Abstract
In September 2001, a waterborne outbreak of gastroenteritis occurred in eastern France. Of 31 fecal samples from symptomatic individuals, 19 tested positive for Cryptosporidium with two PCRs targeting the Hsp70 and the 18S rRNA genes of Cryptosporidium. Sequencing of the PCR fragments produced sequences identical to that of Cryptosporidium parvum genotype 1.
Organisms of the genus Cryptosporidium are widespread protozoans that develop in epithelial cells lining the digestive and respiratory tracts of vertebrates (8). The species Cryptosporidium parvum is the main agent of human cryptosporidiosis. Molecular typing studies have shown that the species can be differentiated into two major genotypes. The human genotype (genotype 1) primarily infects humans, and the zoonotic genotype (genotype 2) is able to infect humans, ruminants, and a variety of other mammals (5). Aside from C. parvum, the following Cryptosporidium species or genotypes have been identified in immunocompetent and immunocompromised humans: C. meleagridis, C. felis, a dog genotype or species (C. canis), a cervine genotype, and C. muris (4, 9, 11, 13, 17-19, 27).
Transmission of the parasite occurs by the fecal-oral route, through the ingestion of oocysts that are shed with the feces of infected hosts (7). Immunocompetent individuals experience short-term gastroenteritis, while immunocompromised patients may suffer from chronic diarrhea (15). Together with the resistance of oocysts to environmental conditions and chlorine (8), a large parasite reservoir and a low infective dose (6) account for the risk of transmission by water. At least 40 waterborne Cryptosporidium outbreaks have been described, mainly in the United States, Canada, the United Kingdom, and Japan (7, 10). Most outbreaks were caused by C. parvum genotype 1, even in areas where the majority of sporadic cases were caused by C. parvum genotype 2 (27). In European countries other than the United Kingdom, few Cryptosporidium outbreaks have been reported. The first outbreak occurred in 1995 in The Netherlands, but the source of transmission was not identified (3, 24). A second outbreak due to the contamination of a water tank was reported in Italy in 1997 (21). In France, Cryptosporidium was the likely causative agent of a waterborne outbreak that occurred in the south of the country in 1998 (12).
The outbreak investigated in the present paper occurred in 2001 in Dracy le Fort, a county of 1,100 inhabitants in eastern France. On 20 September, health authorities were notified of clustered cases of gastroenteritis. Investigations showed that gastroenteritis cases and complaints about the water quality had been reported by a population served by the same water network since 14 September. Contamination of the public water supply was thus suspected, and an outbreak was declared on 20 September. Cryptosporidium oocysts were identified in water samples collected from the public network in Dracy le Fort on 27 September.
Thirty-one samples from symptomatic patients were collected from 28 September to 2 October 2001 and submitted for detection of parasites. The specimens were examined for Cryptosporidium oocysts by using the modified Ziehl-Neelsen stain (14) and an indirect immunofluorescence assay based on a C. parvum oocyst wall antibody (1). For molecular biology experiments, samples were subjected to a phosphate-buffered saline-ether extraction prior to extraction of DNA with a commercially available kit (QIAamp DNA stool minikit; Qiagen). When this method was applied to fecal specimens which tested negative for C. parvum and which were then seeded with purified oocysts, amplification of the expected PCR fragment was achieved for 10 of 10 samples seeded with 1,000 oocysts per g (data not shown). Two diagnostic PCRs were applied to the specimens collected during the outbreak. One assay targeted the region of the Hsp70 gene obtained with the CPHSP2 primer pair (22). This reaction is based on primers designed for identification of a 361-bp DNA fragment specific to C. parvum. However, primer-annealing regions are identical in C. meleagridis and C. wrairi (23), and this assay may also identify these two species. The second reaction targeted a hypervariable region of the 18S rRNA gene that discriminates among all Cryptosporidium species as well as among multiple C. parvum genotypes. This region is flanked by primer-annealing sequences that are common to all known Cryptosporidium 18S rRNA genes (16, 20). Therefore, a positive ∼435-bp fragment at this locus identifies the genus Cryptosporidium, while secondary analysis of a PCR fragment allows for species and genotype identification. In order to detect a false-negative PCR result, we developed a positive internal control of the CPHSP2 region of the Hsp70 gene. Composite primers containing M13 mp18 phage sequences flanked by the C. parvum Hsp70 primer sequences 5′-AAA TGG TGA GCA ATC CTC TGC GCT ACA CTT GCC AG −3′ and 5′-CTT GCT GCT CTT ACC AGT ACC AGC AGG CGA AAA TC-3′ were used to amplify the corresponding region of the M13 mp18 phage DNA (2). This amplification resulted in a 394-bp DNA sequence containing the same primer-annealing sequences as the diagnostic 361-bp C. parvum Hsp70 region. About 10 copies of this control were added to the amplification reactions as a positive control (Fig. 1).
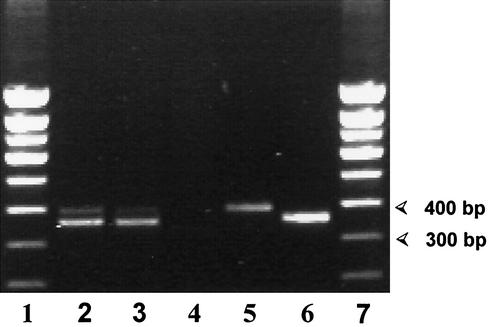
FIG. 1.
Coamplification of the CPHSP2 region of the Cryptosporidium Hsp70 gene together with the corresponding internal control. Ten copies of the internal control were added to the amplification reactions analyzed in lanes 2 to 5. The 394-bp fragment corresponds to the internal control amplification product. The 361-bp fragment corresponds to amplified C. parvum DNA. Lanes 1 and 7, molecular weight markers (Smart ladder; Eurogentec) with apparent molecular sizes given in base pairs. Lanes 2 to 4, PCR products obtained from isolate 15 of the Dracy le Fort outbreak. Two and five microliters of fecal lysates were used as templates of the PCRs displayed in lanes 2 and 3, respectively. For the PCR analyzed in lane 4, 10 μl of the fecal lysate was used as the template; the absence of PCR product is due to the increased amount of PCR inhibitors resulting from the increased fecal template. Lane 5, isolate 1 from the Dracy le Fort outbreak. Amplification of the 394-bp internal control fragment indicates that this result is a true negative. Lane 6, positive control (genomic DNA from purified C. parvum oocysts as a template).
No discrepancy in results was observed between the two PCR methods employed. Altogether, 19 of 31 samples (61.2%) were positive for Cryptosporidium by PCR. Three samples tested negative for Cryptosporidium by microscopy and were positive by PCR. The facts that procedures to avoid false-positive results (work flow organization, a uracyl-N-glycosylase protocol, and negative controls) were utilized and that the two PCR assays targeting distinct genes were positive strongly suggest that the three PCR-positive, microscopy-negative samples were true positives.
Direct sequencing of the PCR products was subsequently performed to identify Cryptosporidium isolates at the species and genotype levels. Primers cphsp2423F and cphsp2764R were used for sequencing the CPHSP2 region of the Hsp70 gene (22). Primers described by Xiao et al. (25) were used for sequencing the polymorphic region of the 18S rRNA gene. At the Hsp70 locus, all 19 samples produced sequences identical to that of C. parvum strain AWS 12 (GenBank accession number AF150827). Comparison of this sequence with homologous regions of C. meleagridis and C. wrairi (GenBank accession numbers AF 221537 and AF 221536, respectively) (23) showed variations at 7 nucleotide positions with C. wrairi and at 14 nucleotide positions with C. meleagridis. In contrast, the sequence from the Dracy le Fort isolate differed at two nucleotide positions with the C. parvum human isolate 497 and at four nucleotide positions with C. parvum bovine isolate 11 (GenBank accession numbers AF 221535 and AF 221528, respectively) (23). At the 18S rRNA locus, sequences were identical to that of C. parvum genotype 1 in the hypervariable regions at nucleotide positions 632 to 667 and 678 to 698 (20). These data indicate that the Dracy le Fort outbreak was due to C. parvum genotype 1.
In the sequencing of the hypervariable18S rRNA locus, a dramatic reduction of signal was observed on both strands after the poly(T) region at nucleotide positions 686 to 696 (20). A similar finding was described previously for this locus in the C. parvum human genotype (26), and it was shown that the signal reduction resulted from heterogeneous copies of the 18S rRNA gene, which differed in the number of T repetitions at nucleotide position 686 and subsequent positions and which generated competing signals. To assess this hypothesis and to ensure that genotype 1 was the sole C. parvum genotype involved in the Dracy le Fort outbreak, PCR products from isolate 7 were cloned, and 10 recombinant clones were selected for sequencing of the insert. All 10 clones showed heterogeneous sequences that were essentially identical to that of C. parvum genotype 1 in the polymorphic regions of nucleotides 632 to 667 and 678 to 698 (20) and that differed in the number of T repetitions at nucleotide position 686 and subsequent positions (20): five clones had 11T repetitions, 2 clones had 10 T repetitions, one clone had 8 T repetitions, and two clones had 12 T repetitions (Table 1). The genotypes with 10 T and 8 T repetitions have been described previously (26). However, the genotype with 12 T repetitions has never been reported. This genotype was confirmed by two separate experiments, as well as by sequencing by an independent laboratory (Genome Express, Grenoble, France). One of the clones with 11 T repetitions also showed a change from C to T at nucleotide position 669, and one of the clones with 12 T repetitions showed a change from T to C at nucleotide position 587 (positions based on the C. parvum reference sequence deposited under GenBank accession number L16996). Interestingly, no reduction of the signals occurred in the poly(T) region upon sequencing of the cloned products, confirming that this phenomenon was caused by the mixture of sequences. Other than C. parvum genotype 1, no clone homologous to the Cryptosporidium species or genotype was obtained.
TABLE 1.
Sequences of the hypervariable regions of the Cryptosporidium 18S rRNA gene from four representative clones of isolate 7
| Clone | Sequencea
|
|
|---|---|---|
| Positions 632 to 667 | Positions 678 to 698 | |
| 1 | ATATAAAATATTTTGATGAATATTTATATAATATTA | TATTACTATTTTTTTTTTT-AGTAT |
| 2 | ATATAAAATATTTTGATGAATATTTATATAATATTA | TATTACTATTTTTTTTTTTTAGTAT |
| 3 | ATATAAAATATTTTGATGAATATTTATATAATATTA | TATTACTATTTTTTTTTT--AGTAT |
| 4 | ATATAAAATATTTTGATGAATATTTATATAATATTA | TATTACTATTTTTTTT----AGTAT |
Sequence numbers based on the sequence of C. parvum 18S rRNA (GenBank accession number L16996).
This was the first waterborne outbreak of Cryptosporidium in France in which a high percentage of the clinical specimens were positive for C. parvum and for which molecular characterization of the isolates was possible. Epidemiologic investigations conducted by public health authorities showed that an estimated 563 individuals presented gastroenteritis symptoms (attack rate, 50.8%). The clinical incidence of patients presenting with diarrhea, vomiting, and abdominal pain spanned a long period. Consumption of tap water before 20 September was the only risk factor associated with the clinical symptoms. Genotyping of Cryptosporidium oocysts recovered from water samples collected in Dracy le Fort could not be performed. However, the identification of C. parvum genotype 1 in all fecal specimens tested, together with the presence of other human pathogens, including group A rotaviruses (20%), enteroviruses (14%), Campylobacter jejuni (14%), enteropathogenic Escherichia coli (12%), and adenoviruses 40 and 41 (6%), indicates that contamination was probably caused by human sewage. Dracy le Fort is located at the end of a water supply network, and no other city served by the same water system was affected by the outbreak. Moreover, water samples collected upstream of Dracy le Fort were negative for Cryptosporidium oocysts and other markers of fecal contamination. These data suggest that the source of the contamination was neither the alluvial water table supply nor the water treatment plant, but, rather, the distribution network upstream of the city of Dracy le Fort. The medical community, public health authorities, and the water treatment industry should be aware of the risk of waterborne cryptosporidiosis in continental Europe and should develop cooperative networks to prevent and treat future outbreaks.
Nucleotide sequence accession number.
The nucleotide sequence for the C. parvum 18S rRNA clone with the 12 T genotype (designated clone 2 in Table 1) has been deposited in the GenBank database under accession number AY 177391.
Acknowledgments
This work was supported by the Institut National de Veille Sanitaire, Saint Maurice, France.
REFERENCES
- 1.Bonnin, A., T. Petrella, J. F. Dubremetz, J. F. Michiels, D. Puygauthier-Toubas, and P. Camerlynck. 1990. Histopathological method for diagnosis of cryptosporidiosis using monoclonal antibodies. Eur. J. Clin. Microbiol. Infect. Dis. 9:664-666. [DOI] [PubMed] [Google Scholar]
- 2.Bretagne, S., J. M. Costa, M. Vidaud, J. Tran, V. Nhieu, and J. Fleury-Feith. 1993. Detection of Toxoplasma gondii by competitive DNA amplification of bronchoalveolar lavage samples. J. Infect. Dis. 168:1585-1588. [DOI] [PubMed] [Google Scholar]
- 3.Caccio, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237-244. [DOI] [PubMed]
- 4.Caccio, S., E. Pinter, R. Fantini, I. Mezzaroma, and E. Pozio. 2002. Human infection with Cryptosporidium felis: case report and literature review. Emerg. Infect. Dis. 8:85-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, D. P. 1999. New insights into human cryptosporidiosis. Clin. Microbiol. Rev. 12:554-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuPont, H. L., C. L. Chappell, C. R. Sterling, P. C. Okhuysen, J. B. Rose, and W. Jakubowski. 1995. The infectivity of Cryptosporidium parvum in healthy volunteers. N. Engl. J. Med. 332:855-859. [DOI] [PubMed] [Google Scholar]
- 7.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 8.Fayer, R., C. A. Speer, and J. P. Dubey. 1997. The general biology of Cryptosporidium, p. 1-41. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Boca Raton, Fla.
- 9.Fayer, R., J. M. Trout, L. Xiao, U. M. Morgan, A. A. Lai, and J. P. Dubey. 2001. Cryptosporidium canis n. sp. from domestic dogs. J. Parasitol. 87:1415-1422. [DOI] [PubMed] [Google Scholar]
- 10.Fricker, C. R., and J. H. Crabb. 1998. Water-borne cryptosporidiosis: detection methods and treatment options. Adv. Parasitol. 40:241-278. [DOI] [PubMed] [Google Scholar]
- 11.Gatei, W., R. W. Ashford, N. J. Beeching, S. K. Kamwati, J. Greensill, and C. A. Hart. 2002. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg. Infect. Dis. 8:204-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyonnet, J. 2000. Pollution des eaux d'alimentation à Sète survenue en Septembre 1998. Rapport final d'enquête épidémiologique. DDASS de l'Hérault, Montpellier, France.
- 13.Guyot, K., A. Follet-Dumoulin, E. Lelièvre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriksen, S., and J. F. Pohlenz. 1981. Staining of cryptosporidia by a modified Ziehl-Neelsen technique. Acta Vet. Scand. 22:594-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter, P. R., and G. Nichols. 2002. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin. Microbiol. Rev. 15:145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type’ from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 19.Pedraza-Diaz, S., C. F. Amar, J. McLauchlin, G. L. Nichols, K. M. Cotton, P. Godwin, A. M. Iversen, L. Milne, J. R. Mulla, K. Nye, H. Panigrahl, S. R. Venn, R. Wiggins, M. Williams, and E. R. Youngs. 2001. Cryptosporidium meleagridis from humans: molecular analysis and description of affected patients. J. Infect. 42:243-250. [DOI] [PubMed] [Google Scholar]
- 20.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pozio, E., G. Rezza, A. Boschini, P. Pezzotti, A. Tamburrini, P. Rossi, M. Di Fine, C. Smacchia, A. Schiesari, E. Gattei, R. Zucconi, and P. Ballarini. 1997. Clinical cryptosporidiosis and human immunodeficiency virus (HIV)-induced immunosuppression: findings from a longitudinal study of HIV-positive and HIV-negative former injection drug users. J. Infect. Dis. 176: 969-975. [DOI] [PubMed] [Google Scholar]
- 22.Rochelle, P. A., D. M. Ferguson, T. J. Handojo, R. De Leon, M. H. Stewart, and R. L. Wolfe. 1997. An assay combining cell culture with reverse transcriptase PCR to detect and determine the infectivity of waterborne Cryptosporidium parvum. Appl. Environ. Microbiol. 63:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulaiman, I. M., U. M. Morgan, R. C. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Asperen, I., C. Stijnen, T. Mank, A. De Boer, J. Groot, G. Medema, P.Ten Ham, J. Sluiters, and M. Borgdorff. 1996. An outbreak investigation of cryptosporidiosis. Report number 215700001. National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
- 25.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao, L., J. R. Limor, L. Li, U. Morgan, R. C. Thompson, and A. A. Lal. 1999. Presence of heterogeneous copies of the small subunit rRNA gene in Cryptosporidium parvum human and marsupial genotypes and Cryptosporidium felis. J. Eukaryot. Microbiol. 46:44S-45S. [PubMed] [Google Scholar]
- 27.Xiao, L., U. M. Morgan, R. Fayer, R. C. Thompson, and A. A. Lal. 2000. Cryptosporidium systematics and implications for public health. Parasitol. Today 16:287-292. [DOI] [PubMed] [Google Scholar]