Abstract
The Western blot for cysticercosis, which uses lentil lectin purified glycoprotein (LLGP) antigens extracted from the metacestode of Taenia solium, has been the “gold standard” serodiagnostic assay since it was first described in 1989. We report that the diagnostic antigens at 14, 18, and 21 kDa, as well as some larger disulfide-bonded antigens, are actually all members of a very closely related family of proteins, the 8-kDa antigens. The genes for 18 unique, mature proteins have been identified. Nine of these were chemically synthesized and tested in an enzyme-linked immunosorbent assay with a battery of defined serum samples, including 32 cysticercosis-positive serum samples reactive with the 8-kDa antigens of LLGP on Western blotting, 34 serum samples from patients with other parasitic infections, and 15 normal human serum samples. One of the 8-kDa antigens, TsRS1, is 100% sensitive and 100% specific. TsRS1 will be one component of a cocktail of three to four synthetic or recombinant antigens, based on the diagnostic bands of the Western blot, which will be used for the serodiagnosis of cysticercosis.
Neurocysticercosis is a debilitating disease caused by the metacestode of the parasitic worm Taenia solium. Humans are an accidental host for the metacestode, or larval form, of T. solium. Infection occurs when eggs are ingested via the fecal-oral route of transmission. Infective eggs hatch, and the liberated oncospheres cross the membrane of the small intestine and migrate in the body, typically ending up in the central nervous system, skeletal muscle, subcutaneous tissue, or ocular tissue (9, 10, 43). Within the host's tissues, the oncosphere matures into a cysticercus and causes the disease cysticercosis. Neurocysticercosis is an infection of the central nervous system. Symptoms are nonspecific and variable and include seizures, headaches, and hydrocephalus, depending upon the numbers, locations, and viabilities of the cysts and the stage of the host immune response (4, 44, 45). Several studies have shown that in countries where the disease is endemic, as many as 18 to 50% of the cases of adult-onset epilepsy are actually cases of neurocysticercosis (11, 22, 26).
In the life cycle of T. solium, humans are the definitive host for the adult worm and pigs are the intermediate host for the metacestode. Maintenance of the life cycle requires a close association between humans and pigs; consequently, this disease is found wherever pigs are allowed to roam freely to scavenge for food and humans lack adequate sanitary facilities for the disposal of feces. Cysticercosis is endemic in Latin America, India, Asia, and Africa (14, 36). A recent estimate of the prevalence of cysticercosis in Latin America indicated that 400,000 people have symptomatic disease (3).
Diagnosis of neurocysticercosis requires either imaging of the brain by computed tomography or magnetic resonance imaging or the demonstration of cysticercosis-specific antibodies in the patient's serum. Brain imaging is expensive and is usually available only in urban centers within the countries where the disease is endemic. In addition, single lesions due to Mycobacterium tuberculosis are often difficult to distinguish from cysts due to T. solium (4, 9). Early antibody detection assays lacked sensitivity and specificity (7, 15, 29, 31), but in 1989, an assay with 98% sensitivity for the detection of cysticercosis cases with two or more cysts, less sensitivity for the detection of cases with single cysts (26, 28, 46), and 100% specificity was developed (40). The assay uses a partially purified preparation of metacestode antigens. Cysts harvested from pigs are solubilized in urea, and the bound fraction from lentil lectin affinity chromatography, the lentil lectin purified glycoproteins (LLGPs), is used as antigen in a Western blot or enzyme-linked immunoelectrotransfer blot assay. Immunoreactivity with any one of seven glycoproteins is diagnostic for cysticercosis. Since 1995, this has been the only assay recognized by the World Health Organization and the Pan American Health Organization for the serodiagnosis of cysticercosis (Pan American Health Organization and World Health Organization informal consultation on the taeniosis/cysticercosis complex, 1997). The assay, however, has some drawbacks. It is dependent upon a supply of naturally infected pigs. Preparation of the antigen and performance of the Western blot require considerable technical expertise. The partially purified LLGP antigen preparation is not suitable for use in an enzyme-linked immunosorbent assay (ELISA) (V. C. W. Tsang, unpublished data); and a Western blot assay is not suitable for field studies, nor is it a suitable or affordable assay for diagnosis in countries where cysticercosis is endemic. To address these issues, we have been systematically characterizing the seven diagnostic LLGP antigens.
The characterization of two LLGP proteins, Ts14 and Ts18, has been reported earlier (16, 17). Here we report on the identification and characterization of a family of diagnostic proteins, the 8-kDa antigens of T. solium metacestodes. The 8-kDa antigens are the diagnostic proteins seen at 14, 18, and 21 kDa on the Western blot and are also found in the bands at 24 and 39 to 42 kDa. Eighteen unique mature proteins have been cloned by us and others (16, 24, 34) and were identified, by phylogenetic analysis, to sort into four clades. Nine were chemically synthesized for use as antigens. Testing of the synthetic proteins in an ELISA identified one 8-kDa protein with 100% sensitivity when it was tested with sera from cysticercosis patients reactive with the 8-kDa protein components of LLGP on Western blot and 100% specificity.
MATERIALS AND METHODS
Parasite material and DNA extraction.
T. solium cysticerci from Peru, India, and China were dissected from surrounding porcine muscle. For each cyst, the protoscolex was removed by dissection and washed with cold phosphate-buffered saline, and the DNA was extracted by using the FastDNA kit with lysing matrix 4 and CLS-TC buffer, according to the instructions of the manufacturer (Qbiogene, Inc., Carlsbad, Calif.). For the cysts from India and China, which had been preserved in 70% ethanol, an overnight incubation step at 37°C was added after the homogenization step to allow rehydration of the DNA. The DNA isolated from the cysts preserved in ethanol was further purified by using the QIAquick PCR purification kit (Qiagen, Carlsbad, Calif.), according to the instructions of the manufacturer, to remove PCR inhibitors.
Amplification, cloning, and sequencing of the 8-kDa diagnostic antigens.
The 8-kDa diagnostic antigens were amplified by using two sets of primers. Primers gTs14F (5′-ATGCGTGCCTACATTGTGCTTCTC-3′) and gTs14R2 (5′-GCAGTTTTTTTCTTAGGACCTTTGCAGTG-3′) amplified the gene for Ts14. The genes for the other 8-kDa proteins were amplified by using primers gTs14F and gTs14R1 (5′-GTGAAGAGAAGAACGCATGAAAGTTG-3′). All PCRs were done with Pfu polymerase (Stratagene, La Jolla, Calif.) at an annealing temperature of 60°C for 40 cycles. The amplicons were cloned into the vector PCR-Script (Stratagene) according to the instructions of the manufacturer. From 4 to 14 clones of each amplicon were sequenced. In addition, the amplicons resulting from amplification of DNA from the Peruvian isolate, the Indian isolate, and the China isolate with gTs14F and gTs14R2 were directly sequenced. In all cases, both strands of DNA were sequenced. All sequencing was done by terminator-based cycle sequencing with BIGDYE fluorescent dye (Applied Biosystems, Foster City, Calif.) (35) and an ABI Prism 377 DNA sequencer (Applied Biosystems).
Sequence data were analyzed with the SeqMan II program (DNASTAR Inc., Madison, Wis.). Sequence homology searches were done by using the BLAST program (1). Signal peptide sequences were predicted by using the SignalP program (23) along with N-terminal sequence data. Alignments were done by using the ClustalX program (39). For the phylogenetic analyses, all sequences were aligned, and the amino acid sequence data common to all sequences were used. The phylogenetic analysis was performed with the PUZZLE program (38), by the FITCH method (a least-squares distance method), and by the protein parsimony (protpars) method (http://evolution.genetics.washington.edu/phylip.html). Phylogenetic trees were displayed and manipulated by using the TreeView program (25).
Nine of the 8-kDa proteins were chemically synthesized, all with a long-chain biotin at the mature N terminus for uniform capture by streptavidin-coated polystyrene “sticks.” Proteins Ts18, Ts18 var1, Ts18 var3, Ts18 var4, Ts18 var6, and TsRS2 var1 were synthesized by SynPep (Dublin, Calif.), and proteins Ts14, Ts18 var8, and TsRS1 were synthesized by AnaSpec Inc. (San Jose, Calif.). All were of at least 95% purity, as determined by high-pressure liquid chromatography. The mass of each synthetic protein, as determined by mass spectrometry, matched the theoretical mass. Synthetic proteins were solubilized in 0.05 M HEPES-0.1 M NaCl-2 mM dithiothreitol (pH 7.0) and stored at −80°C after the addition of glycerol to a final concentration of 43.5%. The protein concentration was determined by the bicinchoninic acid method (Pierce, Rockford, Ill.) (37).
Battery of serum samples.
Our battery of serum samples included 32 T. solium-infected serum samples from patients with neurocysticercosis, 34 serum samples from patients with other parasitic infections, and 15 normal human serum samples (NHSs). All 32 serum samples from patients with T. solium infections were positive for cysticercosis by the LLGP Western blotting assay and all were reactive with the 8-kDa group of antigens (40). Many of the samples were also confirmed to be positive for cysticercosis by brain imaging or biopsy. A positive reference standard was formed by pooling sera from three patients with parasitologically confirmed, immunoblot-positive cysticercosis. The other parasitic infection serum samples were all from parasitologically confirmed cases. Among these serum samples were 4 serum samples from patients with Taenia saginata infections, 3 serum samples from patients with Echinococcus multilocularis infections, 6 serum samples from patients with Echinococcus granulosus infections, 11 serum samples from patients with Ascaris lumbricoides infections, 4 serum samples from patients with Schistosoma mansoni infections, 4 serum samples from patients with Schistosoma haematobium infections, and 2 serum samples from patients with Trichinella sp. infections. The NHSs were obtained from healthy U.S. residents who did not have a history of international travel.
Fast-ELISA.
Fast-ELISA was performed as described previously (18). Streptavidin (recombinant streptavidin; Roche Molecular Biochemicals, Indianapolis, Ind.) was used to coat the polystyrene sticks of the Falcon 3931 assay screening test (Becton Dickinson, Lincoln Park, N.J.) at a saturating concentration of 6.0 μg of streptavidin per ml of 0.05 HEPES- 0.1 M NaCl (pH 7.0). The wash buffer and diluent buffer for the biotinylated proteins and conjugate was 0.05 M HEPES-0.1 M NaCl (pH 7.0) plus 0.3% Tween 20 (Calbiochem-Novabiochem Corp., La Jolla, Calif.). The serum diluent consisted of the same buffer with 5% nonfat dry milk. The biotinylated proteins were diluted to 1.0 μg/ml. The serum samples were assayed in triplicate at 1:100 dilutions. Bound antibodies were detected with a peroxidase-labeled goat anti-human immunoglobulin (41), followed by the substrate SureBlue (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.). All reagents except the sera were present in excess. After 5 min, the A650 was read with a ThermoMax microplate reader (Molecular Devices Corp., Sunnyvale, Calif.). Each plate contained a positive reference standard, which was used to calculate a relative absorbance unit for each sample by dividing the mean A650 for each sample by the mean A650 for the positive reference standard. As a control, the positive reference standard was included as a sample in the assay. If the relative absorbance for the control differed from 1.0 by more than 10%, the data for the plate were discarded.
Nucleotide sequence accession numbers.
The nucleotide sequences identified in this study have been submitted to GenBank, and the accession numbers are given in Table 1.
TABLE 1.
Members of the 8-kDa family of proteins
| GenBank accession no.a | Protein name | Origin | Source | Reference |
|---|---|---|---|---|
| AF082829 | Ts14 | mRNA | Peru | 16 |
| AF158184 | Ts14 | Genome | Peru | 16 |
| AF257776 | Ts14 | mRNA | Mexico | 24 |
| AF356335 | Ts14 | Genome | China | This study |
| AF356336 | Ts14 | Genome | India | This study |
| AF356337 | Ts14 var1 | Genome | India | This study |
| AF356338 | Ts14 var2 | Genome | India | This study |
| AF356339 | Ts14 var3 | Genome | India | This study |
| AF098073 | Ts18 var1 | mRNA | Peru | 16 |
| AF356330 | Ts18 var1 | Genome | Peru | This study |
| AF098074 | Ts18 var2 | mRNA | Peru | 16 |
| AF098075 | Ts18 var3 | mRNA | Peru | 16 |
| AF356331 | Ts18 var4 | Genome | India | This study |
| AF356332 | Ts18 var5 | Genome | China | This study |
| AF350070 | Ts18 variant 1 | mRNA | Mexico | 24 |
| AF350071 | Ts18 variant 2 | mRNA | Mexico | Enciso, direct submission |
| AF082828 | Ts18 | mRNA | Peru | 16 |
| AB044081 | TSOLAg1V1 | mRNA | China | 34 |
| AF356333 | Ts18 var6 | Genome | China | This study |
| AF356334 | Ts18 var7 | Genome | China | This study |
| AF356345 | Ts18 var8 | Genome | India | This study |
| AF356341 | TsRS1 | Genome | Peru | This study |
| AF216695 | Ts21 | mRNA | China | Liu, direct submission |
| AF163972 | Immunogenic protein | mRNA | China | Bin, direct submission |
| AB044083 | TSOLAg2V1 | mRNA | China | 34 |
| AB044082 | TSOLAg2 | mRNA | China | 34 |
| AF082830 | TsRS1 | mRNA | Peru | 16 |
| AF356340 | TsRS1 var1 | Genome | India | This study |
| AF356342 | TsRS1 var2 | Genome | China | This study |
| AB044080 | TSOLAg1 | mRNA | China | 34 |
| AF356344 | TsRS2 | Genome | China | This study |
| AF356343 | TsRS2 var1 | Genome | Peru | This study |
The nucleic acid sequences for the accession number(s) in the same cluster are identical over the shared sequence.
RESULTS
The 8-kDa gene family.
A family of genes encoding low-molecular-weight antigens has been identified in T. solium metacestodes. To date, 32 nucleic acid sequences have been identified as encoding members of this 8-kDa family of proteins (Table 1). The 32 sequences are from T. solium cysts collected in Peru, Mexico, India, and China. A number of these sequences have already been described (16, 24, 34). Additional members of this gene family, described in this paper, were identified by PCR amplification of genomic DNA. Two introns were identified in each of the genomic clones. Screening, at a low stringency, of a bacteriophage lambda T. solium metacestode cDNA library with a mixture of random primed and labeled probes generated from the coding sequences of Ts14, Ts18, and TsRS1, did not identify any additional gene family members (data not shown).
Twenty-six unique sequences that encode 23 unique protein sequences were detected among the 32 nucleic acid sequences. These 8-kDa proteins have a signal sequence, cleavage of which results in a 66- or 67-amino-acid mature protein with a predicted molecular mass of about 8 kDa (x̄ = 7,628 for the 66-amino-acid proteins and 7,746 for the 67-amino-acid proteins) and a predicted pI of about 9 (x̄ = 8.96). There are 18 unique mature protein sequences. Within the mature protein sequence, all of the 8-kDa proteins have from one to three predicted N-linked glycosylation sites, and all of the 8-kDa proteins have one to three cysteines except for the proteins within the TsRS1 clade. These proteins lack N-linked glycosylation sites and cysteines. The 8-kDa proteins are hydrophilic, as determined by hydrophobicity analysis (21). They lack potential transmembrane regions and glycosylphosphatidylinositol-anchor attachment sites (8). These data suggest that the 8-kDa proteins are extracellular secreted proteins that perhaps accumulate in the cyst fluid.
The 8-kDa proteins have been shown by protein sequencing to be the diagnostic proteins found at 14, 18, and 21 kDa in the lentil lectin bound fraction from urea-solubilized cysticerci (17, 40; data not shown). Only 8-kDa proteins have been found in these diagnostic bands. Protein sequencing data also show that the bands at 24 and 39 to 42 kDa contain 8-kDa proteins (27; data not shown). In addition, a polyclonal antibody raised against Ts14 reacts with the diagnostic bands at 14, 18, 21, 24, and 39 to 42 kDa (16). Reduction of the proteins in the diagnostic band from 39 to 42 kDa with dithiothreitol yields proteins migrating at 14, 18, and 21 kDa, suggesting that the 8-kDa proteins in the band from 39 to 42 kDa are disulfide-bonded multimers (17, 27; data not shown).
The 23 sequences were aligned by using the ClustalX program (Fig. 1A). The predicted signal peptide sequences are shown in italics and, as expected, are well conserved. Within the mature sequences, there are 12 absolutely conserved amino acids, 20 positions with more conservative substitutions, and 7 positions with less conservative substitutions. Among these 23 protein sequences, 18% of the amino acids are identical and 68% are similar or identical. Many of these identical and similar amino acids are clustered in the central portion of the mature protein, from position 27 through position 41.
FIG. 1.
Alignments of the T. solium 8-kDa diagnostic antigens obtained with the ClustalX program. (A) Alignments of the 23 unique protein sequences from the 8-kDa gene family, identified by GenBank accession number. The predicted signal peptides are in italics. The lines between accession numbers mark the division of the sequences into four groups, Ts14, Ts18, TsRS1, and TsRS2. Additional protein sequences that are not shown are those with GenBank accession numbers AF257776 and AF158184, which are identical to the sequence with GenBank accession number AF082829; those with GenBank accession numbers AF356366 and AF356338, which are identical to the sequence with GenBank accession number AF356335; that with GenBank accession number AF356330, which is identical to the sequence with GenBank accession number AF098073; that with GenBank accession number AF356345, which is identical to the sequence with GenBank accession number AB044081; and those with GenBank accession numbers AF356342, AF163972, and AF356341, which are identical to the sequence with GenBank accession number AB044083. For this analysis, an intron-like sequence in the sequence with GenBank accession number AB044083 was removed. (B) Alignments of the 8-kDa proteins with the hydrophobic ligand-binding proteins of H. diminuta (GenBank accession number AF249884) and M. expansa (GenBank accession number AF312736). Asterisks mark identical amino acid residues. More and less conservative substitutions are indicated by two dots and one dot, respectively.
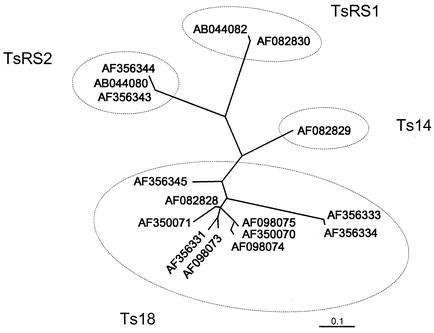
From the alignment of the 8-kDa protein sequences obtained with the ClustalX program (Fig. 1A), the 16 sequences with an alignment over 73 amino acids were analyzed to determine their phylogenetic relationships. Quartet puzzling (a maximum-likelihood method), the FITCH method (a least-squares distance method), and the protpars method (a maximum-parsimony method) gave trees with similar topologies. The quartet puzzling results are shown as a radial tree in Fig. 2. The 8-kDa sequences fall into four clades, with the largest and most diverse clade being Ts18. The other clades are Ts14, TsRS1, and TsRS2. The node support values for the four clades range from 90 to 99%. All four clades contain sequences from cysts from both Latin America and Asia.
FIG. 2.
Phylogenetic tree for the 8-kDa diagnostic antigens of T. solium. The analysis was done by quartet puzzling using a Clustal alignment of 73 amino acids from the 16 8-kDa proteins that are unique over the aligned sequence. The sequences are identified by GenBank accession number. The dashed ovals show the grouping of the sequences into four clades, Ts14, Ts18, TsRS1, and TsRS2. The scale bar indicates an evolutionary distance of 0.1 amino acid per position.
A BLAST search for similar sequences identified 13 homologous proteins. The two proteins with the highest Expect values were cestode hydrophobic ligand-binding proteins, one from Hymenolepis diminuata (GenBank accession number AF249884) and the other from Moniezia expansa (GenBank accession number AF312736). These two proteins were aligned with the 8-kDa proteins by use of the ClustalX program (Fig. 1B). These hydrophobic ligand-binding proteins do not have a signal sequence, but their sequences align with the mature 8-kDa sequences. Among the 8-kDa proteins and the hydrophobic ligand-binding proteins, there are 3 conserved amino acids, 11 positions with more conservative substitutions, and 9 positions with less conservative substitutions. The cluster of identical and similar amino acids seen in the central portion of the 8-kDa proteins is seen here as well.
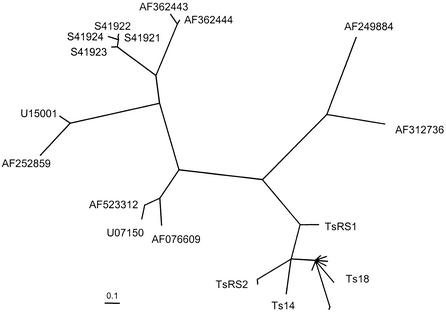
Phylogenetic analysis was done with the 13 homologous proteins plus 13 of the 8-kDa proteins, from all four clades, which aligned over 62 amino acids, as determined with the ClustalX program. The analysis was done by quartet puzzling, the FITCH method, and the protpars method. All methods gave trees with similar topologies. The quartet puzzling results are shown as a radial tree in Fig. 3. The sequences fall into five clades. The T. solium 8-kDa clade is the closest in evolutionary distance to the cestode hydrophobic ligand-binding proteins. The next closest clade consists of two T. solium sequences, one of which encodes a cysticercosis-specific antigen (GenBank accession number AF076609) and the other of which encodes an oncosphere-specific antigen (GenBank accession number AF523312), and one Taenia crassiceps sequence, which encodes an immunodiagnostic antigen (GenBank accession number U07150). The remaining eight sequences, which formed two clades, were all E. granulosus antigen B sequences. The node support values for the five clades ranged from 84 to 100%.
FIG. 3.
Phylogenetic tree for the T. solium 8-kDa proteins and related sequences. The analysis was done by quartet puzzling using a Clustal alignment of 62 amino acids from 26 sequences. The T. solium 8-kDa antigen clades Ts14, Ts18, TsRS1, and TsRS2 are identified. The other sequences are identified by GenBank accession number, as follows: AF249884, H. diminuta hydrophobic ligand-binding protein; AF312736, M. expansa hydrophobic ligand-binding protein; AF362443, AF362444, S41921, S41922, S41923, S41924, U15001, and AF252859, all E. granulosus antigen B; AF523312, T. solium oncosphere-specific antigen; U07150, T. crassiceps immunodiagnostic antigen; and AF076609, T. solium cysticercosis-specific antigen. The scale bar indicates an evolutionary distance of 0.1 amino acid per position.
Immunoreactivities of T. solium 8-kDa proteins.
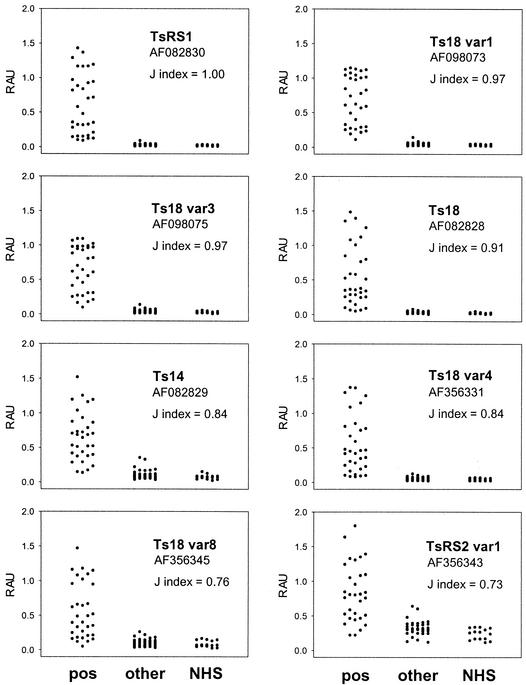
Nine of the T. solium 8-kDa proteins were selected for evaluation of their immunoreactivities with 32 serum samples from patients with cysticercosis, 34 serum samples from patients with other parasitic infections, and 15 NHSs. One sequence was chosen from each of the three clades Ts14, TsRS1, and TsRS2 var1. Six sequences were chosen from the Ts18 clade (Ts18, Ts18 var1, Ts18 var3, Ts18 var4, Ts18 var6, and Ts18 var8). Preliminary studies with Ts18 var6, however, showed that it had minimal reactivity with the positive reference standard and most other positive serum samples (data not shown). Therefore, no further assays were done with this protein. The remaining eight synthetic proteins were assayed, each with 81 serum samples. The Fast-ELISA results are shown in Fig. 4. The cutoff value for each protein was set for 100% specificity, and the J index was calculated (47). The J index is equal to the sensitivity (number positive in the infected group divided by the number of individuals in the infected group) plus the specificity (number negative in the uninfected group divided by the number of individuals in the uninfected group) minus 1. With a J index of 1.00, TsRS1 was 100% specific and 100% sensitive. Ts18 var1 and Ts18 var3 both had J indices of 0.97 and missed 1 of 32 positive serum samples. Ts18 had a J index of 0.91 and missed 3 of 32 positive serum samples. Ts14 and Ts18 var4 both had J indices of 0.84 and missed 6 of 32 positive serum samples. Ts18 var8 had a J index of 0.76 and missed 10 of 32 positive serum samples. TsRS2 var1 had a J index of 0.73 and missed 12 of 32 positive serum samples. The three proteins with J indices ≥0.97 show minimal reactivity with the NHSs and the serum samples from patients with other parasitic infections. Ts14, Ts18 var8, and TsRS2 var1 had higher levels of reactivity with some of the serum samples from patients with other parasitic infections and some of the NHSs. This low level of cross-reactivity with cysticercosis-negative sera prevents the discrimination between positive serum samples with low-level reactivity and negative serum samples. Ts18 and Ts18 var4 had low levels of reactivity with the serum samples from patients with other parasitic infections and the NHSs but lacked reactivity with several cysticercosis-positive serum samples. The best choices for antigens for use in a diagnostic assay are TsRS1, Ts18 var1, and Ts18 var3. All are 100% specific and have sensitivities ranging from 97 to 100%.
FIG. 4.
Antibody reactivities of T. solium 8-kDa proteins. Eight of the T. solium 8-kDa proteins were assayed with a panel of defined serum samples: 32 serum samples from patients positive (pos) for cysticercosis, 34 serum samples from patients with other parasitic infections (other), and 15 NHSs. Relative absorbance units (RAU) are shown on the y axis. The J index for each antigen, calculated at 100% specificity, is shown.
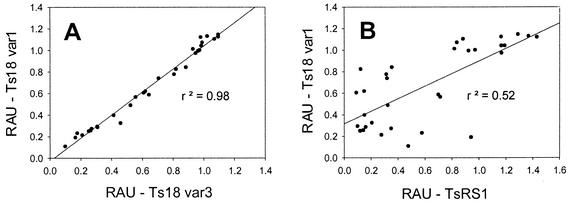
The reactivities of the positive sera with Ts18 var1 were compared with the reactivities of the positive sera with Ts18 var3 (Fig. 5A). The same comparison was made between Ts18 var1 and TsRS1 (Fig. 5B). No difference in the level of reactivity of each serum sample with Ts18 var1 and Ts18 var3 was seen. The coefficient of determination (r2) was 0.98. This result is not surprising, since the two sequences differ by only three amino acids. This result also validates the reliability of the ELISA data. However, when the reactivities with Ts18 var1 and TsRS1 were compared, several positive serum samples had low levels of reactivity with one antigen but high levels of reactivity with the other antigen. The positive serum sample with the lowest level of reactivity with Ts18 var1 and Ts18 var3 had a middle range of reactivity with TsRS1. Conversely, the positive serum sample with the lowest level of reactivity with TsRS1 had a middle range of reactivity with Ts18 var 1 and Ts18 var3. These variations in antibody reactivities between these two antigens are reflected in the low coefficient of determination (r2 = 0.52). Because of these differences in levels of antibody reactivity between Ts18 var1-Ts18 var3 and TsRS1, both Ts18 var1 or Ts18 var3 and TsRS1 will be included in future assays for the detection of immunoreactivities to the T. solium 8-kDa diagnostic antigens.
FIG. 5.
Comparisons of the antibody reactivities of the cysticercosis-positive serum samples. (A) Antibody reactivities of cysticercosis-positive serum samples, in relative absorbance units (RAU), with Ts18 var1 compared to the antibody reactivities of cysticercosis-positive serum samples with Ts18 var3. (B) Antibody reactivities of cysticercosis-positive serum samples with Ts18 var1 compared to the antibody reactivities of cysticercosis-positive serum samples with TsRS1. The r2 value for each line is shown.
DISCUSSION
The proteins in the T. solium 8-kDa family of proteins are important for immunodiagnosis. In the LLGP Western blot for cysticercosis, the diagnostic bands at 14, 18, and 21 kDa are 8-kDa proteins. In addition, the band at 24 kDa contains 8-kDa proteins, and disulfide-bonded multimers of the 8-kDa proteins are found in the 39- to 42-kDa diagnostic band. To date, 18 unique mature 8-kDa protein sequences have been identified. Nine of the 18 protein sequences were synthesized for evaluation in an ELISA. Of these, one (TsRS1) was shown to be 100% sensitive for the detection of cysticercosis-positive serum samples reactive with the 8-kDa bands of LLGP on Western blot and 100% specific. Therefore, TsRS1 will be one component of a cocktail of diagnostic synthetic or recombinant proteins. Another two 8-kDa proteins, Ts18 var1 and Ts18 var3, were shown to be 97% sensitive and 100% specific. Interestingly, there were significant differences between the levels of antibody reactivities of cysticercosis-positive serum samples with TsRS1 compared with their reactivities with Ts18 var1 or Ts18 var3. To guard against the possibility of the lack of detection of a positive serum sample that has low level of reactivity with TsRS1, Ts18 var1 or Ts18 var3 will also be included in the antigen cocktail.
There are at least two additional immunodiagnostic proteins in the LLGP antigen used in the Western blot. These are the protein at 50 kDa (GP50) and the non-8-kDa proteins at 24 and 39 to 42 kDa. While at least 90% of people with cysticercosis have antibodies that recognize more than one band in the immunoblot (40), the sera of some individuals recognize only GP50 and only the 24- or 42-kDa protein. Clearly, an assay that uses synthetic and recombinant antigens and that has the sensitivity of the LLGP Western blot assay must also include GP50 and the 24- and 42-kDa proteins. A recombinant antigen has been developed for GP50, and work is under way to develop a recombinant antigen for the 24- and 42-kDa proteins.
The 8-kDa antigens are not the first cloned proteins to be tested for their immunoreactivities with cysticercosis-infected sera. Hubert et al. (19) identified two proteins, NC-3 and NC-9, by screening a cDNA expression library with cysticercosis-positive sera. They expressed these proteins as glutathione S-transferase fusion proteins and tested them in an ELISA. NC-3 had a sensitivity of 96.3% and a specificity of 91.5%. The sensitivity of NC-9 was only 33.3%. Chung et al. (5) cloned and expressed the T. solium homolog of the T. crassiceps 10-kDa immunodiagnostic antigen (GenBank accession numbers AF076609 and U07150, respectively). These proteins are phylogenetically related to the T. solium 8-kDa antigens (Fig. 3). When expressed as a glutathione S-transferase fusion protein and tested in an immunoblot, the T. solium 10-kDa protein showed a sensitivity of 88% and a specificity of 98%.
Aside from these proteins, two of which are unrelated to the T. solium 8-kDa proteins and one of which is related to the T. solium 8-kDa proteins, Sako et al. (34) cloned and expressed three 8-kDa proteins, Ag1, Ag1V1, and Ag2 (GenBank accession numbers AB044080, AB044081, and AB044082, respectively). Ag1 is a member of the TsRS2 clade, Ag1V1 is in the Ts18 clade, and Ag2 is in the TsRS1 clade (Fig. 2). They used thioredoxin- and histidine-tagged-fusion proteins and reported low levels of antibody reactivity with Ag1 and good antibody reactivities with Ag1V1 and Ag2. For analysis in an ELISA, they expressed a histidine-tagged fusion of a chimera of Ag1V1 and Ag2 and reported a sensitivity of 89.7% and a specificity of 100%. These results are similar to the ones reported here. TsRS2 var1 (GenBank accession number AF356343) was the least sensitive of the synthetic proteins tested, and TsRS1 (GenBank accession number AF082830) was the most sensitive (Fig. 4). However, we report a sensitivity of 100% with synthetic TsRS1.
While other cloned proteins function as immunodiagnostic proteins, such as NC-9 and the T. solium 10-kDa protein, there is an advantage to targeting the diagnostic proteins of an antigen mixture that has been used successfully around the world for more than 10 years. These proteins have a track record of success when used for immunodiagnosis, with a sensitivity of 98% and a specificity of 100% (6, 12, 13, 40, 42, 46). Because it is known that the native proteins are glycoproteins, the one concern is the potential contribution of the carbohydrate moiety to the antibody response observed in cysticercosis patients. This concern was addressed by Obregon-Henao et al. (24), and their data showed diminished antibody activities for the native 8-kDa proteins following deglycosylation. However, our data for the synthetic 8-kDa proteins and the data of Sako et al. (34) for the recombinant 8-kDa proteins clearly show that the majority of the antibody reactivity is retained in proteins lacking glycosylation.
To be able to implement cysticercosis control programs, it is essential to have an accurate and rapid assay for diagnosis. While the LLGP Western blot assay is the “gold standard” assay for sensitivity and specificity (26a, 28), an ELISA format is much preferred because of its ease of use and lower cost. The use of cloned antigens rather than partially purified native proteins makes use of the ELISA format feasible and opens the possibility for the use of other rapid test formats, such as lateral flow assays. In addition, the use of a cloned protein as antigen removes the dependence on a supply of naturally infected pigs as a source of parasite material. The ability to chemically synthesize the antigen rather than express it greatly simplifies the process for obtaining antigen and keeps the cost low. The cost of synthetic TsRS1 antigen required to assay a single sample is $0.03. It may be possible to reduce this cost further by epitope mapping TsRS1 to determine if a smaller peptide(s) retains the same sensitivity for the detection of cysticercosis-positive sera.
The 8-kDa proteins are a family of secreted proteins that elicit an antibody response in infected people. The question arises, what is the function of these proteins in the metacestode? The best clue that we have is that phylogenetically they are the proteins most closely related to the cestode hydrophobic ligand-binding proteins, characterized in M. expansa (GenBank accession number AF312736), the sheep tapeworm, and in H. diminuta (GenBank accession number AF249884), the rat tapeworm (2, 20, 33) (Fig. 3). Like the cestode hydrophobic ligand-binding proteins, the 8-kDa proteins all have a conserved tryptophan that has been shown to be essential to the function of the cestode hydrophobic ligand-binding proteins (2, 32). This conserved tryptophan is not present in the members of the three other clades of phylogenetically related proteins (Fig. 3). On the basis of these similarities, we believe that the T. solium 8-kDa proteins may also be hydrophobic ligand-binding proteins, but ones that function as extracellular carrier proteins for lipids and other hydrophobic ligands. However, the possibility that the 8-kDa proteins may play a role in the host immune response, like the role proposed for the related protein antigen B from E. granulosus, should also be considered (30).
In the host-parasite interactions that occur during infection with T. solium metacestodes, it is clear that the 8-kDa proteins elicit an antibody response that is diagnostic for cysticercosis. In order to determine which flavor of the 18 8-kDa proteins identified to date works best as a diagnostic antigen, 9 of the 66- and 67-mer mature 8-kDa proteins were chemically synthesized and then evaluated in an ELISA. One, TsRS1, was 100% sensitive when it was tested with cysticercosis-positive sera reactive with the 8-kDa proteins on Western blot and was 100% specific. Two others, Ts18 var1 and Ts18 var3, were 97% sensitive and 100% specific and often showed different levels of antibody reactivity with cysticercosis-positive sera compared to the reactivity of TsRS1. TsRS1 and either Ts18 var1 or Ts18 var3 will be components of a diagnostic antigen cocktail of three to four synthetic and recombinant proteins, all derived from the diagnostic proteins used for the LLGP Western blot assay. Identification of these diagnostic antigens and the development of the ELISA are major steps forward for the serodiagnosis of cysticercosis.
Acknowledgments
This work was supported by ICIDR-NIH grant U01 AI35894, NIH NIAID TMRC grant 1 P01 AI51976-01, and Burroughs Wellcome grant 063109.
We thank our collaborators H. Hugo Garcia, A. Emico Gonzalez, and Robert H. Gilman and all members of the Cysticercosis Working Group in Peru for providing the cysts from Peru. We also thank Hai-Chou Xue for providing the cysts from China and Vedantam Rajshekhar for providing the cysts from India. In addition, we are grateful to Norman J. Pieniazek for providing guidance for the phylogenetic analyses.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, J., N. Saghir, A. Timanova, K. Clarke, and P. M. Brophy. 1997. Characterization and properties of an intracellular lipid-binding protein from the tapeworm Moniezia expansa. Eur. J. Biochem. 250:269-275. [DOI] [PubMed] [Google Scholar]
- 3.Bern, C., H. H. Garcia, C. Evans, A. E. Gonzalez, M. Verastegui, V. C. W. Tsang, and R. H. Gilman. 1999. Magnitude of the disease burden from neurocysticercosis in a developing country. Clin. Infect. Dis. 29:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carpio, A., A. Escobar, and W. A. Hauser. 1998. Cysticercosis and epilepsy: a critical review. Epilepsia 39:1025-1040. [DOI] [PubMed] [Google Scholar]
- 5.Chung, J.-Y., Y. Y. Bahk, S. Huh, S.-Y. Kang, Y. Kong, and S.-Y. Cho. 1999. A recombinant 10-kDa protein of Taenia solium metacestodes specific to active neurocysticercosis. J. Infect. Dis. 180:1307-1315. [DOI] [PubMed] [Google Scholar]
- 6.Díaz, J. F., M. Verastegui, R. H. Gilman, V. C. W. Tsang, J. B. Pilcher, C. Gallo, H. H. Garcia, P. Torres, T. Montenegro, E. Miranda, and The Cysticercosis Working Group in Peru (CWG). 1992. Immunodiagnosis of human cysticercosis (Taenia solium): a field comparison of an antibody-enzyme-linked immunosorbent assay (ELISA), an antigen-ELISA, and an enzyme-linked immunoelectrotransfer blot (EITB) assay in Peru. Am. J. Trop. Med. Hyg. 46:610-615. [DOI] [PubMed] [Google Scholar]
- 7.Diwan, A. R., M. Coker-Vann, P. Brown, D. B. Subianto, R. Yolken, R. Desowitz, A. Escobar, C. J. Gibbs, Jr., and D. C. Gajdusek. 1982. Enzyme-linked immunosorbent assay (ELISA) for the detection of antibody to cysticerci of Taenia solium. Am. J. Trop. Med. Hyg. 31:364-369. [DOI] [PubMed] [Google Scholar]
- 8.Eisenhaber, B., P. Bork, and F. Eisenhaber. 1999. Prediction of potential GPI-modification sites in proprotein sequences. J. Mol. Biol. 292:741-758. [DOI] [PubMed] [Google Scholar]
- 9.Flisser, A. 1994. Taeniasis and cysticercosis due to Taenia solium. Prog. Clin. Parasitol. 4:77-116. [PubMed] [Google Scholar]
- 10.Garcia, H. H., and O. H. Del Brutto. 2000. Taenia solium cysticercosis. Emerg. Re-emerg. Dis. Latin Am. 14:97-119. [DOI] [PubMed] [Google Scholar]
- 11.Garcia, H. H., R. Gilman, M. Martinez, V. C. W. Tsang, J. B. Pilcher, G. Herrera, F. Díaz, M. Alvarado, E. Miranda, and The Cysticercosis Working Group in Peru. (CWG). 1993. Cysticercosis as a major cause of epilepsy in Peru. Lancet 341:197-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, H. H., R. H. Gilman, M. A. Tovar, E. Flores, R. Jo, V. C. W. Tsang, F. Díaz, P. Torres, E. Miranda, and The Cysticercosis Working Group in Peru (CWG). 1995. Factors associated with Taenia solium cysticercosis: analysis of nine hundred forty-six Peruvian neurologic patients. Am. J. Trop. Med. Hyg. 52:145-148. [DOI] [PubMed] [Google Scholar]
- 13.Garcia, H. H., M. Martinez, R. Gilman, G. Herrera, V. C. W. Tsang, J. B. Pilcher, F. Díaz, M. Verastegui, C. Gallo, M. Porras, M. Alvarado, J. Naranjo, E. Miranda, and The Cysticercosis Working Group in Peru. 1991. Diagnosis of cysticercosis in endemic regions. Lancet 338:549-551. [PMC free article] [PubMed] [Google Scholar]
- 14.Gemmell, M., Z. Matyas, Z. Pawlowski, and E. J. L. Soulsby. 1983. Guidelines for surveillance prevention and control of taeniasis/cysticercosis, p. 69-75. World Health Organization, Geneva, Switzerland.
- 15.Gottstein, B., D. Zini, and P. M. Schantz. 1987. Species-specific immunodiagnosis of Taenia solium cysticercosis by ELISA and immunoblotting. Trop. Med. Parasitol. 38:299-303. [PubMed] [Google Scholar]
- 16.Greene, R. M., K. Hancock, P. P. Wilkins, and V. C. W. Tsang. 2000. Taenia solium: molecular cloning and serologic evaluation of 14- and 18-kDa related, diagnostic antigens. J. Parasitol. 86:1001-1007. [DOI] [PubMed] [Google Scholar]
- 17.Greene, R. M., P. P. Wilkins, and V. C. W. Tsang. 1999. Diagnostic glycoproteins of Taenia solium cysts share homologous 14- and 18-kDa subunits. Mol. Biochem. Parasitol. 99:257-261. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, K., and V. C. W. Tsang. 1986. Development and optimization of the FAST-ELISA for detecting antibodies to Schistosoma mansoni. J. Immunol. Methods 92:167-176. [DOI] [PubMed] [Google Scholar]
- 19.Hubert, K., A. Andriantsimahavandy, A. Michault, M. Frosch, and F. A. Mühlschlegel. 1999. Serological diagnosis of human cysticercosis by use of recombinant antigens from Taenia solium cysticerci. Clin. Diagn. Lab. Immunol. 6:479-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssen, D., and J. Barrett. 1995. A novel lipid-binding protein from the cestode Moniezia expansa. Biochem. J. 311:49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 22.Medina, M., E. Rosas, F. Rubio, and J. Sotelo. 1990. Neurocysticercosis as the main cause of late-onset epilepsy in Mexico. Arch. Intern. Med. 150:325-327. [PubMed] [Google Scholar]
- 23.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Obregon-Henao, A., D. L. Gil, D. I. Gomez, F. Sanzon, J. M. Teale, and B. I. Restrepo. 2001. The role of N-linked carbohydrates in the antigenicity of Taenia solium metacestode glycoproteins of 12, 16 and 18 kD. Mol. Biochem. Parasitol. 114:209-215. [DOI] [PubMed] [Google Scholar]
- 25.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 26.Palacio, L. G., I. Jiménez, H. H. Garcia, M. E. Jiménez, J. L. Sánchez, J. Noh, L. Ahn, O. Mora, M. Giraldo, V. C. W. Tsang, and The Neuroepidemiological Research Group of Antioquia. 1998. Neurocysticercosis in epileptic persons in Medellín, Colombia. Epilepsia 39:1334-1339. [DOI] [PubMed] [Google Scholar]
- 26a.Pan American Health Organization. 1997. PAHO/WHO informal consultation on the taeniosis/cysticercosis complex. Pan American Health Organization, Washington, D.C.
- 27.Plancarte, A., C. Hirota, J. Martínez-Ocaña, G. Mendoza-Hernández, E. Zenteno, and A. Flisser. 1999. Characterization of GP39-42 and GP24 antigens from Taenia solium cysticerci and of their antigenic GP10 subunit. Parasitol. Res. 85:680-684. [DOI] [PubMed] [Google Scholar]
- 28.Proaño-Narvaez, J. V., A. Meza-Lucas, O. Mata-Ruiz, R. C. García-Jerónimo, and D. Correa. 2002. Laboratory diagnosis of human neurocysticercosis: double-blind comparison of enzyme-linked immunosorbent assay and electroimmunotransfer blot assay. J. Clin. Microbiol. 40:2115-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos-Kuri, M., R. M. Montoya, A. Padilla, T. Govezensky, M. L. Díaz, E. Sciutto, J. Sotelo, and C. Larralde. 1992. Immunodiagnosis of neurocysticercosis. Disappointing performance of serology (enzyme-linked immunosorbent assay) in an unbiased sample of neurological patients. Arch. Neurol. 49:633-636. [DOI] [PubMed] [Google Scholar]
- 30.Riganò, R., E. Profumo, F. Bruschi, G. Carulli, A. Azzarà, S. Ioppolo, B. Buttari, E. Ortona, P. Margutti, A. Teggi, and A. Siracusano. 2001. Modulation of human immune response by Echinococcus granulosus antigen B and its possible role in evading host defenses. Infect. Immun. 69:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosas, N., J. Sotelo, and N. Dionisio. 1986. ELISA in the diagnosis of neurocysticercosis. Arch. Neurol. 43:353-356. [DOI] [PubMed] [Google Scholar]
- 32.Saghir, N., P. J. Conde, P. M. Brophy, and J. Barrett. 2001. Biochemical characterization of a hydrophobic ligand binding protein from the tapeworm Hymenolepis diminuta. Int. J. Parasitol. 31:653-660. [DOI] [PubMed] [Google Scholar]
- 33.Saghir, N., P. J. Conde, P. M. Brophy, and J. Barrett. 2000. A new diagnostic tool for neurocysticercosis is a member of a cestode specific hydrophobic ligand binding protein family. FEBS Lett. 487:181-184. [DOI] [PubMed] [Google Scholar]
- 34.Sako, Y., M. Nakao, T. Ikejima, X. Z. Piao, K. Nakaya, and A. Ito. 2000. Molecular characterization and diagnostic value of Taenia solium low-molecular-weight antigen genes. J. Clin. Microbiol. 38:4439-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schantz, P. M., P. P. Wilkins, and V. C. W. Tsang. 1998. Immigrants, imaging, and immunoblots: the emergence of neurocysticercosis as a significant public health problem, p. 213-242. In W. M. Scheld, W. A. Craig, and J. M. Hughes (ed.), Emerging infections 2. ASM Press, Washington, D.C.
- 37.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 38.Strimmer, K., and A. Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsang, V. C., J. A. Brand, and A. E. Boyer. 1989. An enzyme-linked immunoelectrotransfer blot assay and glycoprotein antigens for diagnosing human cysticercosis (Taenia solium). J. Infect. Dis. 159:50-59. [DOI] [PubMed] [Google Scholar]
- 41.Tsang, V. C. W., R. M. Greene, and J. B. Pilcher. 1995. Optimization of the covalent conjugating procedure (NaIO4) of horseradish peroxidase to antibodies for use in enzyme-linked immunosorbent assay. J. Immunoassay 16:395-418. [DOI] [PubMed] [Google Scholar]
- 42.Tsang, V. C. W., and M. Wilson. 1995. Taenia solium cysticercosis: an under-recognized but serious public health problem. Parasitol. Today 11:124-126. [Google Scholar]
- 43.White, A. C., Jr. 1997. Neurocysticercosis: a major cause of neurological disease worldwide. Clin. Infect. Dis. 24:101-115. [DOI] [PubMed] [Google Scholar]
- 44.White, A. C., Jr., P. Robinson, and R. Kuhn. 1997. Taenia solium cysticercosis: host-parasite interactions and the immune response. Chem. Immunol. 66:208-230. [PubMed] [Google Scholar]
- 45.White, A. C., Jr. 2000. Neurocysticercosis: updates on epidemiology, pathogenesis, diagnosis, and management. Annu. Rev. Med. 51:187-206. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, M., R. T. Bryan, J. A. Fried, D. A. Ware, P. M. Schantz, J. B. Pilcher, and V. C. W. Tsang. 1991. Clinical evaluation of the cysticercosis enzyme-linked immunoelectrotransfer blot in patients with neurocysticercosis. J. Infect. Dis. 164:1007-1009. [DOI] [PubMed] [Google Scholar]
- 47.Youden, W. J. 1950. Index for rating diagnostic tests. Cancer 3:32-35. [DOI] [PubMed] [Google Scholar]