Abstract
A multiplex PCR assay for the identification of human diarrheagenic Escherichia coli was developed. The targets selected for each category were eae for enteropathogenic E. coli, stx for Shiga toxin-producing E. coli, elt and est for enterotoxigenic E. coli, ipaH for enteroinvasive E. coli, and aggR for enteroaggregative E. coli. This assay allowed the categorization of a diarrheagenic E. coli strain in a single reaction tube.
Escherichia coli is the most important etiologic agent of childhood diarrhea and represents a major public health problem in developing countries (8). Identification of diarrheagenic E. coli strains requires that these organisms be differentiated from nonpathogenic members of the normal flora. Serogrouping of O antigen is not sufficient to identify a strain as diarrheagenic, because it does not correlate, in most cases, with the presence of virulence factors (18). Thus, identification of diarrheagenic E. coli strains needs to detect factors that determine the virulence of these organisms. With the advent of PCR, it has become possible to detect pathogenic genes in bacterial isolates, allowing the rapid diagnosis of diarrheagenic E. coli. PCR methods using single primer sets have been reported elsewhere (5, 10, 17, 20), but screening of bacterial isolates requires a large number of individual PCRs if single primer sets are used in separate reactions. To reduce the number of tests needed for diagnosis of diarrheagenic E. coli, several multiplex PCR systems have been reported previously (7, 9, 12, 13, 15). However, usually more than one multiplex PCR is required for identification of a diarrheagenic E. coli strain. Recently, Pass et al. (11) reported a multiplex PCR to detect 11 virulence genes, but it has not been fully evaluated against a large panel of isolates. This study attempted to develop a multiplex PCR for identification of enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), and Shiga toxin-producing E. coli (STEC).
Thirteen E. coli control strains were used in this study (Table 1). EPEC, STEC, and EIEC strains were characterized in previous studies and confirmed to have the relevant gene by single PCRs and phenotypic assays (1, 6). For ETEC strains, the production of heat-labile enterotoxin was determined by a reversed passive latex agglutination test (Denka Seiken, Co., Ltd., Tokyo, Japan), and the production of heat-stable enterotoxin (ST) was determined by an enzyme immunoassay kit (Denka Seiken Co., Ltd.). For EAEC strains, the HEp-2 cell adherence assay was performed as described by Cravioto et al. (2).
TABLE 1.
Control strains used in this study
| Strain | Description | Serogroup | Positive gene(s) | Reference |
|---|---|---|---|---|
| E22-5 | EPEC | O152 | eae | 6 |
| E23-8 | EPEC | O164 | eae | 6 |
| O111-19 | STEC | O111 | eae, stx1 | 6 |
| O157-29 | STEC | O157 | eae, stx2 | 6 |
| O157-6 | STEC | O157 | eae, stx1, stx2 | 6 |
| EBa 35 | ETEC | O167 | elt | This study |
| O126-53 | ETEC | O126 | est | This study |
| KNH172 | ETEC | O148 | elt, est | This study |
| O126-32 | EAEC | O126 | aggR, aspU, CVD432 | This study |
| O111-15 | EAEC | O111 | aggR, aspU, CVD432 | This study |
| C-481 | EIEC | O143 | ipaH | 1 |
| D36f | EIEC | O124 | ipaH | 1 |
| O115-3 | None | O115 | This study |
The targets selected for each category were eae for EPEC, stx for STEC, elt and est for ETEC, and ipaH for EIEC. The primers to detect the bfpA (bundle-forming pilus) gene, which is present in typical EPEC, were not included in this multiplex PCR since the presence of eae is sufficient to define EPEC. On the other hand, atypical EPEC strains which do not possess bfpA and a high rate of spontaneous cure of the EAF plasmid have been reported previously (8). For each of the target genes, different pairs of primers were selected from the literature (Table 2) and tested in a single PCR. Universal primers were selected when different alleles could be present to reduce the number of primer sets. Therefore, primer set SK1-SK2 (10), which can detect all the intimin variants, was used for detection of eae; primer set VTcom-u-VTcom-d, which allows amplification of stx1, stx2, and its variants (21), was selected for stx; and the primer set AL65-AL125 (4), which reacts with the two ST-I toxin genes (ST-Ia and ST-Ib), was used for detection of the est gene. Primers sets LTL-LTR (20) and ipaIII-ipaIV (17) were selected to detect elt and ipaH, respectively, so that PCR products were sufficiently different in size to be distinguishable by agarose gel electrophoresis.
TABLE 2.
PCR primers used in this study
| Designation | Sequence (5′ to 3′) | Target gene | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| SK1 | CCCGAATTCGGCACAAGCATAAGC | eae | 881 | 10 |
| SK2 | CCCGGATCCGTCTCGCCAGTATTCG | |||
| VTcom-u | GAGCGAAATAATTTATATGTG | stx | 518 | 21 |
| VTcom-d | TGATGATGGCAATTCAGTAT | |||
| AL65 | TTAATAGCACCCGGTACAAGCAGG | est | 147 | 4 |
| AL125 | CCTGACTCTTCAAAAGAGAAAATTAC | |||
| LTL | TCTCTATGTGCATACGGAGC | elt | 322 | 20 |
| LTR | CCATACTGATTGCCGCAAT | |||
| ipaIII | GTTCCTTGACCGCCTTTCCGATACCGTC | ipaH | 619 | 17 |
| ipaIV | GCCGGTCAGCCACCCTCTGAGAGTAC | |||
| aggRks1 | GTATACACAAAAGAAGGAAGC | aggR | 254 | 14 |
| aggRkas2 | ACAGAATCGTCAGCATCAGC | |||
| Eaggfp | AGACTCTGGCGAAAGACTGTATC | CVD432 | 194 | 11 |
| Eaggbp | ATGGCTGTCTGTAATAGATGAGAAC | |||
| aspU-3 | GCCTTTGCGGGTGGTAGCGG | aspU | 282 | This study |
| aspU-2 | AACCCATTCGGTTAGAGCAC |
As EAEC strains are heterogenous (3, 16, 19) and no DNA sequence was demonstrated to be present in all strains, the HEp-2 cell adherence assay is still the best method of defining this category. To determine the appropriate target gene to define EAEC, we investigated the presence of aggR (transcriptional activator of AAF/I and AAF/II), CVD432 probe (cryptic open reading frame), and aspU (EAEC-secreted protein U) genes described as prevalent in this E. coli category by Czeczulin et al. (3). Twenty EAEC strains, defined by HEp-2 cell adherence pattern, were studied by single PCR with use of the primer pairs aggRks1-aggRkas2 for aggR (14), Eaggfp-Eaggbp for the CVD432 probe (11), and aspU-3-aspU-2 for aspU (Table 2). Ten strains (50%) were positive for aggR and the CVD432 probe, while 11 strains (55%) were positive for aspU. Nine strains (45%) were negative for the three sets of primers. According to the results obtained, each primer set was tested in combination with the primer sets to define EPEC, ETEC, EIEC, and STEC.
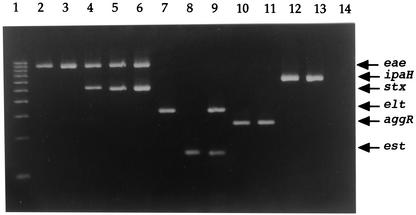
For the PCR, DNAs were extracted from control strains by the method described by Yokoyama (22). Briefly, control strains were cultured in 2 ml of Luria-Bertani broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) and incubated overnight at 37°C with shaking. Thirty-six microliters of broth culture was added to 4 μl of 10× Tris-EDTA buffer (100 mM Tris-HCl, 10 mM EDTA, pH 8.3), and 60 μl of 2× proteinase K buffer (100 mM KCl, 20 mM Tris-HCl, 5 mM MgCl2, 1% Tween 20, 800 μg of proteinase K/ml, pH 8.3) was added. After incubation for 90 min at 56°C and 10 min at 95°C, the sample was centrifuged at 10,000 × g for 1 min, and the supernatant was used as DNA template. Having confirmed the specificity of each primer set by single PCR, we combined six primer sets in different ratios and tested the control strains in several PCR cycling protocols. The optimized protocol was carried out with a 50-μl mixture containing 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 0.1% Triton X-100; 1.5 mM MgCl2; 2.5 U of Taq DNA polymerase (Toyobo, Osaka, Japan); 0.2 mM deoxynucleoside triphosphate; a 0.125 μM concentration (each) of primers SK1, SK2, ipaIII, and ipaIV; a 0.25 μM concentration (each) of primers VTcom-u, VTcom-d, LTL, LTR, aggRks1, and aggRkas2; a 0.5 μM concentration (each) of primers AL65 and AL125; and 5 μl of the DNA template. The PCR program was 95°C for 1 min, 52°C for 1 min, and 72°C for 1 min, for 30 cycles, and 72°C for 10 min. PCR products were then electrophoresed on a 2.5% agarose gel (AmpliSize; Bio-Rad Laboratories), stained with ethidium bromide, and visualized by UV transillumination. The buffer in the electrophoresis chamber and in the agarose gel was 0.5× Tris-borate-EDTA (11). The strains shown in Fig. 1 gave PCR products of the expected sizes for eae (881 bp, lanes 2 to 6), stx (518 bp, lanes 4 to 6), elt (322 bp, lanes 7 and 9), est (147 bp, lanes 8 and 9), aggR (254 bp, lanes 10 and 11), and ipaH (619 bp, lanes 12 and 13). PCR products were obtained for all six genes, and no PCR product was detected in the negative control (lane 14). To assess the sensitivity of the multiplex PCR, overnight cultures of control strains were serially 10-fold diluted and DNA was extracted as described above. Extracts of these samples were then subjected to multiplex PCR. The sensitivity of detection was 103 CFU per assay for ipaH and 104 CFU per assay for eae, elt, est, aggR, and stx (data not shown). Therefore, the presence of 104 CFU per assay must be ensured for detection of all the categories.
FIG. 1.
Agarose gel electrophoresis of products from multiplex PCR with control strains. Lanes: 1, 100-bp DNA ladder; 2, E22-5 (EPEC); 3, E23-8 (EPEC); 4, O111-19 (STEC); 5, O157-29 (STEC); 6, O157-6 (STEC); 7, EBa 35 (ETEC); 8, O126-53 (ETEC); 9, KNH172 (ETEC); 10, O126-32 (EAEC); 11, O111-15 (EAEC); 12, C-481 (EIEC); 13, D36f (EIEC); 14, O115-3 (negative control).
To demonstrate the utility of the multiplex PCR assay, 156 strains isolated from diarrheic patients were subjected to the optimized protocol, and the results were compared with those obtained by single PCR (Table 3). The EPEC strains tested included representatives of serogroups O55 (3 strains), O111 (11 strains), O119 (11 strains), O114 (1 strain), and O26 (1 strain). The STEC strains tested belonged to serogroups O26 (8 strains); O111 (10 strains); O157 (16 strains); O145 (2 strains); and O15, O121, O171, and OX3 (1 strain each). The EIEC strains tested belonged to serogroups O28ac (eight strains), O112ac (one strain), O124 (three strains), and O143 (one strain). The ETEC strains were of serogroups O26 (1 strain), O27 (2 strains), O126 (13 strains), and O128 (6 strains), and there were six strains that were O nontypeable or of unknown type. The EAEC strains belonged to serogroups O44 (six strains); O111 (eight strains); O126 (seven strains); O128 (two strains); O144 (two strains); and O127a, O157, O159, O164, and O166 (one strain each). There was agreement between the single and multiplex PCRs for all the strains.
TABLE 3.
Results of single and multiplex PCRs
| Category | No. of strains examined | Gene | No. of positives by PCR
|
|
|---|---|---|---|---|
| Single | Multiplex | |||
| EPEC | 27 | eae | 27 | 27 |
| STEC | 40 | eae | 37 | 37 |
| stx | 40 | 40 | ||
| EIEC | 13 | ipaH | 13 | 13 |
| ETEC | 28 | elt | 7 | 7 |
| est | 22 | 22 | ||
| EAEC | 30 | aggR | 30 | 30 |
| None | 18 | 0 | 0 | |
Our multiplex PCR could identify EPEC, STEC, EIEC, and ETEC strains because the virulence markers for these four categories are well defined. To include the identification of EAEC strains in the multiplex PCR, we selected the primer set aggRks1-aggRkas2, which gave the best result when combined with the other five sets of primers. Some aggR-negative strains with aggregative adherence will not be detected by this assay. However, considering the difficulty of performing phenotypic assays in some laboratories, the multiplex PCR presented here is a practical and rapid diagnostic tool for identification of diarrheagenic E. coli in a single reaction tube.
Acknowledgments
The work was partly supported by the Uruma Trust Fund for Research into Science and Humanity of Japan.
REFERENCES
- 1.Chinen, I., M. Rivas, M. I. Caffer, R. O. Cinto, and N. Binsztein. 1993. Diagnosis of enteroinvasive Escherichia coli associated with diarrhea. Rev. Argent. Microbiol. 25:27-35. [PubMed] [Google Scholar]
- 2.Cravioto, A., R. J. Gross, S. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 3.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hornes, E., Y. Wasteson, and Ø. Olsvik. 1991. Detection of Escherichia coli heat-stable enterotoxin genes in pig stool specimens by an immobilized, colorimetric, nested polymerase chain reaction. J. Clin. Microbiol. 29:2375-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karch, H., and T. Meyer. 1989. Single primer pair for amplifying segments of distinct Shiga-like toxin genes by polymerase chain reaction. J. Clin. Microbiol. 27:2751-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu, Y., C. Toma, Y. Honma, and M. Iwanaga. 2002. Detection of EspB using reversed passive latex agglutination: application to determination of enteropathogenic Escherichia coli. Diagn. Microbiol. Infect. Dis. 43:7-12. [DOI] [PubMed] [Google Scholar]
- 7.Nagano, I., M. Kunishima, Y. Itoh, Z. Wu, and Y. Takahashi. 1998. Detection of verotoxin-producing Escherichia coli O157:H7 by multiplex polymerase chain reaction. Microbiol. Immunol. 42:371-376. [DOI] [PubMed] [Google Scholar]
- 8.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osek, J. 2001. Multiplex polymerase chain reaction assay for identification of enterotoxigenic Escherichia coli strains. J. Vet. Diagn. Investig. 13:308-311. [DOI] [PubMed] [Google Scholar]
- 10.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marchès, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pass, M. A., R. Odedra, and R. M. Batt. 2000. Multiplex PCRs for identification of Escherichia coli virulence genes. J. Clin. Microbiol. 38:2001-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rappelli, P., G. Maddau, F. Mannu, M. M. Colombo, P. L. Fiori, and P. Cappuccinelli. 2001. Development of a set of multiplex PCR assays for the simultaneous identification of enterotoxigenic, enteropathogenic, enterohemorrhagic and enteroinvasive Escherichia coli. Microbiologica 24:77-83. [PubMed] [Google Scholar]
- 14.Ratchtrachenchai, O. A., S. Subpasu, and K. Ito. 1997. Investigation on enteroaggregative Escherichia coli infection by multiplex PCR. Bull. Dep. Med. Sci. 39:211-220. [Google Scholar]
- 15.Rich, C., A. Alfidja, J. Sirot, B. Joly, and C. Forestier. 2001. Identification of human enterovirulent Escherichia coli strains by multiplex PCR. J. Clin. Lab. Anal. 15:100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaletsky, I. C. A., S. H. Fabbricotti, K. R. Aranda, M. B. Morais, and U. Fagundes-Neto. 2002. Comparison of DNA hybridization and PCR assays for detection of putative pathogenic enteroadherent Escherichia coli. J. Clin. Microbiol. 40:1254-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sethabutr, O., M. Venkatesan, G. S. Murphy, B. Eampokalap, C. W. Hoge, and P. Echeverria. 1993. Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J. Infect. Dis. 167:458-461. [DOI] [PubMed] [Google Scholar]
- 18.Sunabe, T., and Y. Honma. 1998. Relationship between O-serogroup and presence of pathogenic factor genes in Escherichia coli. Microbiol. Immunol. 42:845-849. [DOI] [PubMed] [Google Scholar]
- 19.Suzart, S., B. E. C. Guth, M. Z. Pedroso, U. M. Okafor, and T. A. T. Gomes. 2001. Diversity of surface structures and virulence genetic markers among enteroaggregative Escherichia coli (EAEC) strains with and without the EAEC DNA probe sequence. FEMS Microbiol. Lett. 201:163-168. [DOI] [PubMed] [Google Scholar]
- 20.Tamanai-Shacoori, Z., and A. Jolivet-Gougeon. 1994. Detection of enterotoxigenic Escherichia coli in water by polymerase chain reaction amplification and hybridization. Can. J. Microbiol. 40:243-249. [DOI] [PubMed] [Google Scholar]
- 21.Yamasaki, S., Z. Lin, H. Shirai, A. Terai, Y. Oku, H. Ito, M. Ohmura, T. Karasawa, T. Tsukamoto, H. Kurazono, and Y. Takeda. 1996. Typing of verotoxins by DNA colony hybridization with poly- and oligonucleotide probes, a bead-enzyme-linked immunosorbent assay, and polymerase chain reaction. Microbiol. Immunol. 40:345-352. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama, T. 1993. Study on mec gene in methicillin-resistant staphylococci. Kansenshogaku Zasshi 67:1203-1210. [DOI] [PubMed] [Google Scholar]