | v | ||||||||
| 9 (2c) | X | X | X | X | ||||||||||
| 10 (3a) | X | X | X | X | v | v | ||||||||
| 11 (1b) | v | v | X | X | X | |||||||||
| 12 (2c) | v | v | X | X | X | |||||||||
| 13 (3a) | v | v | X | X | ||||||||||
| 14 (4a) | X | X | ||||||||||||
Three independent batches of serum samples were designated A, B, and C. Sample numbers in each batch are shown, with corresponding subtypes listed in parentheses. Each letter in the table indicates a combination of the samples listed above and to the left. X indicates a combination of two samples from the same batch; v indicates a combination of two samples from different batches.
Final HCV (sub)type assignment was performed by phylogenetic analysis of sequences obtained from coding (core or NS5B) regions. Interestingly, all five HCV isolates ascribed to type 2 based on 5′ UTR analysis (by RFLP and LiPA) were finally ascribed to HCV type 2c by phylogenetic analysis of the NS5B region. Four out of five HCV type 1 samples were characterized as subtype 1b. Three out of four isolates initially characterized as HCV type 3a were finally assigned to the same subtype by cDNA sequencing.
As shown in Table 1, the remaining two (discrepant) samples exhibited infection with either the 1a or, unexpectedly, the 4a HCV (sub)type. Since no further sera with these viral subtypes were available at the time of the mixing experiments, a comprehensive evaluation was not carried out with 1a and 4a subtypes. Thus, minor discrepancies in HCV (sub)type assignment were observed for just one subtype 1a isolate when results of LiPA and 5′ UTR RFLP assays were compared with those of sequence and phylogenetic analyses of viral genomic coding regions. It is reportedly known that LiPA does not reliably subtype HCV, and up to 10% of viral 1b isolates are misclassified as 1a when subtype assignment is based on 5′ UTR RFLP analysis (2).
There was general agreement among the data when viral loads were determined based on limiting dilution PCR units and results of HCV Monitor assay version 2.0. Sample 10 showed the lowest titer by both methodologies.
Level of in vitro detection of subtype-specific HCV RNAs in prepared mixtures.
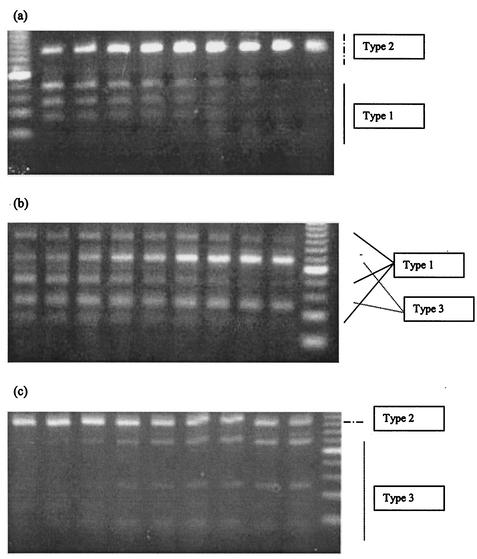
Interesting results appeared when three independent researchers analyzed gel photographs of RFLP studies (Fig. 1). At first sight, it appeared that the limit of detection of cleaved amplicons largely exceeded the equimolar concentrations of contributing RNAs within each mix.
FIG. 1.
Assessment of detection levels. Two different HCV types were analyzed by RFLP in each mixing experiment. The HCV types mixed are indicated at the right of each panel.
Computerized analysis allowed us to improve the limit of detection of very faint bands, since contributions under approximately 15% of total pixels in a given mix had been missed by visual observation of gel photographs.
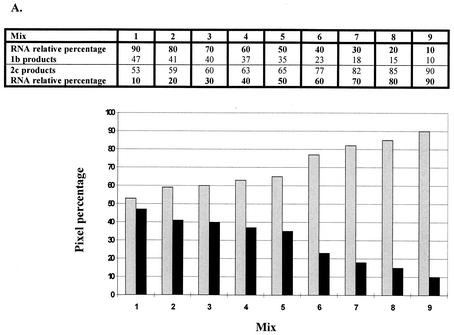
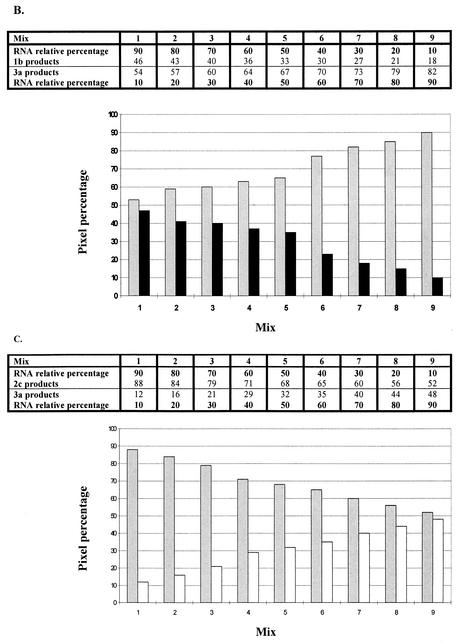
The ability to detect a given HCV type within each mix was different for most cases, as shown in Fig. 2 and Table 3. In general, types 2 and 3 were detectable when either of them contributed up to 10% of a mixture with the 1b subtype. In contrast, subtype 1b was less evident when lower contributing percentages (10 to 30%) were assayed, whichever the combination used (Fig. 2); i.e., when subtype 1b was mixed with subtypes 2c or 3a, it could be visually detected through its 10% (with 3a) or 40% (with 2c) RNA contribution. HCV subtype 2c was similarly detected when mixed with subtype 3a, with a slight predominance in sensitivity shown for the former.
FIG. 2.
Results of mixing assays. Scale and bar graphs show the quantitative contributions of each subtype (measured as percentages of pixels) to whole 5′ UTR RT-nested PCR products obtained by mixing different amounts of (sub)type-specific HCV RNAs. (A) Subtypes 1b (black bars) and 2c (gray bars). (B) Subtypes 1b and 3a (white bars). (C) Subtypes 2c and 3a.
TABLE 3.
Quantitative contributions of each type of HCV to total 5′ UTR RT-nested PCR products obtained by mixing different amounts of type-specific HCV RNAsa
| Virus types mixed | Ratio of types in mix | Amount of product (percentage of pixels ± SD) from type:
|
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 1 and 2 | 90:10 | 48.7 ± 5.0 | 51.2 ± 5.0 | |
| 50:50 | 30.0 ± 3.3 | 70.0 ± 3.3 | ||
| 10:90 | 12.7 ± 7.5 | 87.2 ± 7.5 | ||
| 1 and 3 | 90:10 | 53.3 ± 12.1 | 46.7 ± 12.1 | |
| 50:50 | 53.7 ± 15.6 | 46.2 ± 15.6 | ||
| 10:90 | 25.2 ± 5.1 | 74.8 ± 5.1 | ||
| 2 and 3 | 90:10 | 90.5 ± 4.1 | 9.5 ± 4.1 | |
| 50:50 | 73.7 ± 8.2 | 26.2 ± 8.2 | ||
| 10:90 | 59.7 ± 8.7 | 40.3 ± 8.7 | ||
Data shown are the averages of results from six (for mixtures of types 1 and 3 and 2 and 3) or eight (for mixture of types 1 and 2) independent experiments with 13 viremic serum samples. MMLV reverse transcriptase and 5′ UTR primer sets 1 and 2 were used for experiments. Reverse transcription performed by using Tth reverse transcriptase with another batch of five serum samples (not described in this study) rendered similar results. P values for comparisons between product ratios were as follows: for types 1 and 2 versus types 2 and 3, <0.0001 with all mix ratios; for types 1 and 2 versus types 1 and 3, not significant with a 90:10 mix ratio, 0.0060 with a 50:50 mix ratio, and 0.0044 with a 10:90 mix ratio; for types 2 and 3 versus types 1 and 3, <0.0001 with 90:10 and 10:90 mix ratios and not significant with a 50:50 mix ratio.
As a preliminary observation in mixing assays, no significant differences in the levels of detection were observed when the sole 1a isolate was compared with 1b isolates. In this case, a quite exact contribution (with respect to RNA viral input) was detected when cDNA-cleaved amplicons were analyzed. Likewise, the sole 4a isolate mixed with 1b or 2c isolates exhibited a behavior resembling that of 3a isolates when mixed similarly (data not shown). Further studies conducted with more HCV subtype 1a and 4a isolates are obviously needed to confirm these preliminary results.
HCV RNA secondary structures of the genomic 5′ UTRs depicted under computerized analysis exhibited no significant differences among the HCV subtypes included in the study.
RFLP detects nonequimolar concentrations of artificially mixed subtype-specific HCV RNAs in vitro: epidemiological, clinical, and virological impact.
Multiple molecular epidemiological studies of HCV have been carried out around the world, considering the implications of HCV types for pathogenesis, diagnosis, therapy, and prophylaxis. The superinfection of chimpanzees with heterologous subtypes has been previously described (12), as well as acute exacerbations in patients infected with mixed subtypes during the course of chronic HCV infection (6, 7, 14). Possible pathogenic features associated with HCV genotypes have been recently reviewed elsewhere (21).
The rate of HCV mixed infections detected in epidemiological studies varies according to the methodology employed. In Argentina, several populations with increased risk of exposure to diverse potential infectious sources (intravenous drug users, hemophiliacs, and dialyzed and polytransfused patients) have shown a high rate of mixed HCV subtype infections (15) as detected by the RFLP methodology.
When the results of all experiments (six to eight for each type of mix) performed in this study were grouped according to HCV type, significant differences in type-specific RT-nested PCR products were observed among artificially mixed RNAs, in spite of the use of equivalent amounts of template input. Interestingly, no statistically significant difference was found in results obtained with the use of either set 1 or set 2 of 5′ UTR primers when the primers were tested in independent experiments. Moreover, to rule out the possibility of obtaining results that could be ascribed to the mere chance of combining certain pairs of sera, further combinations were examined by using RNAs from samples 7, 8, and 10, properly mixed with that from either 11, 12, or 13 (Table 2). Results obtained provided further support for those of the initial experiments, since no statistically significant differences in values were observed (two-sided t test). Due to this fact, a global view is shown in Table 3. Moreover, most of the experiments with the same pairs of HCV types rendered similar results, in spite of the inclusion of different subtypes (i.e., 1a and 2c or 1b and 2c). This trend was reflected in the small SD observed throughout (Table 3).
Several controls were performed to obtain reliable conclusions. First, to make sure that adequate amounts of RNAs were added, HCV serum titration was performed both by a fivefold limiting dilution titration assay and by means of a commercially available kit (HCV Roche Monitor version 2.0). A reasonable concordance was observed between results from both methodologies. However, due to the narrow quantitative range of the HCV Monitor assay (11), quantities of the artificially mixed RNA templates used for RT-nested PCR assays in our study were based upon values calculated by the limiting dilution method. Second, primer annealing sites employed for 5′ UTR HCV sequence amplification failed to show significant differences for the studied subtypes, although experiments were carried out with two independent sets of widely used primers (2, 13, 15). Third, comparative analysis of the secondary structures of the 5′ UTRs (including the internal ribosome entry sites) of GenBank sequences from HCV RNAs belonging to subtypes assayed in this work revealed no variations inter-se, although there are point mutations that do not disturb the secondary structures due to the appearance of covariant substitutions (18). Changes in G-C content modify the free energy of the system, and accordingly, a higher percentage of G-C could lead to less-efficient RNA reverse transcription when thermolabile reverse transcriptases—i.e., those of MMLV and avian myeloblastosis virus—are used, despite initial RNA denaturing at 95°C. However, since we had previously carried out reverse transcription (data not shown) by using Tth thermostable enzyme at 70°C for 20 min as previously described (13), the possibility that in such experiments any residual native viral RNAs remained in the reaction mix seems to be negligible, and our results with Tth suggest that the observed difference in detection sensitivities is independent of the reverse transcriptase used. These preliminary results with another batch of five samples (including sera with HCV types 1, 2, and 3) are not fully depicted in this study because the final subtype assignment was not available from cDNA sequences of coding regions. Nevertheless, a definitive (sub)type assignment may be obtained only after complete HCV genomic sequencing, since it has been recently shown that HCV intergenotypic recombination is possible (5).
This study shows the RFLP typing method to have dissimilar degrees of detection ability when two HCV (sub)types are involved in a single in vitro assay.
Although no obvious explanations appear to justify these results, future experiments should investigate other potential factors, such as thermodynamic conditions of reverse transcription associated with a putative dissimilar secondary or tertiary full-length HCV RNA type-specific structure (8, 18, 20, J. Wood, A. Tuplin, A. H. Patel, and P. Simmonds, 8th Int. Symp. Hepat. C Virus Relat. Viruses, abstr. O-03, p. 29, 2001). In this regard, it has been previously reported that the 5′ UTR main stem-loops of genotypes 2, 3, and 4 exhibit a more open structure than that of subtype 1a (18). Thus, it would be worthwhile to analyze the behavior of artificial RNA transcripts corresponding to previously sequenced cDNA HCV clones.
In summary, in contrast with previous assumptions and supporting recent data observed among hemodialyzed patients (14), this work shows that RFLP is able to detect in vitro dissimilar amounts of HCV (sub)type-specific RNA molecules from a wider range of contributing subtypes in mixes prepared from sera of persons infected with a single HCV genotype than originally described for naturally occurring mixed HCV infections (10). Moreover, this study demonstrated that the true contribution of each virus type in mixed infections may be underestimated when only visual photograph analysis is carried out. These results may be useful for comparing data obtained from this and other currently used methodologies.
Nucleotide sequence accession numbers.
Nucleotide sequence data reported in this paper were deposited in the GenBank database with the following accession numbers: AY172636 to AY172639 (5′ UTR-core sequences) and AY172640 to AY172649 (NS5B sequences).
Acknowledgments
We are grateful for useful comments from Stanley Lemon (University of Texas). We thank María Victoria Illas for enhancing readability.
This work was partly supported by grants BID 802/OC-AR-PICT 04977/99 from the National Agency for Scientific and Technological Promotion and PIP 842/98 from CONICET and by the Oñativia Carrillo Fellowship from the Argentine Ministry of Health, Argentina.
REFERENCES
- 1.Chan, S.-W., F. McOmish, E. C. Holmes, B. Dow, J. F. Peutherer, E. Follett, P. L. Yap, and P. Simmonds. 1992. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J. Gen. Virol. 73:1131-1141. [DOI] [PubMed] [Google Scholar]
- 2.Davidson, F., P. Simmonds, J. Ferguson, L. Jarvis, B. Dow, E. Follet, C. Seed, T. Krusius, C. Lin, G. Medgyesi, H. Kiyokawa, G. Olim, G. Duraisamy, T. Cuypers, A. Saeed, D. Teo, J. Conradie, M. Kew, M. Lin, C. Nuchaprayoon, O. Ndimbie, and P.-L. Yap. 1995. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J. Gen. Virol. 76:1197-1204. [DOI] [PubMed] [Google Scholar]
- 3.Furione, M., L. Simoncini, M. Gatti, F. Baldanti, M. Grazia Revello, and G. Gerna. 1999. HCV genotyping by three methods: analysis of discordant results based on sequencing. J. Clin. Virol. 13:121-130. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins, A., F. Davidson, and P. Simmonds. 1997. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J. Clin. Microbiol. 35:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 1993. Superinfection of heterologous hepatitis C virus in a patient with chronic type C hepatitis. Gastroenterology 105:583-587. [DOI] [PubMed] [Google Scholar]
- 7.Kao, J. H., P. J. Chen, M. Y. Lai, P. M. Yang, J. C. Sheu, T. H. Wang, and D. S. Chen. 1994. Mixed infections of hepatitis C virus as a factor in acute exacerbations of chronic type C hepatitis. J. Infect. Dis. 170:1128-1133. [DOI] [PubMed] [Google Scholar]
- 8.Kieft, J. S., K. Zhou, R. Jubin, M. G. Murray, J. Y. N. Lau, and J. A. Doudna. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292:513-529. [DOI] [PubMed] [Google Scholar]
- 9.Matzura, O., and A. Wennborg. 1996. RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biosci. 12:247-249. [DOI] [PubMed] [Google Scholar]
- 10.McOmish, F., P. L. Yap, B. C. Dow, A. C. Follett, C. Seed, A. J. Keller, T. J. Cobain, T. Krusius, E. Kolho, R. Naukkarinen, C. Lin, C. Lai, S. Leong, G. A. Medgyesi, M. Hejjas, H. Hiyokawa, K. Fukada, T. Cuypers, A. A. Saeed, A. M. Al-Rasheed, M. Lin, and P. Simmonds. 1994. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J. Clin. Microbiol. 32:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellors, J., A. Hawkins, and P. Simmonds. 1999. Genotype dependence of hepatitis C virus load measurement in commercially available quantitative assays. J. Clin. Microbiol. 37:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto, H., S. Mishiro, H. Tokita, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1994. Superinfection of chimpanzees carrying hepatitis C virus of genotype II/1b with that of genotype III/2a or I/1a. Hepatology 20:1131-1136. [PubMed] [Google Scholar]
- 13.Oubiña, J. R., J. Quarleri, M. Rudzinski, C. Parks, I. Badía, and S. González Cappa. 1995. Genomic characterization of hepatitis C virus from Argentina. J. Med. Virol. 47:97-104. [DOI] [PubMed] [Google Scholar]
- 14.Qian, K. P., S. N. Natov, B. J. Pereira, and J. Y. Lau. 2000. Hepatitis C virus mixed genotype infection in patients on haemodialysis. J. Viral Hepat. 7:153-160. [DOI] [PubMed] [Google Scholar]
- 15.Quarleri, J. F., B. H. Robertson, V. L. Mathet, M. Feld, L. Espínola, M. P. Requeijo, O. Mandó, G. Carballal, and J. R. Oubiña. 2000. Genomic and phylogenetic analysis of hepatitis C virus isolates from Argentine patients: a six-year retrospective study. J. Clin. Microbiol. 38:4560-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin i, P. Simmonds, D. Smith, L. Stuyver, A. Weiner, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposal for standarization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 17.Simmonds P. 1999. Viral heterogeneity of the hepatitis C virus. J. Hepatol. 31(Suppl. 1):54-60. [DOI] [PubMed] [Google Scholar]
- 18.Smith, D. B., J. Mellor, L. M. Jarvis, F. Davidson, J. Kolberg, M. Urdea, P. L. Yap, and P. Simmonds. 1995. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J. Gen. Virol. 76:1749-1761. [DOI] [PubMed] [Google Scholar]
- 19.Stuyver, L., R. Rossau, A. Wyseur, M. Duhamel, B. Vanderborght, H. van Heuverswyn, and G. Maertens. 1993. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J. Gen. Virol. 74:1093-1102. [DOI] [PubMed] [Google Scholar]
- 20.Wang, C., S. Y. Le, N. Ali, and A. Siddiqui. 1995. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA 1:526-537. [PMC free article] [PubMed] [Google Scholar]
- 21.Zein, N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]