Abstract
Nocardia veterana is a recently characterized species within the genus Nocardia, and only three human clinical isolates have been reported for this species. We describe a case of ascitic fluid infection in an immunocompromised patient due to N. veterana. To our knowledge, this is the first report of a Nocardia sp. strain from ascitic fluid and the fourth report of N. veterana isolated from human samples. Chemotaxonomic methods showed the strain to belong to the genus Nocardia, and identification to the species level was done by 16S ribosomal DNA gene sequencing. The antibiotic susceptibility profile of N. veterana is reported here for the second time. The strain was deposited in the Collection of the Pasteur Institute and in the Culture Collection of the University of Göteborg (CIP 107497 and CCUG 46576). The corresponding 16S ribosomal DNA gene sequence is available from the GenBank database under accession number AY149599. A phylogenetic analysis was conducted and showed that N. veterana was most closely related to the recently characterized species Nocardia africana rather than to Nocardia vaccinii, as previously reported.
Nocardiosis is a localized or disseminated infection caused by soilborne aerobic actinomycetes. Infection is commonly introduced through the respiratory tract, and pulmonary disease is the most common presentation. Hematogenous dissemination can occur, mainly involving the nervous system and skeletal soft-tissue structures (10, 20). However, some unusual locations of nocardial infection have been reported (5, 16, 17). Nocardiosis is chiefly an opportunistic infection, particularly in patients with underlying malignancies, chronic lung disease, or disorders of cell-mediated immunity, including human immunodeficiency virus (HIV) infection, or those who have received long-term immunosuppressive therapy. Although Nocardia asteroides complex and Nocardia brasiliensis are the species most frequently involved in human infections, numerous new species recently characterized within the genus Nocardia have been reported in human clinical samples. Among these, Nocardia veterana was characterized in 2001, and only three clinical isolates have been reported in the literature (11, 14, 28).
We describe here a case of ascitic fluid infection in an immunocompromised patient due to N. veterana. To our knowledge, this is the first report of a Nocardia species isolated from ascitic fluid and the fourth report of N. veterana isolated from human samples.
Case report.
A 40-year-old man with AIDS was hospitalized in April 2001 in the palliative-care department of the Montpellier University Hospital for a terminal state of lymphoma of serosa discovered 6 months before after a pleural effusion infected by Salmonella sp. His past medical history included HIV infection since 1990 treated by quadritherapy (lamivudine, stavudine, didanosine, and efavirenz) and chronic hepatitis B infection. The initial cytotoxic chemotherapy of the lymphoma, including vinorelbine and prednisone, was not effective, and a new treatment with a combination of cyclophosphamide, oncovin, prednisone, and doxorubicin was instituted. This new treatment did not improve the clinical and biological states of the patient. In April 2001, following an important degradation of the health status of the patient and a repetition of the hemorrhagic ascitic effusions with bacteriology and mycology cultures always negative, the patient was admitted to the palliative-care department. After 3 weeks of hospitalization, the patient presented an episode of fever and a new ascitic effusion. The ascitic fluid was obtained by puncture and sent to the bacteriology laboratory, where the microbiological diagnosis of Nocardia infection was made, but the patient died from acute multivisceral failure before antibiotic treatment was started.
Laboratory identification and antibiotic susceptibility testing. (i) Conventional laboratory identification.
The microbiological diagnosis was made by isolation of a strain of Nocardia after puncture of ascitic fluid. The examination of this sample by direct microscopy showed a large number of leukocytes with a majority of lymphomatous cells and the presence of some polymorphonuclear leukocytes. Direct examination of the fluid did not reveal any bacteria. The specimen was cultured on Trypticase soy agar, Trypticase soy broth, Schaedler broth with vitamin K3 and 0.2% agar (bioMérieux, Marcy l’Etoile, France) and blood-chocolate agar incubated at 37°C in an atmosphere of 5% (vol/vol) CO2. In contrast with previous ascitic fluid samples, which were repeatedly negative, all four of the media were positive after 3 or 4 days of incubation. The puncture fluid yielded the growth of a gram-positive rod-shaped organism in pure culture. It was coryneform and strictly aerobic. Catalase production and urease activity were noted. On the basis of the colony aspect and the Gram morphology, an API CORYNE kit (bioMérieux) was used as recommended by the manufacturer. Esculin was metabolized, and phosphatase alkaline and α-glucosidase activities were noted. However, the profile was incompatible with any species of the genus Corynebacterium. Moreover, the macroscopic and microscopic pictures we observed after subculturing the strain were typical of the genus Nocardia. Cultures on blood agar plates or blood-chocolate agar plates showed a beige substrate mycelium and a white aerial mycelium. By Gram staining, many irregular, gram-positive, branching filamentous rods were noted. Identification of the isolate, named B430, to the genus level was achieved by using standard manual methods. The isolate was affiliated with the genus Nocardia on the basis of nitrate reductase and β-d-galactosidase (o-nitrophenyl-β-d-galactopyranoside) reactions, which were both positive. Biochemical tests revealed that adenine, casein, tyrosine, xanthine, and hypoxanthine were not degraded (Table 1), as previously described for several other nocardial species, such as N. asteroides, Nocardia farcinica, and Nocardia nova (Table 2) (3). The strain was able to grow at 45°C. Enzymatic tests were performed with the API ZYM system (bioMérieux) as recommended by the manufacturer after homogenization of the bacterial suspension by shaking it with glass beads. The following enzymatic activities were detected: alkaline phosphatase, caprylate esterase, leucine arylamidase, acid phosphatase, phosphohydrolase, α-glucosidase, and β-glucosidase. This profile was not specific for any of the previously described nocardial species. However, some of the species most frequently involved in human infections, like N. asteroides and N. nova, which possess valine arylamidase, and N. brasiliensis, which usually displays N-acetyl-β-glucosaminidase and α-mannosidase activities, could be excluded. Finally, all the tests and characteristics performed did not allow us to identify the Nocardia isolate to the species level, since (i) the same biochemical profile is shared by several species within the genus Nocardia and (ii) the enzymatic-activity profile obtained for strain B430 was not specific for a particular Nocardia species. Growth on various carbon sources was not performed. Molecular identification of the isolate B430 was decided on, and it was performed at the Centre d'Identification Moléculaire des Bactéries of the Pasteur Institute in Paris, France, by 16S ribosomal DNA (rDNA) sequencing and phylogenetic analysis.
TABLE 1.
Characteristics of N. veterana isolates compared with the recently characterized nocardial species N. abscessus, N. africana, N. cyriacigeorgici, and N. paucivorans and the phylogenetically related N. vaccinii
| Characteristic | Valuea
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Isolate B430b | N. veterana from Japan | N. veterana DSM 44445T | N. abscessus DSM 44432T | N. africana DSM 44491T | N. cyriacigeorgici DSM 44484T | N. paucivorans DSM 44386T | N. vaccinii DSM 43285T | |
| Biochemical | ||||||||
| Esculin hydrolysis | + | + | ND | − | − | + | − | + |
| Urea hydrolysis | + | ND | ND | + | − | + | + | + |
| Decomposition | ||||||||
| Adenine | − | − | ND | − | − | − | − | − |
| Casein | − | − | ND | − | + | − | − | − |
| Hypoxanthine | − | − | ND | − | − | − | − | − |
| Tyrosine | − | − | ND | − | − | − | − | − |
| Xanthine | − | − | ND | − | − | − | − | − |
| 16S rDNA similarity with isolate B430 (%) | 99.7 | 100 | 96.4 | 99.2 | 96.7 | 96.8 | 98.0 | |
| Clinical isolate source | Ascitic fluid | Mycetoma | BAL | Abcesses | Sputum | Bronchial secretions | Sputum, CSF | −c |
| Reference | This study | 14 | 11 | 31 | 12 | 32 | 30 | 7 |
+, positive for characteristic; −, negative for characteristic; ND, not determined; BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid.
N. veterana isolate B430 (equivalent to CIP 107497 and CCUG 46576).
Currently not reported from human clinical isolate.
TABLE 2.
Comparison of phenotypic properties of N. veterana isolates and of type strains of more common Nocardia species encountered in clinical samplesa
| Test | Resulta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| N. veteranab | N. asteroides ATCC 19247T | N. farcinica ATCC 3318T | N. nova JCM 6044T | N. brasiliensis ATCC 19296T | N. otitidiscaviarum NCTC 1934T | N. carnea DSM 43397T | N. tranvalensis DSM 43405T | N. pseudobrasiliensis ATCC 51512T | |
| Biochemical | |||||||||
| Esculin hydrolysis | + | + | + | + | + | + | + | + | + |
| Nitrate reduction | + | + | + | + | + | + | + | + | − |
| Urea hydrolysis | + | + | + | + | + | + | − | + | + |
| Decomposition (% [wt/vol]) | |||||||||
| Adenine (0.4) | − | − | − | − | − | − | − | − | + |
| Casein (1.0) | − | − | − | − | + | − | − | − | + |
| Hypoxanthine (0.4) | − | − | − | − | + | + | − | + | + |
| Tyrosine (0.5) | − | − | − | − | + | − | − | − | + |
| Xanthine (0.4) | − | − | − | − | − | + | − | − | − |
| Assimilation (% [wt/vol]) | |||||||||
| d-Mannitol (1.0) | + | + | − | + | + | + | + | + | + |
| l-Rhamnose (1.0) | + | − | + | − | − | − | − | + | − |
| d-Sorbitol (1.0) | + | − | − | + | − | − | + | + | + |
| Sodium acetate (0.1) | + | + | + | + | + | + | + | + | + |
| Sodium citrate (0.1) | + | + | − | − | + | − | − | − | + |
| Growth at 45°C | + | − | + | − | + | + | − | − | − |
Data for type strains of the more common Nocardia species encountered in clinical specimens were derived from Hamid et al. (12) and Yassin et al. (31, 32).
Data for N. veterana were either from the clinical isolate B430 (equivalent to CIP 107497 and CCUG 46576) (this study) or from the Japanese isolate (14) or the type strain, DSM 44445T (11).
+, positive for characteristic; −, negative for characteristic.
(ii) Antibiotic susceptibility testing.
Antibiotic susceptibility testing was performed by disk diffusion assay on Mueller-Hinton blood agar plates incubated for 48 h at 37°C in an atmosphere enriched with 5% CO2 (2). The results were interpreted according to the recommendations of the Comité Français de l'Antibiogramme (25). Isolate B430 was susceptible to amoxicillin, amoxicillin-clavulanic acid, imipenem, trimethoprim-sulfamethoxasole, amikacin, and erythromycin. It was resistant to cefotaxime, gentamicin, tobramycin, pefloxacin, ciprofloxacin, and vancomycin. This antibiotic susceptibility profile was not characteristic of a particular Nocardia species, and therefore it could not contribute to the identification of the isolate, as previously reported for some nocardial species (27, 29). However, it seemed to indicate that the strain was not affiliated with N. brasiliensis or Nocardia otitidiscaviarum, on the basis of the gentamicin resistance observed, or to N. asteroides, considering its resistance to tobramycin. N. nova identification could also be excluded because of the results obtained for amoxicillin-clavulanic acid and tobramycin (3, 12).
(iii) Molecular identification of the organism and phylogenetic analysis.
Strain B430 was cultured overnight in brain heart infusion broth. After centrifugation, the cells were suspended in 500 μl of 1× Tris-EDTA with 150 μl of lysozyme (100 mg/ml) and 15 μl of mutanolysine (5 U/ml). After incubation at 37°C overnight, 150 μl of proteinase K (20 mg/ml) was added, and the suspension was incubated at 37°C for 3 h; 150 μl of sodium dodecyl sulfate (25% [wt/vol]) was added, and the mixture was incubated at 55°C for 60 min. DNA was extracted and purified with the Wizard Genomic DNA Purification kit (Promega, Madison, Wis.). Amplification of the rrs gene encoding 16S rRNA was performed as follows: the extracted DNA was amplified by using the universal 16S rRNA primers A (corresponding to positions 8 to 28 of Escherichia coli 16S rRNA) (9) and rJ (corresponding to the complement of positions 1510 to 1492) (13). The amplification reaction was carried out in a final volume of 100 μl, with 1 μl of DNA template; 20 pmol of each primer; 200 μM (each) dATP, dTTP, dGTP, and dCTP; 50 μM MgCl2; 2.5 U of Taq polymerase; and 10 μl of 10× reaction buffer (Amersham International, Amersham, England). Initial denaturation was carried out for 4 min at 94°C. Thirty-five cycles of amplification were performed in a DNA Thermal Cycler 9700 (PE Applied Biosystems, Foster City, Calif.). Each cycle consisted of three steps: denaturation at 94°C for 1 min, annealing at 49°C for 1 min, and elongation at 72°C for 2 min. The final extension was performed at 72°C for 5 min. Amplification products were detected by electrophoresis on a 0.8% (wt/vol) agarose gel in TBE buffer (0.089 M Tris, 0.089 M borate, 0.018 M EDTA), with a 1-kb DNA Ladder (Gibco BRL, Gaithersburg, Md.) as a molecular size marker. The amplified product was sequenced by Genome Express (Meylan, France) with three primers, E, rE, and D, in conserved regions of E. coli 16S rRNA. Primer E corresponds to positions 787 to 806, rE corresponds to the complement of the E primer, and D corresponds to positions 519 to 536 (numbering according to Brosius et al. [4]). The modules EditSeq and SeqMan were used to assemble ∼1,300 bp of the rrs gene, and BLAST (Basic Local Alignment Search Tool) program analysis (http://www.ncbi.nlm.nih.gov:80/BLAST/) of the sequence gave the maximum identity (100%) with the 16S rDNA sequence of the N. veterana type strain, M157222T (equivalent to DSM44445T and NRRL B-24136T). A second strain of N. veterana has been isolated in Japan; its 16S rDNA sequence is not available in the data banks, but a partial sequence of 942 bp was provided by R. Kano (14). Alignment by LALIGN revealed 99.7% homology between the sequences obtained from the Japanese strain and isolate B430. The third isolate of N. veterana was not included in this study, but a 16S rRNA gene identity of 99.1% was observed by the authors with the sequence of the type strain (28). High 16S rDNA sequence similarity values were observed with other species belonging to the genus Nocardia, particularly with Nocardia africana isolates (99.2 and 98.9% for strains with the accession numbers AF277198 and AF302232 and accession numbers AF302230 and AF302231, respectively) and with the Nocardia vaccinii type strain, DSM 43285T (accession number Z36927; 98%).
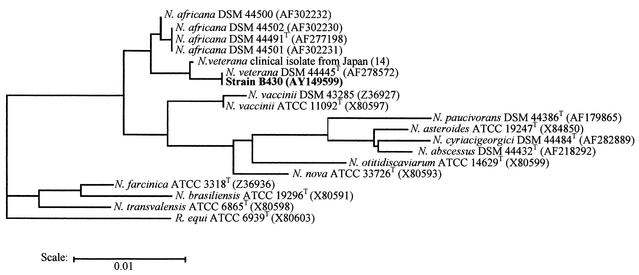
In two previous studies (11, 14), N. veterana was shown to be phylogenetically most closely related to N. vaccinii. When N. africana was characterized (12), it was also described as the species most closely related to N. vaccinii. However, the phylogenetic relationship between N. veterana and N. africana could not be established, since the two species were both characterized in 2001. Therefore, we conducted a phylogenetic analysis as follows. Modules of the Lasergene software (DNAstar, London, United Kingdom) were used in all computations on a Macintosh G4 (Apple Computer, Cupertino, Calif.). Sequences related to that of strain B430 were searched in GenBank using the BLAST program. The closest sequences were imported from the Ribosomal Database Project II database (http: //rdp.cme.msu.edu/htlm/) and aligned with that of strain B430 (22). Pairwise evolutionary distances were computed using the Jukes- Cantor equation, and a neighbor-joining tree (24) was drawn (Fig. 1). The topology of the phylogenetic tree confirmed that (i) isolate B430 and the two other strains of N. veterana grouped together, (ii) N. africana was the species most closely related to N. veterana, and (iii) N. vaccinii was more deeply branched than N. africana in the phylogenetic tree.
FIG. 1.
Phylogenetic tree based on 16S rDNA gene sequences indicating the relationships between clinical isolate B430, N. veterana strains, related species N. africana and N. vaccinii, and other type species of the genus Nocardia. The sequence of Rhodococcus equi was used as an outgroup. The scale bar indicates 0.01 substitution per nucleotide position. Accession numbers are given in parentheses.
Finally, 16S rDNA sequence analysis and phylogenetic investigations indicated that we had isolated the fourth representative strain of the species N. veterana. Moreover, biochemical characteristics observed for isolate B430 were consistent with those of the Japanese isolate, although they are not specific for the species (Table 1). Comparison with the N. veterana type strain, DSM44445T, could not be performed, since it was not tested for the decomposition of the substrates we had analyzed. Phenotypic properties that distinguish validly described Nocardia species have been largely reported before (for a recent review, see reference 12). Therefore, we chose to summarize in Table 1 the characteristics of the N. veterana isolates for which they are available, compared to the recently described species Nocardia abscessus, N. africana, Nocardia cyriacigeorgici, and Nocardia paucivorans and to the phylogenetically related species N. vaccinii. A comparison of the phenotypic properties that distinguish N. veterana from the Nocardia species most commonly encountered in clinical samples is given in Table 2.
Discussion.
Systemic Nocardia infections occur frequently in immunocompromised patients, particularly in patients with lymphoreticular neoplasms (leukemia and lymphoma). Therapy with cytotoxic agents, alone or in combination with steroids, is also an important risk factor. In spite of deep immunodepression, nocardiosis is an uncommon complication in patients with AIDS. This could result from the therapeutic efficiency against Nocardia spp. of trimethoprim-sulfamethoxazole, used in primary prophylaxis of toxoplasmosis in patients with HIV infection. On the other hand, the frequency of nocardiosis is probably underestimated due to the difficulties of clinical and biological diagnosis of nocardiosis and the fact that the infection is not considered a sickness indicative of AIDS. With regard to the medical history, the patient presented an important risk for developing nocardiosis. The ascites fluid infection could be due to the hematogenous spread of Nocardia sp. from the lungs. Indeed, according to the literature, because lung lesions may be small or obscured by underlying pulmonary abnormalities, infections with no known primary site may be of pulmonary origin (1). Moreover, the pleural effusion infected by Salmonella sp., supported by the report of some coinfections of the lung by Salmonella sp. and Nocardia sp., favors the pulmonary portal of entry for the infection (6). Although many unusual nocardial infections have been reported in the literature (5, 16, 17), including two cases of peritonitis (21, 23), no case of ascites fluid infection caused by Nocardia sp. has been reported. We describe here the first case of ascitic fluid infection caused by Nocardia sp., subsequently identified as N. veterana. The N. veterana strain displayed relative susceptibility to antibiotics, particularly to trimethoprim-sulfamethoxazole, which is the drug of choice for the treatment of nocardiosis (15). However, the patient died rapidly before any treatment was started. The nocardial infection was probably not the direct cause of death but may have accelerated it.
Identification of the nocardial isolate was complicated in this case by the initial coryneform aspect of the strain, probably due to fragmentation of the mycelium into rod-shaped elements. This microscopic aspect is in contrast to those observed for at least two of the previously published isolates of N. veterana. Indeed, the type strain was described as having a quite stable mycelium that did not fragment, and the Japanese isolate was described as being rod shaped to coccoid (11, 14). The macroscopic aspects of the N. veterana isolates could not be compared, since the strains were cultured on different media and therefore displayed various characteristics: beige substrate mycelium, scant dirty-white aerial mycelium, and a yellowish reverse side of the culture for the type strain on glucose-yeast extract-malt extract agar medium (DSMZ no. 65); orange, wrinkled colonies for the Japanese isolate on Sabouraud medium; and beige substrate mycelium and white aerial mycelium for isolate B430 on blood agar or blood-chocolate agar plates (11, 14). The biochemical characteristics of the isolate were not discriminating enough to allow identification to the species level, whereas the enzymatic activity profile, together with the antibiotic susceptibility profile, was not consistent with validly described species within the genus Nocardia. Moreover, most of these data have not been reported for the previously described N. veterana isolates, precluding comparison between strains. Antimicrobial susceptibility data for N. veterana were reported only once before; these data were consistent with those obtained for strain B430, with the exception of amoxicillin-clavulanate and gentamicin, reported to have intermediate and susceptible MICs, respectively, for the isolate of N. veterana reported by Wellinghausen et al. (28).
Finally, identification of isolate B430 as N. veterana could be accomplished only after 16S rDNA gene sequencing. The difficulties in identifying the isolate to the species level can be explained by the numerous taxonomic changes that occurred recently in the genus Nocardia. Indeed, five new Nocardia species recovered from human samples have been characterized since 2000: N. abscessus (31), N. africana (12), N. cyriacigeorgici (32), N. paucivorans (30), and N. veterana (11). Most of them are implicated in infectious processes: strains belonging to N. africana, N. cyriacigeorgici, and N. paucivorans were isolated from sputa or bronchial secretions of patients with pulmonary infections; N. paucivorans was also reported to have been isolated from the cerebrospinal fluid of a patient with relapse of cerebral nocardiosis (10) and from the central nervous system biopsy specimen of a patient with brain abscesses (28), and N. abscessus was isolated from various abscesses (31) and from a pericardial aspirate (28). Little is known about N. veterana, since only three isolates have been published. The first strain was recovered from bronchial lavage fluid and supported the primary description of the species, but the strain was thought by the authors to be of no clinical significance (11). The second isolate was recovered in a subcutaneous biopsy of a mycetoma (14), and the third was cultured from a respiratory specimen from a patient with pulmonary nocardiosis (28). Consequently, the infectious spectrum of N. veterana has to be considered relatively large.
Most often, traditional identification methods are inadequate for the identification of these recently characterized species, for which only a few isolates are reported in the literature and in the databases. Only biochemical and enzymatic profiles can be determined in routine practice, and they are not discriminating enough to identify these new species within the genus Nocardia. Moreover, the conventional identification to the genus level based on chemotaxonomic characteristics—biochemical tests and enzymatic activities—is time-consuming and relatively slow (1 to 2 weeks). In these cases, molecular methods represent a simple and rapid means to make or to complete identification to the species level. The available molecular methods allow identification to the genus or species level. They are based on 16S rDNA amplification followed by restriction endonuclease analysis (8, 19), on ribotyping (18), or on the Hsp65-restriction fragment length polymorphism technique (26). As previously emphasized by other authors, identification of clinical isolates of Nocardia to the species level is important, not only for defining the spectrum of disease produced by these species, but also for predicting antimicrobial susceptibility (8). In the present case, identification of the isolate as N. veterana was achieved only after 16S rDNA gene sequencing. Therefore, we concluded that 16S rDNA-based identification is a useful tool for the identification of the recently characterized species within the genus Nocardia for which the sequences for the 65-kDa heat-shock protein are not yet available.
In conclusion, we report the first case of ascitic fluid infection caused by Nocardia sp. and the fourth clinical strain of N. veterana. Since only three isolates of N. veterana have been described, the differentiation of N. veterana from the other Nocardia species was not achievable on the basis of the phenotypic traits usually tested for nocardial identification but could be achieved only by 16S rDNA gene sequencing. The antibiotic susceptibility profile of N. veterana is reported here for the second time, but it was not helpful for the identification of the isolate to the species level.
Nucleotide sequence accession number.
A partial 16S rDNA sequence of 1,272 bp for strain B430 was deposited in the GenBank database under accession number AY149599. The strain was deposited in the Collection of the Pasteur Institute (CIP 107497) and the Culture Collection of the University of Göteborg (CCUG 46576).
Acknowledgments
We are very grateful to R. Kano for his help with collecting the data about the N. veterana strain isolated in Japan and to C. Bizet for her help with depositing the strains in the CIP and CCUG collections. We thank E. Jumas-Bilak for critical reading of the manuscript and help with phylogenetic analysis.
REFERENCES
- 1.Agarwal, A., S. K. Mishra, and A. K. Sharma. 1998. Acute suppurative thyroiditis with demonstrable distant primary focus: a report of two cases. Thyroid 8:399-401. [DOI] [PubMed] [Google Scholar]
- 2.Ambaye, A., P. C. Kohner, P. C. Wollan, K. L. Roberts, and F. R. Cokerill. 1997. Comparison of agar dilution, broth microdilution, disk diffusion, E-test, and BACTEC radiometric methods for antimicrobial susceptibility testing of clinical isolates of the Nocardia asteroides complex. J. Clin. Microbiol. 35:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boiron, P., F. Provost, and B. Dupont (ed.). 1993. Laboratory methods for the diagnosis of nocardiosis. Institut Pasteur, Paris, France.
- 4.Brosius, J., M. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from E. coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrière, C., H. Marchandin, J. M. Andrieu, A. Vandome, and C. Perez. 1999. Nocardia thyroiditis: unusual location of infection. J. Clin. Microbiol. 37:2323-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casado, J. L., E. Navas, B. Frutos, A. Moreno, P. Martin, J. M. Hermida, and A. Guerrero. 1997. Salmonella lung involvement in patients with HIV infection. Chest 112:1197-1201. [DOI] [PubMed] [Google Scholar]
- 7.Chun, J., and M. Goodfellow. 1995. A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:240-245. [DOI] [PubMed] [Google Scholar]
- 8.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenblatter, M., U. Disko, G. Stoltenburg-Didinger, H. Scherubl, K. P. Schaal, A. Roth, R. Ignatius, M. Zeitz, H. Hahn, and J. Wagner. 2002. Isolation of Nocardia paucivorans from the cerebrospinal fluid of a patient with relapse of cerebral nocardiosis. J. Clin. Microbiol. 40:3532-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurtler, V., R. Smith, B. C. Mayall, G. Potter-Reinemann, E. Stackebrandt, and R. M. Kroppenstedt. 2001. Nocardia veterana sp. nov., isolated from human bronchial lavage. Int. J. Syst. E vol. Microbiol. 51:933-936. [DOI] [PubMed] [Google Scholar]
- 12.Hamid, M. E., L. Maldonado, G. S. Sharaf Eldin, M. F. Mohamed, N. S. Saeed, and M. Goodfellow. 2001. Nocardia africana sp. nov., a new pathogen isolated from patients with pulmonary infections. J. Clin. Microbiol. 39:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janvier, M., and P. A. D. Grimont. 1995. The genus Methylophaga, a new line of descent within phylogenetic branch γ of Proteobacteria. Res. Microbiol. 146:543-550. [DOI] [PubMed] [Google Scholar]
- 14.Kano, R., Y. Hattori, N. Murakami, N. Mine, M. Kashima, R. M. Kroppenstedt, M. Mizoguchi, and A. Hasegawa. 2002. The first isolation of Nocardia veterana from a human mycetoma. Microbiol. Immunol. 46:409-412. [DOI] [PubMed] [Google Scholar]
- 15.Khardori, N., R. Shawar, R. Gupta, B. Rosenbaum, and K. Rolston. 1993. In vitro antimicrobial susceptibilities of Nocardia species. Antimicrob. Agents Chemother. 37:882-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kontoyiannis, D. P., K. L. Jacobson, E. E. Whimbey, K. V. Rolston, and I. I. Raad. 2000. Central venous catheter-associated Nocardia bacteremia: an unusual manifestation of nocardiosis. Clin. Infect. Dis. 31:617-618. [DOI] [PubMed] [Google Scholar]
- 17.Lanotte, P., S. Watt, R. Ruimy, P. Boiron, A. Robier, and R. Quentin. 2001. Nocardia farcinica infection of a cochlear implant in an immunocompetent boy. Eur. J. Clin. Microbiol. Infect. Dis. 20:880-882. [DOI] [PubMed] [Google Scholar]
- 18.Laurent, F., A. Carlotti, and P. Boiron. 1996. Ribotyping: a tool for taxonomy and identification of the Nocardia asteroides complex species. J. Clin. Microbiol. 34:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurent, F. J., F. Provost, and P. Boiron. 1999. Rapid identification of clinically relevant Nocardia species to genus level by 16S rRNA gene PCR. J. Clin. Microbiol. 37:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerner, P. I. 1996. Nocardiosis. Clin. Infect. Dis. 22:891-905. [DOI] [PubMed] [Google Scholar]
- 21.Liassine, N., and K. Rahal. 1992. Peritonitis caused by Nocardia farcinica in a patient undergoing continuous ambulatory peritoneal dialysis. Arch. Inst. Pasteur Alger. 58:95-102. [PubMed] [Google Scholar]
- 22.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez, M., M. Forné, J. Viver, J. Lite, and J. Garau. 1994. Nocardia asteroides peritonitis in a patient with cirrhosis and human immunodeficiency virus infection. Clin. Infect. Dis. 18:1010-1011. [DOI] [PubMed] [Google Scholar]
- 24.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 25.Soussy, C. J., G. Carret, J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M.-H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, and J. Sirot. 2000. Antibiogram Committee of the French Microbiology Society. Report 2000-2001. Pathol. Biol. (Paris) 48:832-871. [PubMed] [Google Scholar]
- 26.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. C. Jost, Jr., Z. Blacklock, J. L. Gibson, and R. J. Wallace, Jr. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallace, R. J., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 23:19-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wellinghausen, N., T. Pietzcker, W. V. Kern, A. Essig, and R. Marre. 2002. Expanded spectrum of Nocardia species causing clinical nocardiosis detected by molecular methods. Int. J. Med. Microbiol. 292:277-282. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, R. W., V. A. Steingrube, B. A. Brown, Z. Blacklock, K. C. Jost, Jr., A. McNabb, W. D. Colby, J. R. Biehle, J. L. Gibson, and R. J. Wallace, Jr. 1997. Recognition of a Nocardia transvalensis complex by resistance to aminoglycosides, including amikacin, and PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 35:2235-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yassin, A. F., F. A. Rainey, J. Burghardt, H. Brzezinka, M. Mauch, and K. P. Schaal. 2000. Nocardia paucivorans sp. nov. Int. J. Syst. E vol. Microbiol. 50:803-809. [DOI] [PubMed] [Google Scholar]
- 31.Yassin, A. F., F. A. Rainey, U. Mendrock, H. Brzezinka, and K. P. Schaal. 2000. Nocardia abscessus sp. nov. Int. J. Syst. E vol. Microbiol. 50:1487-1493. [DOI] [PubMed] [Google Scholar]
- 32.Yassin, A. F., F. A. Rainey, and U. Steiner. 2001. Nocardia cyriacigeorgici sp. nov. Int. J. Syst. E vol. Microbiol. 51:1419-1423. [DOI] [PubMed] [Google Scholar]