Abstract
We conducted a nationwide survey of the variable 5′ emm (M protein gene) sequences from 614 pharyngeal Streptococcus pyogenes isolates susceptible (299 isolates) and resistant (315 isolates) to erythromycin that were isolated in Spain from 1996 to 1999. Almost 98% of these isolates had emm sequences in agreement with previously recorded M antigen association. We only identified a new 5′ emm sequence in 17 isolates. Nine different emm types accounted for 85% of the S. pyogenes isolates susceptible to erythromycin. By contrast, only 3 emm types accounted for 70% of the erythromycin-resistant isolates. Further characterization of these isolates by ribotyping and pulsed-field gel electrophoresis indicated that high frequency of erythromycin resistance in Spain is due to few clones.
Streptococcus pyogenes or group A streptococci (GAS) is an important human pathogen as the etiological agent of streptococcal sore throat, skin and soft tissue infections, and the postinfectious syndromes of glomerulonephritis and acute rheumatic fever.
Penicillin remains the drug of choice for the treatment of streptococcal pharyngitis (7); however, an increased failure of this treatment due to copathogenicity with β-lactamase-producing microorganisms has been reported (3). In these cases and in cases where patients are allergic to penicillin, other antibiotics not subject to inactivation by β-lactamases, i.e., amoxicillin-clavulanate, oral cephalosporins, or erythromycin, have been substituted for penicillin (3).
Resistance to erythromycin remained at low levels among S. pyogenes in most countries of the world (14); however, in the last years, a significant increase in erythromycin-resistant isolates in different countries has been reported (15, 20, 22, 23). In most of these studies, resistance to erythromycin was caused by a few clones and the mechanisms of resistance were mainly based on the presence of an active drug efflux by pumps encoded by the gene mefA (M phenotype) that take out of the cells 14- and 15-membered macrolides (25). By contrast, the other strategy described for erythromycin resistance, target site modification by erm methylase strains that express the macrolide-lincosamide-streptogramin B (MLS) phenotype, was not very common among the erythromycin-resistant S. pyogenes isolates.
In the last few years, in two different nationwide antimicrobial surveillance studies, an increasing resistance to erythromycin has been reported in S. pyogenes isolated in Spain, where the erythromycin resistance frequency has increased from 12 to 26% in the last 10 years, reaching 60% in some regions of the country (2, 19). Therefore, we decided to conduct an epidemiologic investigation to determine the reasons for the high frequency of erythromycin resistance in S. pyogenes observed in our country.
A serologic test for M protein antigens has long been a primary method for the identification and epidemiological study of S. pyogenes isolates. However, this method is dependent on the preparation of type-specific antisera and extraction of a protein identified as M protein on the surface of S. pyogenes, a process that is difficult and specialized. The 5′ ends of emm genes are highly heterogeneous and encode the serotype specificity used for the M typing system developed by Lancefield in 1928. Since, in 1995, Beall et al. demonstrated the usefulness of emm gene sequence analysis for the routine typing of GAS (5), few studies have used this typing method for monitoring S. pyogenes diversity (4, 5, 8, 9, 27).
In this study, we conducted an epidemiological survey of emm gene sequences from pharyngeal S. pyogenes isolates isolated over a period of 4 years in 8 Spanish hospitals and studied their relationship to erythromycin resistance.
MATERIALS AND METHODS
Strains.
S. pyogenes clinical strains isolated from patients with community-acquired acute pharyngitis were collected at 8 different hospitals geographically distributed in Spain and belonging to the surveillance collections SAUCE I (May 1996 to April 1997) (2) and SAUCE II (November 1998 to October 1999) (19). For this study, we selected 315 S. pyogenes isolates resistant to erythromycin (MIC ≥ 1 μg/ml) from both surveillance collections (145 isolates from 1996 to 1997 and 170 isolates from 1998 to 1999). Additional 1:1 matched erythromycin-susceptible S. pyogenes strains were also selected. Hence, for every resistant isolate, the immediate next susceptible strain isolated in the same hospital from a patient belonging to the same age group was chosen in order to minimize temporal, geographical, or age-related bias. Two hundred ninety-nine isolates (133 from 1996 to 1997 and 166 from 1998 to 1999) fulfilled the above criteria. The organisms were isolated from patients from 7 cities (number of isolates and period of isolation): Córdoba (30 isolates, 1996 to 1997; 15 isolates, 1998 to 1999), Seville (26 isolates, 1996 to 1997; 47 isolates, 1998 to 1999), Granada (24 isolates, 1996 to 1997; 34 isolates, 1998 to 1999), Valencia (117 isolates, 1996 to 1997), Madrid (81 isolates, 1996 to 1997; 174 isolates, 1998 to 1999), San Sebastián (32 isolates, 1998 to 1999), and Santander (34 isolates, 1998 to 1999). Identification of S. pyogenes isolates was confirmed by serogroup A immunoagglutination assays (Streptex; Murex, Chantillon, France).
Susceptibility tests.
Susceptibility testing was performed on all isolates by double dilution with the semiautomated microdilution method with custom-dried 96-well trays and Mueller-Hinton broth supplemented with 3% lysed horse blood according to the guidelines of the NCCLS (18) and with a final inoculum of 5 × 105 CFU/ml. Cultures were incubated for 24 h at 35°C in ambient air with erythromycin. The breakpoint used for erythromycin resistance was ≥1 μg/ml.
The mechanism of resistance to erythromycin was evaluated with a double-diffusion disk test as described elsewhere (24) with erythromycin (15 μg) and clindamycin (2 μg) disks placed 20 mm (edge to edge) apart on 5% defibrinated horse blood agar. After overnight incubation at 35°C, the presence of blunting in the zone of inhibition of the clindamycin disk was recorded (21). If the clindamycin inhibition zone was blunted toward the erythromycin disk, the strain was interpreted as clindamycin inducible. Resultant phenotype patterns were clindamycin-sensitive strains (M phenotype) and clindamycin-resistant or -inducible strains (MLS phenotype).
emm gene typing.
The emm gene type of S. pyogenes isolates was determined by amplification and sequencing of the emm gene as described by Beall et al. (5). Lysates of the S. pyogenes isolates were prepared with mutanolysin as described previously (1). Primers GASM1 and GASM2 were used in PCRs carried out according to the method described previously (5). PCR products were sequenced with primer GASM1 with the dye terminator mix (Applied Biosystems, Foster City, Calif.) and were subjected to automated sequence analysis on a 377 DNA sequencer (Applied Biosystems). DNA sequences were subjected to homology searches against the bacterial DNA database with BLASTN. Sequences were given the GenBank emm designations following the criteria previously described (4, 10).
Ribotyping analysis.
Ten micrograms of purified genomic DNA, isolated following the protocol previously described (1), were digested with HindIII, SacI, or XhoI according to the specifications of the manufacturer (Amersham-Pharmacia, Uppsala, Sweden). DNA fragments were separated on a 1% agarose gel. Southern blot analysis and S. pyogenes 16S rRNA gene probe labeling and detection were carried out with the ECL kit (Amersham-Pharmacia). Differences in banding patterns were documented by visual examination and indexed by small lettering.
PFGE analysis.
Analysis of chromosomal DNA was carried out by pulsed-field gel electrophoresis (PFGE) analysis by following standard procedures (16). Chromosomal DNA was digested with SfiI (Amersham-Pharmacia), and fragments were separated in a CHEF-DRIII apparatus (Bio-Rad Laboratories, Barcelona, Spain). Electrophoretic pulses were linearly distributed from 20 to 70 s, for a run time of 22 h. The voltage was 6 V/cm, and the temperature of the electrophoresis chamber was kept at 14°C. The gels were stained with ethidium bromide and photographed.
Differences in banding patterns were documented by visual examination and indexed by capital lettering. The interpretation of restriction fragment patterns was performed in accordance with recent consensus publications (28).
Nucleotide sequence accession number.
The sequence of emmst2002 obtained in this study has been given GenBank accession number AJ515525.
RESULTS
Prevalent emm genes.
Overall, 597 (97.2%) of 614 S. pyogenes clinical isolates included in our study had 5′ emm sequences ≥95% identical to the 160 first bases of one of the emm or emm-like genes deposited in GenBank. For most of these sequences, this high level of identity actually extended to 200 to 450 bases, without diminishing. The sequences of 522 (85%) isolates were ≥95% identical to the sequence of standard M type reference strain emm genes, and the sequences of 75 isolates were ≥95 identical to the sequence of st1815, a sequence not linked with known M typing specificity. The remaining 17 of 614 isolates had an undocumented emm gene sequence. This sequence was provisionally designated as st2002 and was only 85% identical to the emm104 (formerly st2035) sequence over the first 160 bases.
emm1, emm3, emm12, and emm9 were the most prevalent emm sequences among S. pyogenes isolates susceptible to erythromycin (Table 1). They accounted for 12.4, 11.4, 10, and 9.4%, respectively, of these isolates. Besides these emm sequences, the following most common sequences were emm28, emm44, emm4, emm6, and the new emm gene sequence designated emmst2002, each one of which accounted for approximately ≤6% of the erythromycin-susceptible isolates. We did not detect substantial differences between both periods (1996 to 1997 versus 1998 to 1999) in the distribution of the prevalent emm sequences among GAS erythromycin-susceptible isolates (Table 1). Perhaps the most interesting difference was that most of the M type st2002 isolates were collected in the first period and their number decreased markedly in the second period.
TABLE 1.
emm sequences of pharyngeal S. pyogenes isolated in Spain (1996 to 1999)
| No. of isolates | emm type | No. (%) of isolates resistant to erythromycina collected in:
|
No. % of isolates susceptible to erythromycin collected in:
|
||||
|---|---|---|---|---|---|---|---|
| 1996-1997 (n = 145) | 1998-1999 (n = 170) | Total (n = 315) | 1996-1997 (n = 133) | 1998-1999 (n = 166) | Total (n = 299) | ||
| 144 | 4 | 72 (49.7) | 57 (33.5) | 129 (41.0) | 8 (6.0) | 7 (4.2) | 15 (5.0) |
| 75 | st1815 | 42 (29.0) | 23 (13.5) | 65 (20.6) | 8 (6.0) | 2 (1.2) | 10 (3.3) |
| 67 | 12 | 15 (10.3) | 22 (12.9) | 37 (11.7) | 19 (14.3) | 11 (6.6) | 30 (10.0) |
| 39 | 75 | 2 (1.4) | 35 (20.6) | 37 (11.7) | 1 (0.8) | 1 (0.6) | 2 (0.7) |
| 38 | 1 | 1 (0.6) | 1 (0.3) | 11 (8.3) | 26 (15.7) | 37 (12.4) | |
| 34 | 9 | 1 (0.7) | 5 (2.9) | 6 (1.9) | 8 (6.0) | 20 (12.0) | 28 (9.4) |
| 34 | 3 | 11 (8.3) | 23 (13.9) | 34 (11.4) | |||
| 25 | 28 | 3 (2.1) | 3 (1.8) | 6 (1.9) | 6 (4.5) | 13 (7.8) | 19 (6.4) |
| 24 | 6 | 9 (5.3) | 9 (2.9) | 8 (6.0) | 7 (4.2) | 15 (5.0) | |
| 22 | 22 | 1 (0.7) | 8 (4.7) | 9 (2.9) | 3 (2.3) | 10 (6.0) | 13 (4.3) |
| 17 | st2002 | 14 (10.5) | 3 (1.8) | 17 (5.7) | |||
| 16 | 2 | 5 (3.4) | 5 (1.6) | 6 (4.5) | 5 (3.0) | 11 (3.7) | |
| 16 | 44 | 6 (4.5) | 10 (6.0) | 16 (5.4) | |||
| 12 | 11 | 8 (6.0) | 4 (2.4) | 12 (4.0) | |||
| 10 | 77 | 6 (3.5) | 6 (1.9) | 4 (2.4) | 4 (1.3) | ||
| 8 | 78 | 8 (4.8) | 8 (2.7) | ||||
| 6 | 13 | 5 (3.8) | 1 (0.6) | 6 (2.0) | |||
| 5 | 50 | 1 (0.7) | 1 (0.3) | 4 (3.0) | 4 (1.3) | ||
| 3 | 73 | 3 (1.8) | 3 (1.0) | ||||
| 2 | 67 | 1 (0.6) | 1 (0.3) | 1 (0.6) | 1 (0.3) | ||
| 2 | 87 | 2 (1.5) | 2 (0.7) | ||||
| 2 | 19 | 2 (1.2) | 2 (0.7) | ||||
| 2 | 66 | 2 (1.5) | 2 (0.7) | ||||
| 2 | 8 | 2 (1.2) | 2 (0.7) | ||||
| 2 | 41 | 2 (1.4) | 2 (0.6) | ||||
| 2 | fcrV | 1 (0.7) | 1 (0.3) | 1 (0.8) | 1 (0.3) | ||
| 2 | 89 | 1 (0.8) | 1 (0.6) | 2 (0.7) | |||
| 1 | 61 | 1 (0.6) | 1 (0.3) | ||||
| 1 | 58 | 1 (0.6) | 1 (0.3) | ||||
| 1 | 33 | 1 (0.8) | 1 (0.3) | ||||
The breakpoint for erythromycin was ≥1 μg/ml.
The prevalent emm sequence encountered in the S. pyogenes isolates resistant to erythromycin was emm4 followed by st1815, emm12, and emm75 (Table 1). Altogether, these four M types represented 85% of the S. pyogenes isolates resistant to erythromycin while they only represented 19% of the susceptible isolates. Interestingly, M type 75 resistant isolates were almost absent in 1996 to 1997 and became predominant, together with M type 4 resistant isolates, in 1997 to 1998. By contrast, the frequency of M type st1815 resistant isolates was significantly reduced in 1998 to 1999 compared to the first time period. We found a strong association between the resistance to erythromycin and three different M types (Table 1). Among the erythromycin-resistant strains, up to 40.9% belonged to M type 4 isolates versus 5.0% among susceptible strains (odds ratio [OR], 13.13; 95% confidence interval [CI], 7.25 to 24.14). Likewise, 20.6% of the resistant strains were type st1815 versus only 3.3% among erythromycin-susceptible isolates (OR, 7.51; 95% CI, 3.64 to 15.92). For M type 75 isolates, this association was even stronger since this type comprised up to 11.7% of the overall resistant isolates (but 20.6% in the period 1998 to 1999) versus only 0.7% of the susceptible strains (OR, 19.76; 95% CI, 5.00 to 170.29).
We found that 298 (94.6%) of the erythromycin-resistant isolates expressed the M phenotype of resistance. All the M type 4, type st1815, and type 75 (except one M75 isolate) erythromycin-resistant isolates presented this phenotype of resistance. Only 17 (5.4%) resistant isolates (M types 22, 6, 77, and 28) displayed the classical MLS phenotype, including constitutive (5 isolates) and inducible (12 isolates) resistance.
Clonal diversity of the erythromycin-resistant S. pyogenes isolates.
We investigated whether the erythromycin-resistant isolates represent only a few clonal types, since they belong mainly to only three different M types (M4, st1805, and M75), and all of them displayed the M phenotype. For this purpose, we randomly selected at least one S. pyogenes erythromycin-resistant isolate of each one of these M types from each hospital and from both national surveillance collections.
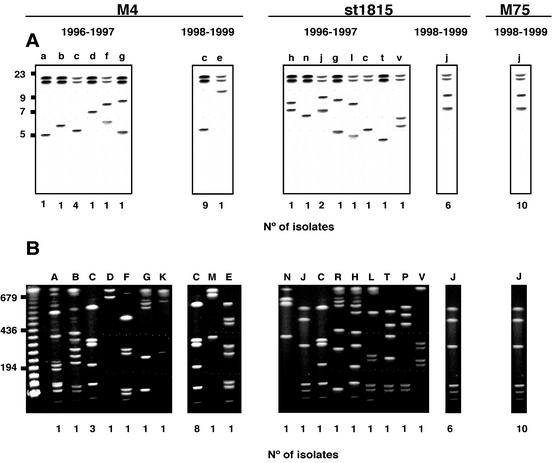
Ribotyping analysis with genomic DNA from S. pyogenes isolates digested with SacI and probed with an internal fragment of the 16S rRNA gene led to 13 different patterns, which are illustrated in Fig. 1A. Each strain was tested at least twice with SacI and with two additional enzymes (XhoI and HindIII); however, SacI-digested fragments were more discriminatory than the XhoI- and HindIII-digested fragments that only rendered two and three different patterns, respectively (data not shown). Type c and j were the most common types among M type 4 and M type st1815 isolates, respectively. For both M types, we observed more variability between the S. pyogenes isolated from 1996 to 1997 than in those isolated from 1998 to 1999, where types c and j were almost exclusive (Table 2). Similarly, all M type 75 S. pyogenes isolates that were collected only between 1998 and 1999 presented pattern j. All the other patterns were represented by only one strain.
FIG. 1.
Representative ribotypes (A) and PFGE types (B) of erythromycin-resistant S. pyogenes M types 4, st1815, and 75 isolated between 1996 to 1997 and 1998 to 1999 in Spain. (A) Southern blot of genomic S. pyogenes DNA digested with SacI and probed with a GAS 16S rRNA gene probe as described in Materials in Methods. (B) PFGE analysis of genomic DNA restricted with SfiI. Letters on the lanes refer to types, and numbers below each lane refer to the number of isolates of each type. Molecular size markers (in kilobases) are indicated on the left.
TABLE 2.
Correlation between emm typing, ribotyping, and PFGE typing of erythromycin-resistant S. pyogenes isolated in two different time periods in Spain
| emm type | Type (no.) of isolates collected in:
|
|||
|---|---|---|---|---|
| 1996-1997
|
1998-1999
|
|||
| Ribotype | PFGE type | Ribotype | PFGE type | |
| 4 | a (1) | A (1) | ||
| b (1) | B (1) | |||
| c (4) | C (3), K (1) | c (9) | C (8), E (1) | |
| d (1) | D (1) | |||
| f (1) | F (1) | |||
| g (1) | G (1) | |||
| e (1) | M (1) | |||
| st1815 | h (1) | H (1) | ||
| n (1) | N (1) | |||
| j (2) | J (1), P (1) | j (6) | J (6) | |
| g (1) | R (1) | |||
| l (1) | L (1) | |||
| c (1) | C (1) | |||
| t (1) | T (1) | |||
| v (1) | V (1) | |||
| 75 | j (10) | J (10) | ||
Further genotypic characterization was carried out by genomic DNA macrorestriction with SfiI and PFGE. Representative PFGE profiles of the erythromycin-resistant S. pyogenes isolates are shown in Fig. 1B. We identified 17 distinct PFGE types, one (type C) of them divided in 3 subtypes (C.1, C.2, and C.3; data not shown). We found a good correlation between ribotyping and PFGE patterns (Table 2). Thus, ribotypes c and j corresponded, for most of the isolates, to PFGE types C and J, respectively. Again, PFGE types C and J were prevalent, and all other ribotypes corresponded to PFGE types represented, again, by a single isolate.
In summary, results obtained by either ribotyping or PFGE suggest that S. pyogenes resistance to erythromycin in Spain between 1996 to 1999 was caused by a few clones belonging to three M types. For M types 4 and st1815, we observed more variability between the S. pyogenes isolated from 1996 to 1997 than in those isolated from 1998 to 1999, where all the isolates presented the same genotyping profile. Interestingly, clones c and j, prevalent from 1998 to 1999, were also present from 1996 to 1997.
DISCUSSION
The purpose of this study was to survey the genetic diversity of pharyngeal S. pyogenes isolates by using emm gene sequence analysis to better understand the increased level of resistance to erythromycin in Spain and to further investigate the epidemiology of these S. pyogenes erythromycin-resistant isolates.
In our study, we have determined the 5′ emm sequence from 614 pharyngeal S. pyogenes isolates taken from two nationwide surveillance collections conducted from 1996 to 1999. To our knowledge, this is the first survey study of emm typing conducted in Spain. As expected, nearly 100% of the strains could be genotyped by the emm typing system as described recently Facklam et al. (10). Only one newly encountered emm gene, provisionally designated emm st2002, from 17 isolates had a 5′ emm sequence with an identity ≤95% over the first 160 bases to emm sequences deposited in GenBank. However, according to Facklman et al. (10), further studies are required to validate this new emm type that shows only 85% identity over the first 160 bases with emm104 (formerly st2035) (11).
In our survey, M type 1 and M type 3 were the most prevalent M types among S. pyogenes isolates susceptible to erythromycin. Since resistance to erythromycin represents approximately 20 to 30% of the isolates (2, 19), we can conclude that these M types are probably the most common in Spain. This result is in agreement with other M typing studies conducted by other authors (4, 8, 13).
To investigate the reasons for the increased levels of resistance to erythromycin detected in Spain, we analyzed the emm gene sequence of 315 GAS isolates resistant to erythromycin collected simultaneously and at the same site to that of the erythromycin-susceptible isolates. Our results show that a few M types were responsible for up to 85% of the erythromycin-resistant isolates, suggesting that few clones caused this phenomenon. This hypothesis was supported by the fact that the M phenotype was the most common resistance mechanism, a feature previously reported by other authors (20, 23). However, genetic divergence among strains sharing the same emm gene sequence has been reported (5, 6, 17, 29). For this reason, emm sequencing must be supplemented with other approaches such as PFGE to identify related GAS isolates (4). To further investigate the clonal diversity of the erythromycin-resistant GAS isolates, we randomly selected strains from each hospital geographically distributed all over Spain and from both surveillance collections. We used two genotyping techniques that yielded similar results. Ribotyping analysis of the GAS genomic DNA digested with HindIII and XhoI was poorly discriminatory as previously described (data not shown) (26). However, ribotyping by SacI restriction polymorphism showed equivalent power for strain typing to that of PFGE in favor of labor and time benefit.
The data obtained by both approaches indicate that during the first period (1996 to 1997) many different clones sharing the same M type (M type 4 or M type st1815) were distributed all over Spain and responsible for the resistance to erythromycin detected between 1996 to 1997. By contrast, in the following two years (1997 to 1998), clonal diversity was reduced to a few clones that were the major clones identified in the years 1996 to 1997. These results suggest that these clones have been selected over a short period of time (only 2 years) and have become predominant in Spain. It is reasonable to believe that selection of these clones is related to the consumption of macrolides and that this may account for the geographical differences in the prevalence of resistance to erythromycin in Spain (12, 19).
Our results are in agreement with those of a previous study conducted by Pérez-Trallero et al. in Spain (20). In that study, erythromycin-resistant GAS isolates from Spain (n = 437, 1988 to 1997) were composed mainly of two clones designated clone B (emm type 4) and clone D (emm type 75) that were already isolated in Spain in 1991. Those clonal types showed same lineage to our emm types, corresponding to PFGE C and J types, respectively, suggesting that they are probably the same strains. Apart from the strains described in Spain, M type 4 strains with the M phenotype were isolated in Finland and Great Britain before 1991 (22, 23). We do not know whether the strains of clone C isolated in our study are the same as the erythromycin-resistant type 4 found in Finland and Great Britain, but it is probable. In addition to emm type 4, emm type st1815 accounted for 20% of the GAS resistant to erythromycin. This emm type was not detected by Pérez-Trallero et al. in their study, where they assayed only 14 emm types (20); however, it was one of the prevalent types in our study. Interestingly, the most common PFGE and ribotype pattern among st1815 emm type strains was identical to that of emm type 75 strains. A similar observation was previously reported by Whatmore et al. that demonstrated that highly divergent emm sequences were present in strains with identical multilocus electrophoretic types, suggesting horizontal transfer of emm genes between unrelated strains (29). Alternatively, it might be that emm75 and emmst1815 share identical PFGE types because emm sequence st1815 was generated by homologous excision between the tandem emm and enn sequences in an emm75 parental strain, as suggested in the emm sequence database (http://www.cdc.gov/ncidod/biotech/strep/emmtypes.htm).
In summary, our results show that monitoring of GAS isolate diversity by emm gene typing is a useful approach for a better understanding of the epidemiology and origins of specific GAS strains. The application of this typing technique to nationwide multicenter surveillance has revealed that a few clones cause GAS erythromycin resistance in Spain.
Acknowledgments
This study was supported by a grant from GlaxoSmithKline, Madrid, Spain.
REFERENCES
- 1.Albertí, S., C. D. Ashbaugh, and M. R. Wessels. 1998. Structure of the has operon promoter and regulation of hyaluronic acid capsule expression in group A Streptococcus. Mol. Microbiol. 28:343-353. [DOI] [PubMed] [Google Scholar]
- 2.Baquero, F., J. A. García-Rodríguez, J. García de Lomas, L. Aguilar, and the Spanish Surveillance Group for Respiratory Pathogens.1999. Antimicrobial resistance of 914 beta-hemolytic streptococci isolated from pharyngeal swabs in Spain: results of a 1-year (1996-1997) multicenter surveillance study. Antimicrob. Agents Chemother. 43:178-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass, J. W. 1991. Antibiotic management of group A streptococcal pharyngotonsillitis. Pediatr. Infect. Dis. J. 10:543-549. [DOI] [PubMed] [Google Scholar]
- 4.Beall, B., R. Facklam, T. Hoenes, and B. Schwartz. 1997. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J. Clin. Microbiol. 35:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., R. Facklam, and T. Thompson. 1995. Sequencing emm-specific polymerase chain reaction products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessen, D. E., C. M. Sotir, T. L. Readdy, and S. K. Hollingshead. 1996. Genetic correlates of throat and skin isolates of group A streptococci. J. Infect. Dis. 17:896-900. [DOI] [PubMed] [Google Scholar]
- 7.Betriu, C., A. Sánchez, M. Gómez, A. Cruceyra, and J. J. Picazo. 1993. Antibiotic susceptibility of group A streptococci: a 6-year follow-up study. Antimicrob. Agents Chemother. 37:1717-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandt, C. M., B. Spellerberg, M. Honscha, N. D. Truong, B. Hoevener, and R. Lutticken. 2001. Typing of Streptococcus pyogenes strains isolated from throat infections in the region of Aachen, Germany. Infection 29:163-165. [DOI] [PubMed] [Google Scholar]
- 9.Dicuonzo, G., G. Gherardi, G. Lorino, S. Angeletti, M. de Cesaris, E. Fiscarelli, D. E. Bessen, and B. Beall. 2001. Group A streptococcal genotypes from pediatric throat isolates in Rome, Italy. J. Clin. Microbiol. 39:1687-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facklam, R., B. Beall, A. Efstratiou, V. Fischetti, D. Johnson, E. Kaplan, P. Kriz, M. Lovgren, D. Martin, B. Schwartz, A. Totolian, D. Bessen, S. Hollingshead, F. Rubin, J. Scott, and G. Tyrrell. 1999. emm typing and validation of provisional M types for group A streptococci. Emerg. Infect. Dis. 5:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Facklam, R. F., D. R. Martin, M. Lovgren, D. R. Johnson, A. Efstratiou, T. A. Thompson, S. Gowan, P. Kriz, G. J. Tyrrell, E. Kaplan, and B. Beall. 2002. Extension of the Lancefield classification for group A sreptococci by addtion of 22 new M protein gene sequence types from clinical isolates: emm103 to emm124. Clin. Infect. Dis. 34:28-38. [DOI] [PubMed] [Google Scholar]
- 12.Granizo, J. J., L. Aguilar, J. Casal, R. Dal-Ré, and F. Baquero. 2000. Streptococcus pyogenes resistance to erythromycin in relation to macrolide consumption in Spain (1986-1997). J. Antimicrob. Chemother. 46:959-964. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, D. R., D. L. Stevens, and E. L. Kaplan. 1992. Epidemiologic analysis of group A streptococcal serotypes associated with severe systemic infections, rheumatic fever, or uncomplicated pharyngitis. J. Infect. Dis. 166:374-382. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, E. L. 1997. Recent evaluation of antimicrobial resistance in beta-hemolytic streptococci. Clin. Infect. Dis. 24(Suppl. 1):S89-S92. [DOI] [PubMed] [Google Scholar]
- 15.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2002. Erythromycin-resistant group A streptococci in schoolchildren in Pittsburgh. N. Engl. J. Med. 346:1200-1206. [DOI] [PubMed] [Google Scholar]
- 16.Maslow, J. N., A. M. Slutsky, and R. D. Arbeit. 1993. The application of pulsed field gel electrophoresis to molecular epidemiology, p. 563-571. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 17.Musser, J. M., V. Kapur, J. Szeto, X. Pan, D. S. Swanson, and D. R. Martin. 1995. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect. Immun. 6:994-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard. NCCLS document M7-45. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Pérez-Trallero, E., C. Fernández-Mazarrasa, C. García-Rey, E. Bouza, L. Aguilar, J. García de Lomas, F. Baquero, and the Spanish Surveillance Group for Respiratory Pathogens. 2001. Antimicrobial susceptibilities of 1,684 Streptococcus pneumoniae and 2,039 Streptococcus pyogenes isolates and their ecological relationships: results of a 1-year (1998-1999) multicenter surveillance study in Spain. Antimicrob. Agents Chemother. 45:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pérez-Trallero, E., M. J. Marimón, M. Montes, B. Orden, and M. de Pablos. 1999. Clonal differences among erythromycin-resistant Streptococcus pyogenes in Spain. Emerg. Infect. Dis. 5:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Trallero, E., M. Urbieta, M. Montes, I. Ayestarán, and J. M. Marimón. 1998. Emergence of Streptococcus pyogenes strains resistant to erythromycin in Gipuzkoa, Spain. Eur. J. Clin. Microbiol. Infect. Dis. 17:25-31. [DOI] [PubMed] [Google Scholar]
- 22.Scott, R. J. D., J. Naidoo, N. F. Lightfoot, and R. C. George. 1989. A community outbreak of group A beta-haemolytic streptococci with transferable resistance to erythromycin. Epidemiol. Infect. 102:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seppala, H., A. Nissinen, H. Jarvinen, S. Houvinen, T. Henriksson, and E. Herva. 1992. Resistance to erythromycin in group A streptococci. N. Engl. J. Med. 326:292-297. [DOI] [PubMed] [Google Scholar]
- 24.Seppala, H., Q. Y. Nissinen, and P. Huovinen. 1993. Three different phenotypes of erythromycin resistance in group A streptococci in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]
- 25.Seppala, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley, J., D. Linton, M. Desai, A. Efstratiou, and R. George. 1995. Molecular subtyping of prevalent M serotypes of Streptococcus pyogenes causing invasive disease. J. Clin. Microbiol. 33:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka, D., Y. Gyobu, H. Kodama, J. Isobe, S. Hosorogi, Y. Hiramoto, T. Karasawa, and S. Nakamura. 2002. Emm typing of group A streptococcus clinical isolates: identification of dominant types for throat and skin isolates. Microbiol. Immunol. 46:419-423. [DOI] [PubMed] [Google Scholar]
- 28.Tenover, R. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whatmore, A. M., V. Kapur, D. J. Sullivan, J. M. Musser, and M. A. Kehoe. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol. 14:619-631. [DOI] [PubMed] [Google Scholar]