Abstract
Species of the Burkholderia cepacia complex cause chronic and life-threatening infections in persons with cystic fibrosis. Epidemic strains infect multiple patients, reside primarily in genomovar III, and have an apparent enhanced capacity for human infection and/or interpatient transmission. By using subtractive hybridization, a novel insertion element, designated IS1363, was identified in epidemic strain PHDC, known to infect many cystic fibrosis patients in the mid-Atlantic region of the United States. IS1363 was also found in most isolates of the ET12 lineage, responsible for infecting large numbers of patients in Ontario, Canada, and the United Kingdom. Southern blot analysis demonstrated that whereas multiple copies of IS1363 were present in strain PHDC, only one copy was present in ET12 isolates. IS1363 was used to probe a collection of 943 B. cepacia complex isolates, representing all nine genomovars, recovered from 761 cystic fibrosis patients or the natural environment. IS1363 was not found in other genomovar III strains and, with the exception of B. ambifaria, was absent from other B. cepacia complex species. Genotyping analyses of all IS1363-positive isolates demonstrated that strain PHDC was more widely distributed in the United States than previously appreciated; 212 cystic fibrosis patients in 24 states were identified as being infected with PHDC.
The Burkholderia cepacia complex consists of nine closely related bacterial species (or genomovars) (8, 35). Although generally not pathogenic for healthy persons, these species can cause life-threatening pulmonary infection in persons with cystic fibrosis (17). Most of the species in this group have received formal binomial designations, including B. multivorans, B. stabilis, B. vietnamiensis, B. ambifaria, B. anthina, and B. pyrrocinia. The name B. cepacia will be reserved for genomovar I, while genomovars III and VI await formal names pending the identification of distinguishing phenotypes. Recent analyses of large numbers of isolates recovered from cystic fibrosis patients indicate that although all nine species are capable of causing infection in cystic fibrosis, some are much more commonly involved than others. In fact, in the United States, approximately half of infected cystic fibrosis patients harbor genomovar III (19); in Canada and parts of Europe, the proportion is even higher (1, 32).
A number of studies employing isolate genotyping analyses have also demonstrated the existence of specific B. cepacia complex strains, or clonal lineages, that infect multiple cystic fibrosis patients. Such so-called epidemic strains reside primarily (although not exclusively) in genomovar III. The best studied of these is strain ET12, which predominates among cystic fibrosis patients in Ontario, Canada, and the United Kingdom (12, 13, 26). Another genomovar III strain, designated PHDC, has been described more recently as the dominant strain among infected cystic fibrosis patients in the mid-Atlantic region of the United States, where it has been endemic for at least the past 20 years (5). Isolates belonging to this lineage have also been found recently in agricultural soil in New York State (20). Another genomovar III strain, referred to as the Midwest clone, infects multiple cystic fibrosis patients in Ohio and Michigan (7, 14).
The factors that account for the apparent enhanced capacity of these strains for pulmonary infection and/or interpatient spread in cystic fibrosis remain to be elucidated. An approach that may be useful in this regard involves the use of subtractive hybridization, a method by which DNA that is present in one bacterial genome (the tester strain) but absent in another (the driver strain) is identified (3). In ongoing studies, we have used this approach to identify genetic elements present in certain epidemic B. cepacia strains and absent from strains infrequently recovered from cystic fibrosis patients (unpublished data).
In the study reported here, we describe the use of subtractive hybridization to identify a novel putative insertion element, designated IS1363, which is found in B. cepacia genomovar III epidemic strains PHDC and ET12. Screening of a large collection of sputum isolates from cystic fibrosis patients for the presence of IS1363 together with isolate genotyping analyses indicated that strain PHDC was much more common and widely distributed among cystic fibrosis patients in the United States than was previously appreciated.
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
Isolates were recovered from sputum cultures from cystic fibrosis patients and sent by referring clinical microbiology laboratories to the Burkholderia cepacia Research Laboratory and Repository at the University of Michigan. These included B. cepacia genomovar III isolates AU1482 and PC8, which represent strain PHDC (5), and PC184, representing the Midwest clone (7, 14). Several sputum isolates representing the ET12 strain were kindly provided by D. Henry (Vancouver, Canada) (7). Other isolates, including B. cepacia genomovar III strains ES0222 and ES0263, were recovered from agricultural soil as described previously (24). P. Vandamme (Ghent, Belgium) kindly provided additional isolates from soil and cystic fibrosis patients. Species identification (i.e., assignment to one of the nine species [or genomovars] within the B. cepacia complex) was performed by using ribosomal DNA (rDNA)- and recA-directed PCR assays as previously described (18, 22). Isolates were recovered from frozen stocks maintained at −80°C by aerobic growth on Mueller-Hinton broth (Becton Dickinson) supplemented with 1.8% (wt/vol) agar following incubation overnight at 32°C.
Escherichia coli TOP10 (Invitrogen), XL1-Blue MRF′, and XLOLR (Stratagene) were used for cloning, genomic library propagation, and phagemid excision, respectively. E. coli strains were grown in Luria-Bertani (LB) medium supplemented with appropriate antibiotics. Kanamycin and tetracycline (Sigma) were used at 50 μg/ml and 12.5 μg/ml, respectively, as needed.
Subtractive hybridization and DNA sequence analysis.
Subtractive hybridization was performed essentially as described by Akopyants et al. (3), except that the amounts of both tester and driver DNA were doubled and HaeIII was used instead of AluI. AU1482 was used as the tester DNA, and a pool of equal amounts of ES0222 and ES0263 DNA was used as the driver. A hybridization temperature of 65°C was used. After the second round of PCR, the subtractive hybridization products were cloned into pCR-Blunt II-TOPO (Invitrogen) and transformed into E. coli TOP10. For DNA sequence determination, plasmid DNA was purified from host bacteria by using the Qiagen mini spin kit (Qiagen). DNA sequence was determined by using an Applied Biosystems 3700 DNA sequencer (PE Applied Biosystems) with the BigDye terminator cycle sequencing ready reaction kit. Sequence analyses were performed by using EditSeq (DNAStar).
Genomic library construction and screening, Southern blot, and dot blot assays.
Twenty micrograms of genomic DNA from strain PC8 was partially digested by incubating with 0.4 U of Sau3AI at 37°C for 30 min. DNA fragments between 5 kb and 10 kb in size were purified by using the Qiagen gel extraction kit (Qiagen), and a genomic library was constructed in ZAP Express (Stratagene) according to the manufacturer's instructions.
The PC8 genomic library was screened by transfecting E. coli XL1 Blue MRF′ with approximately 5,000 PFU/plate (132-mm diameter). After incubation at 37°C overnight, phage DNA was transferred onto positively charged nylon membranes (Roche) and denatured. Southern blot analysis was performed by transferring 2 μg of EcoRV-digested genomic DNA fragments from a 0.8% agarose gel onto a positively charged nylon membrane. For dot blot analysis, DNA was prepared from each bacterial isolate, and approximately 1 μg was applied to a nylon membrane as described previously (19). For Southern blot, dot blot, and library screening, a digoxigenin-labeled 235-bp DNA fragment within IS1363 was generated by PCR (below) by employing the PCR DIG probe synthesis kit (Roche) and used as a probe. After hybridization at 42°C overnight in Easy Hyb solution (Roche), membranes were washed twice (15 min each wash) in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 55°C prior to detection.
IS1363-specific PCR and reverse transcription-PCR.
The oligonucleotide primers and conditions for the PCR assays used in this study are provided in Table 1. Template DNA was prepared from bacterial isolates by using a colony lysis method as described previously (7). IS1363-specific PCR (IS-PCR) was performed with oligonucleotide primers (P1 and P2) that target sequences within IS1363. PCR was performed in 25-μl reactions containing 2 μl of bacterial cell lysate (7), 1 U of Taq polymerase (Gibco-BRL), 1.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates, 0.4 μM each primers P1 and P2, and 1× PCR buffer (Gibco-BRL). Amplification was carried out by using a PTC-100 programmable thermal cycler (MJ Research). The resulting 235-bp amplicon was visualized after gel electrophoresis and staining with ethidium bromide. Genomic DNAs prepared from strains PC8 and PC184 were used as positive and negative PCR controls, respectively.
TABLE 1.
Primers and PCR conditions
| Assay | Primer | Primer sequence | PCR conditions | Amplicon size (bp) |
|---|---|---|---|---|
| IS-PCR | P1 P2 | 5′-GCTTAATAGGATGGTCAG-3′ 5′-TCCATGACCACCGTACAACTC-3′ | 95°C for 2 min, 1 cycle; 95°C for 45 s, 55°C for 45 s, 72°C for 45 s, 30 cycles; 72°C for 10 min, 1 cycle | 235 |
| Full-length PCR | P1 P5 | As above 5′ -TCTATGGTCAGCCCGATTTTTG-3′ | 98°C for 3 min, 1 cycle; 98°C for 30 s, 56°C for 30 s, 72°C for 2 min, 30 cycles; 72°C for 10 min, 1 cycle | 1,363 |
| RT-PCR | P3 P4 | 5′-TCCGGCAAGGCAACAAGAACGAT-3′ 5′-GCCCGACCGACGCCGCAAACTG-3′ | 95°C for 2 min, 1 cycle; 95°C for 45 s, 61°C for 45 s, 72°C for 45 s, 40 cycles; 72°C for 10 min, 1 cycle | 495 |
For PCR amplification of the entire IS1363 sequence, primers P1 and P5 were used. The PCR mixture and thermal cycler were as described previously for the IS-PCR (above) except that ThermalAce DNA polymerase (Invitrogen) was used.
Reverse transcription-PCR (RT-PCR) was performed by using the Superscript One-Step RT-PCR kit (Invitrogen). RNA was isolated from strains AU1482 and PC8 grown under in vitro growth conditions (above) by using the SNAP total RNA isolation kit (Invitrogen). Oligonucleotide primers P3 and P4, targeting sequences within the IS1363 open reading frame (ORF), were used. RT-PCR was carried out in a 50-μl reaction containing 250 ng of RNA template, 0.2 μM each oligonucleotide primer, and 1 μl of RT-Platinum Taq mix. cDNA was synthesized at 50°C for 30 min followed by the amplification program detailed in Table 1. Absence of genomic DNA in RNA preparations was verified by omitting the RT-Platinum Taq mix and substituting 2 U of Taq DNA polymerase in the reaction.
Isolate genotyping analyses.
Many bacterial isolates included in this study had been genotyped in the course of previous studies by using either random amplified polymorphic DNA typing (RAPD), rep-PCR typing by using a BOX-A1R primer (BOX-PCR), macrorestriction analysis by using SpeI digestion and pulsed-field gel electrophoresis (PFGE), or multilocus restriction typing as described previously (5, 7, 9, 20). For RAPD, BOX-PCR, and PFGE, gel images were digitized by using a GelDoc2000 gel analyzer (Bio-Rad), stored as TIF files, and analyzed by using Molecular Analyst Fingerprinting Plus software (Bio-Rad), also as described previously (5). Similarity matrices of densitometric curves of the gel tracks were calculated by using Pearson's product-moment correlation coefficient, and cluster analyses were performed by using the unweighted pair group method with arithmetic averages. Isolates clustering with similarity coefficients of 80%, 70%, and 65% for RAPD, BOX-PCR, and PFGE analyses, respectively, were considered the same strain (9).
Nucleotide sequence accession numbers.
The nucleotide sequences of the IS1363 elements cloned from B. cepacia genomovar III PHDC strain PC8 and PCR-amplified from B. ambifaria strain ES0607 were deposited in GenBank under accession numbers AY148030 and AY148031, respectively.
RESULTS
Subtractive hybridization and library screening.
Subtractive hybridization was performed by using genomic DNA from AU1482 as the tester DNA and a pool of DNAs from non-PHDC genomovar III strains ES0222 and ES0263 as the driver DNA. Several hundred clones were obtained in the resultant subtraction library. PCR analysis by using vector-specific primers identified several clones with different sizes of inserts. From among these, eight distinct clones were selected for DNA sequence determination. Based on these sequences, insert-specific PCR assays were designed. These assays were used with tester or driver DNA as the template to confirm that six of the eight clones contained tester-specific DNA. The DNA inserts of these six clones ranged from 159 to 318 bp in length and were found to have a G+C content ranging from 47 to 58%. The DNA sequences were analyzed by using the NCBI BlastX program, and four were found to have homology to predicted proteins present in the GenBank database (Table 2).
TABLE 2.
Tester-specific clones analyzed from the subtractive hybridization library
| Clone | Insert size (bp) | % G+C | Similar protein
|
||||
|---|---|---|---|---|---|---|---|
| Organism | Putative function | % Identity | % Similarity | NCBI accession no. | |||
| GIII-35 | 318 | 47 | Pseudomonas fluorescens | Hypothetical protein | 64 | 88 | ZP_00088075 |
| GIII-44 | 181 | 54 | Deinococcus radiodurans | Transposase | 55 | 68 | NP_293902 |
| GIII-56 | 202 | 56 | Pseudomonas aeruginosa | Transcriptional regulator | 86 | 90 | NP_250517 |
| GIII-66 | 159 | 47 | |||||
| GIII-76 | 278 | 54 | |||||
| GIII-78 | 244 | 58 | Pseudomonas sp. | Putative transposase | 57 | 73 | AAF80258 |
One of the subtraction clones, designated GIII-78, had a 244-bp insert with homology to a Pseudomonas putative transposase. This insert was used as a probe to screen the PC8 genomic library, and four probe-positive clones, each with a different-size insert, were selected for further analysis.
IS1363 characterization.
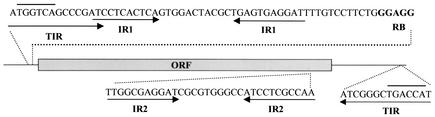
DNA sequence analysis showed that each of the four GIII-78 probe-positive library clones had an identical 1.4-kb fragment containing a 1,026-bp ORF, flanked by perfect 14-bp terminal inverted repeats. Additional perfect 12-bp inverted repeats (IR1) and perfect 11-bp inverted repeats (IR2) were also identified (Fig. 1). Potential −35 promoter regions (TGACCA) were imbedded in the terminal inverted repeats, and a putative ribosome-binding site (GGAGG) was identified between the downstream IR1 and the ORF. The G+C content was 58%. The ORF encoded a putative polypeptide of 341 amino acids, the deduced sequence of which had 61.1% homology to the IS1328 transposase of Yersinia enterocolitica and 57.8% homology to TnpA of Enterobacter aerogenes. The deduced protein had a high isoelectric point (10.01) characteristic of transposases. This 1,363-bp insertion element was designated IS1363. RT-PCR analysis with total RNA from PHDC strains AU1482 and PC8 grown in vitro indicated that the putative transposase of IS1363 was transcribed.
FIG. 1.
Schematic map and partial nucleotide sequence of IS1363. Sequences of perfect terminal inverted repeats (TIR) and additional inverted repeats (IR1 and IR2) are underlined with arrowheads. The putative ribosome-binding site (RB) is in bold. Potential −35 promoter regions in the terminal inverted repeats (oriented outward from the IS element) are indicated with overlining.
Southern blot.
Southern blot analysis of 30 B. cepacia genomovar III isolates representing the ET12, PHDC, and Midwest lineages was performed by using the IS-PCR-generated probe. Twelve of the 14 ET12 isolates examined had a single copy of IS1363, while the remaining two had none. Each of the 14 PHDC isolates examined contained four copies of IS1363. In contrast, IS1363 was not found in two isolates belonging to the Midwest clone. The presence or absence of IS1363 in all these isolates was consistent with the dot blot and IS-PCR results (below).
Dot blot and IS-PCR.
A total of 943 B. cepacia complex isolates were examined by dot blot analysis employing the digoxigenin-labeled 235-bp IS-PCR probe. The isolates represented all nine species of the B. cepacia complex and were recovered from either soil or cultures of sputum from cystic fibrosis patients. The 761 cystic fibrosis sputum-derived isolates were recovered from 761 cystic fibrosis patients attending 154 cystic fibrosis treatment centers in 133 cities in North America; the 182 environmental isolates were recovered from soil as previously described (20, 24). All dot blot-positive isolates also tested positive with the IS-PCR assay. The results are shown in Table 3. While 269 (55%) of the 489 genomovar III isolates examined were dot blot and IS-PCR positive, only 2 (0.4%) of the 454 non-genomovar III isolates were positive. Both of these isolates, ES0607 and ES0864, were B. ambifaria recovered from soil.
TABLE 3.
Dot blot analysis for presence of IS1363
| Species or genomovar | No. dot blot positive/total no. tested
|
||
|---|---|---|---|
| Sputum isolates | Soil isolates | Total | |
| Genomovar I | 0/17 | 0/20 | 0/37 |
| B. multivorans | 0/224 | 0/0 | 0/224 |
| Genomovar III | 238/450 | 31/39 | 269/489 |
| B. stabilis | 0/13 | 0/3 | 0/16 |
| B. vietnamiensis | 0/30 | 0/0 | 0/30 |
| Genomovar VI | 0/20 | 0/0 | 0/20 |
| B. ambifaria | 0/4 | 2/48 | 2/52 |
| B. anthina | 0/1 | 0/18 | 0/19 |
| B. pyrrocinia | 0/2 | 0/54 | 0/56 |
| Total | 238/761 | 33/182 | 271/943 |
IS1363 sequence analyses.
The IS1363 cloned and sequenced from PHDC isolate PC8 showed 92.5% nucleotide identity to the IS1363 PCR-amplified from B. ambifaria strain ES0607. An IS1363 element with 94.3% nucleotide identity to the IS1363 cloned from PC8 was found in the genome sequence database of strain J2315, the B. cepacia genomovar III ET12 lineage isolate currently being sequenced by the Sanger Institute (http://www.sanger.ac.uk/Projects/B_cepacia). In contrast, IS1363 was not found in the genomes of either the B. pseudomallei strain sequenced by the Sanger Institute (http://www.sanger.ac.uk/Projects/B_pseudomallei) or the Burkholderia sp. strain LB400 sequenced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/JGI_microbial/html/burkholderia/burk_homepage.html).
Isolate genotypes.
Among the 489 B. cepacia genomovar III isolates screened for the presence of IS1363 by dot blot and IS-PCR, 355 (73%) had been genotyped by using RAPD, BOX-PCR, PFGE, or multilocus restriction typing in previous studies (5, 7, 9, 20); 28 of these were previously determined to be of the ET12 lineage. In the present study, any isolate testing positive for the presence of IS1363 and not previously genotyped was typed by using BOX-PCR. The cumulative genotyping analyses demonstrated that among the 269 IS1363-positive B. cepacia genomovar III isolates were 26 ET12 strains and 243 isolates belonging to the PHDC lineage. Among the 243 PHDC isolates, 31 were recovered from soil (20) and 212 were cultured from 212 cystic fibrosis patients (below). One hundred forty-four (65%) of the 220 IS1363-negative genomovar III isolates were genotyped. Among these were two ET12 isolates and 88 isolates belonging to the Midwest clone; 40 different genotypes were detected among the remaining 54 isolates. The specificity of IS1363 for PHDC is summarized in Table 4. This also shows that 26 of 28 ET12 isolates tested were also IS1363 positive.
TABLE 4.
IS1363 PCR assay for 413 genomovar III strains for which genotype was determined
| Strains tested | No. of strains for which PCR was:
|
|
|---|---|---|
| Positive | Negative | |
| PHDC (n = 243) | 243a | 0 |
| All others (n = 170) | 26b | 144c |
Includes 212 isolates recovered from 212 CF patients and 31 isolates from soil.
All 26 are of the ET12 lineage.
Includes 2 ET12 isolates, 88 isolates belonging to the Midwest clone, and 54 isolates representing an additional 40 genotypes.
Distribution of strain PHDC.
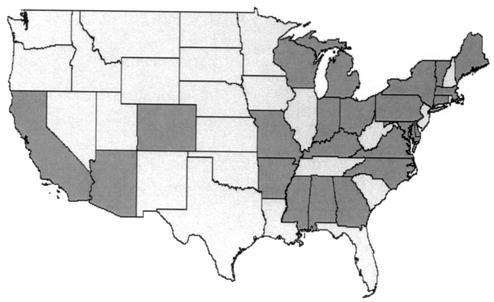
The PHDC soil isolates were obtained from agricultural fields in New York State as previously described (20). The sputum-derived PHDC isolates were recovered from cystic fibrosis patients between 1977 and 2001, with the majority (58%) being recovered between 1997 and 2001. The majority (83%) of PHDC-infected cystic fibrosis patients received care in treatment centers in the mid-Atlantic region of the United States. Most (65%) of these individuals attended a single treatment center where this strain has been endemic for at least the past 20 years (5). In total, cystic fibrosis patients infected with PHDC were found in 24 U.S. states and the District of Columbia (Fig. 2).
FIG. 2.
Distribution of strain PHDC. Shaded U.S. states are those in which at least one cystic fibrosis patient infected with strain PHDC receives care. PHDC-infected patients also received care in Alaska and the District of Columbia (not depicted).
DISCUSSION
The observation that many persons with cystic fibrosis are infected with the same B. cepacia genomovar III strain implies that certain strains have an enhanced capacity for pulmonary infection and/or interpatient spread in cystic fibrosis. The bacterial factors that may account for this, however, remain to be defined. The ET12 strain, which infects a large number of cystic fibrosis patients in Canada and the United Kingdom, elaborates distinctive “cable” pili and an associated adhesin that mediates adherence to respiratory epithelial cells (28). Although likely important in the pathogenesis of ET12, this phenotype has not been found in other epidemic B. cepacia complex lineages. Most of the B. cepacia genomovar III strains found by Mahenthiralingam and colleagues (23) to infect multiple cystic fibrosis patients contained a common 1.4-kb DNA segment termed the B. cepacia epidemic strain marker. Within this segment resides esmR, an ORF with homology to negative transcriptional regulators; the role that this might play in contributing to human infection is not clear. In previous work we demonstrated that strain PHDC, common among patients in cystic fibrosis treatment centers in the mid-Atlantic region of the United States, neither expresses cable pili nor contains the B. cepacia epidemic strain marker (5).
In the present study we sought to identify other genomic elements that may be specific for epidemic B. cepacia complex strains. To do this we employed subtractive hybridization, a method that has been used successfully in a number of recent studies to discern genetic differences between bacterial strains exhibiting a differential capacity for human infection (15, 29, 31). This strategy has also been used to identify differences between closely related bacterial species (2, 11). Particularly relevant to our study is the recent use of subtractive hybridization between the nonpathogenic species B. thailandensis and the pathogens B. mallei and B. pseudomallei. This allowed the identification of novel insertion sequences in B. pseudomallei (4), as well as the characterization of loci encoding capsular polysaccharides, believed to contribute to the virulence of B. pseudomallei and B. mallei (10, 27).
In our study, analysis of the library generated by subtractive hybridization between epidemic strain PHDC and two B. cepacia genomovar III isolates recovered from soil revealed several clones with insert DNA comprising a lower G+C content than would be expected from the B. cepacia genomovar III genome, suggesting possible acquisition of these determinants by horizontal transfer. We used one of these inserts, showing high homology to IS1328 of Y. enterocolitica, as a probe to recover from a genomic library of PHDC strain PC8 a DNA segment containing the novel insertion sequence IS1363.
A number of other insertion sequences have been described previously in B. cepacia complex and are believed to contribute to the genomic plasticity of these species (16, 33). This is attributed to the ability of these elements to integrate at various sites within a recipient genome as discrete DNA segments through nonhomologous recombination. The consequences of this include not only inactivation of disrupted genes at the integration site, but also activation/inactivation of downstream genes (30, 37). For example, the insertion of any one of several elements, including IS402, IS403, IS404, and IS405, was sufficient for activation of an otherwise poorly expressed β-lactamase gene in B. cepacia (30). IS406 appears to activate lac gene expression by generating a hybrid promoter with a new −35 region provided by the element (16, 36), while IS407 contains an outward-directed σ70-like promoter that may serve to activate foreign genes in B. cepacia (16, 36).
We are currently working to characterize the genes flanking the multiple copies of IS1363 in PHDC in order to assess the potential role that this element may have in activating gene expression in this strain. In these efforts, we have identified a large (27-kb) genomic cluster containing 19 ORFs, many of which have high homology to the recently described capsular polysaccharide biosynthesis genes of B. pseudomallei (27) and B. mallei (10; L. Liu and J. J. LiPuma, unpublished data).
Although the role that IS1363 might have in contributing to virulence requires additional study, it is clear that this element is quite specific for strains ET12 and PHDC, both of which infect a great many cystic fibrosis patients in North America and Europe (1, 5, 26). Interestingly, this element is absent from the Midwest clone, another B. cepacia genomovar III strain responsible for infecting a significant number of cystic fibrosis patients in the United States, Also of considerable interest is our finding that two different strains of B. ambifaria also contain IS1363, suggesting possible horizontal genetic transfer between these closely related yet taxonomically distinct members of the B. cepacia complex. This observation has particular relevance in light of the consideration given to the commercial use of B. ambifaria strains (e.g., AMMD, RAL-3, and MC17) in agricultural and bioremedial applications (6, 25). Other insertion elements, such as IS406 and IS407, have similarly been found in multiple Burkholderia species, including B. cepacia genomovar III, B. multivorans, and B. pseudomallei (21). In contrast, IS1356 and an IS402-IS1356 hybrid element appear to be specific for strain ET12 (33).
The finding in our initial analyses that IS1363 was restricted almost exclusively to isolates previously identified as either strain PHDC or ET12 prompted us to assess a much larger collection of sputum isolates. This collection consisted primarily of isolates recovered between 1997 and 2001 from cystic fibrosis patients residing throughout the United States. It also included many previously uncharacterized isolates from cystic fibrosis patients who had received care between 1977 and 1997 at the cystic fibrosis center where strain PHDC was initially identified (5). From this collection we identified several dozen patients infected with B. cepacia genomovar III containing IS1363. Genotyping analyses identified each of these as strain PHDC. In contrast, this strain type was not found by genotyping analyses of 147 IS1363-negative genomovar III isolates. Although the isolates available to us for analysis do not necessarily represent the first B. cepacia complex isolate cultured from each patient, our data indicate that not only has PHDC been a pathogen in cystic fibrosis patients for the past 20 years, but that this strain continues to account for significant infection in this population. In total, by using IS1363 as a probe, we were able to identify 212 cystic fibrosis patients in 31 cities in 24 U.S. states who were or are currently infected with this strain.
In summary, we employed subtractive hybridization to identify a novel insertion sequence that is specific for at least two major epidemic B. cepacia genomovar III strains. By using this element to screen a large collection of B. cepacia complex isolates, we demonstrated that the geographic range of cystic fibrosis patients infected with strain PHDC is much broader than previously appreciated. The role that this insertion element might play in contributing to the apparent increased capacity of B. cepacia genomovar III strains ET12 and PHDC for infection in cystic fibrosis patients is the subject of ongoing efforts.
Acknowledgments
This work was supported by a grant from the Cystic Fibrosis Foundation (to J.J.L.). L.L. and T.C. were supported by the Carroll Haas Research Fund in Cystic Fibrosis.
We gratefully acknowledge the generosity and cooperation of participating cystic fibrosis care centers and microbiology laboratories for submission of clinical isolates.
REFERENCES
- 1.Agodi, A., E. Mahenthiralingam, M. Bachitta, V. Giannino, A. Sciacca, and S. Stefani. 2001. Burkholderia cepacia complex infection in Italian patients with cystic fibrosis: prevalence, epidemiology, and genomovar status. J. Clin. Microbiol. 39:2891-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agron, P. G., R. L. Walker, H. Kinde, S. J. Sawyer, D. C. Hayes, J. Wollard, and G. L. Anderson. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl. Environ. Microbiol. 67:4984-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, N. F., A. E. Lew, and I. R. Beacham. 2000. Identification of new transposable genetic elements in Burkholderia pseudomallei using subtractive hybridisation. FEMS Microbiol. Lett. 183:73-79. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. S., K. Witzmann, T. Spilker, R. Fink, and J. J. LiPuma. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 39:643-649. [DOI] [PubMed] [Google Scholar]
- 6.Ciccillo, F., A. Fiore, A. Bevivino, C. Dalmastri, S. Tabacchioni, and L. Chiarini. 2002. Effects of two different application methods of Burkholderia ambifaria MC17 on plant growth and rhizospheric bacterial diversity. Environ. Microbiol. 4:238-245. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing, a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185: 1454-1462. [DOI] [PubMed] [Google Scholar]
- 8.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye, T., T. Spilker, A. Martin, and J. J. LiPuma. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeShazer, D., D. M. Waag, D. L. Fritz, and D. E. Woods. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253-269. [DOI] [PubMed] [Google Scholar]
- 11.Emmerth, M., W. Goebel, S. I. Miller, and C. J. Hueck. 1999. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J. Bacteriol. 181:5652-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govan, J. R. W., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographical and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar, A., S. Dietrich, W. Schneider, R. Jacobson, F. P. Downes, B. E. Robinson-Dunn, R. Honicky, J. Smith, and R. Martin. 1997. Genetic relatedness of Burkholderia (Pseudomonas) cepacia isolates from five cystic fibrosis centers in Michigan. Respir. Med. 91:485-492. [DOI] [PubMed] [Google Scholar]
- 15.Lai, Y. C., S. L. Yang, H. L. Peng, amd H. Y. Chang. 2000. Identification of genes present specifically in a virulent strain of Klebsiella pneumoniae. Infect. Immun. 68:7149-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 17.LiPuma, J. J. 1998. Burkholderia cepacia: management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 18.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LiPuma, J. J., T. Spilker, L. Gill, P. W. Campbell, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility factors in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 20.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 21.Mack, K., and R. W. Titball. 1998. The detection of insertion sequences within the human pathogen Burkholderia pseudomallei which have been identified previously in Burkholderia cepacia. FEMS Microbiol. Lett. 162:69-74. [DOI] [PubMed] [Google Scholar]
- 22.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahenthiralingam, E., D. A. Simpson, and D. P. Speert. 1997. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 35:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, S. C. M., J. J. LiPuma, and J. L. Parke. 2002. Culture-based and growth-independent detection of the Burkholderia cepacia complex in soil environments. Appl. Environ. Microbiol. 68:3750-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 26.Pitt, T. L., M. E. Kaufmann, P. S. Patel, L. C. Benge, S. Gaskin, and D. M. Livermore. 1996. Type characterisation and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J. Med. Microbiol. 44:203-210. [DOI] [PubMed] [Google Scholar]
- 27.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sawada, K., S. Kokeguchi, H. Hongyo, S. Sawada, M. Miyamoto, H. Maeda, F. Nishimura, S. Takashiba, and Y. Murayama. 1999. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect. Immun. 67:5621-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scordilis, G. E., H. Ree, amd T. G. Lessie. 1987. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J. Bacteriol. 169:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smoot, L. M., D. D. Franke, G. McGillivary, and L. A. Actis. 2002. Genomic analysis of the F3031 Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization. Infect. Immun. 70:2694-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyler, S. D., K. R. Rozee, and W. M. Johnson. 1996. Identification of IS1356, a new insertion sequence, and its association with IS402 in epidemic strains of Burkholderia cepacia infecting cystic fibrosis patients. J. Clin. Microbiol. 34:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 35.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. W. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound test results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 36.Wood, M. S., A. Byrne, and T. G. Lessie. 1991. IS406 and IS407, two gene-activating insertion sequences from Pseudomonas cepacia. Gene 105:101-105. [DOI] [PubMed] [Google Scholar]
- 37.Wood, M. S., C. Lory, and T. G. Lessie. 1990. Activation of the lac genes of Tn951 by insertion sequences from Pseudomonas cepacia. J. Bacteriol. 172:1719-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]