Abstract
Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification method in which reagents react under isothermal conditions with high specificity, efficiency, and rapidity. We used LAMP for detection of Mycobacterium tuberculosis complex, Mycobacterium avium, and Mycobacterium intracellulare directly from sputum specimens as well as for detection of culture isolates grown in a liquid medium (MGIT; Nippon Becton Dickinson Co., Ltd., Tokyo, Japan) or on a solid medium (Ogawa's medium). Species-specific primers were designed by targeting the gyrB gene, and their specificities were validated on 24 mycobacterial species and 7 nonmycobacterial species. The whole procedure is quite simple, starting with the mixing of all reagents in a single tube, followed by an isothermal reaction during which the reaction mixture is held at 63°C. The resulting amplicons are visualized by adding SYBR Green I to the reaction tube. The only equipment needed for the amplification reaction is a regular laboratory water bath or heat block that furnishes a constant temperature of 63°C. The assay had a detection limit of 5 to 50 copies of purified DNA with a 60-min incubation time. The reaction time could be shortened to 35 min for the species identification of M. tuberculosis complex, M. avium, and M. intracellulare from a solid-medium culture. Residual DNA lysates prepared for the Amplicor assay (Roche Diagnostics GmbH) from 66 sputum specimens were tested in the LAMP assay. Although the sample size used for the latter assay was small, 2.75 μl of the DNA lysates, it showed a performance comparable with that of the Amplicor assay, which required 50 μl of the lysates. This LAMP-based assay is simple, rapid, and sensitive; a result is available in 35 min for a solid-medium culture and in 60 min for a liquid-medium culture or for a sputum specimen that contains a corresponding amount of DNA available for testing.
Because of their slow growth rate, identification of mycobacteria is a notorious problem for public health and clinical laboratories. To address the need for rapid and sensitive identification of Mycobacterium tuberculosis and other mycobacteria, various genotyping methods for routine diagnosis have been introduced during the past decade (1, 2, 4, 9, 10, 21-23, 25). The recent trend in genetic testing is to make systems fully automatic with high-throughput analysis. Although this may be an ideal approach, it requires expensive equipment as well as a huge amount of space in routine diagnostic laboratories. As a result, it centralizes genetic testing in highly sophisticated facilities. If this trend continues, escalated pressure for transportation of clinical specimens will pose another problem. On the other hand, there are pressing needs for point-of-care testing at hospitals and primary care facilities.
Loop-mediated isothermal amplification (LAMP) is a novel nucleic acid amplification method which relies on autocycling strand displacement DNA synthesis performed by the Bst DNA polymerase large fragment (13, 15-17). The amplification products are stem-loop DNA structures with several inverted repeats of the target and cauliflower-like structures with multiple loops. LAMP has the following characteristics: (i) all reactions can be conducted under isothermal conditions ranging from 60 to 65°C by using only one type of enzyme; (ii) the specificity of the reaction is extremely high because it uses four primers recognizing six distinct regions on the target DNA; (iii) amplification can be performed in a shorter time than amplification by PCR because there is no time loss due to thermal cycling; and (iv) it produces extremely large amounts of amplified products and enables simple detection methods such as visual judgment by the turbidity or fluorescence of the reaction mixture, which is kept in the reaction tube (13). With all these characteristics, LAMP of DNA has emerged as a powerful tool to facilitate point-of-care genetic testing at the bedside. Recently, Nagamine et al. (14) reported that when two more primers, termed loop primers, were added, the LAMP reaction time could be even less than half of that for the original LAMP method. In their procedure, six primers recognize eight distinct regions on the targeted DNA. In the present study, we used LAMP technology with a modification of the detection system for diagnosis of mycobacteria in sputum samples. The sensitivity, specificity, and applicability of this method for direct detection of M. tuberculosis complex, Mycobacterium avium, and Mycobacterium intracellulare from sputum samples were evaluated.
MATERIALS AND METHODS
DNA preparation.
Genomic DNAs used for evaluation of primer specificity were prepared from 27 reference strains and 8 clinical isolates representing 24 mycobacterial species and 7 nonmycobacterial species (Table 1). One to two colonies from each strain were suspended in 100 μl of lysis buffer (20 mM Tris · HCl [pH 8.0], 2 mM EDTA, 1.2% Triton X-100) and boiled for 20 min. The crude lysates were used as LAMP templates. Genomic DNAs of M. tuberculosis H37Rv, M. tuberculosis H37Ra, Mycobacterium bovis BCG Tokyo, M. avium ATCC 15769, M. avium ATCC 25291, M. avium ATCC 35718, M. intracellulare ATCC 13950, M. intracellulare ATCC 35847, and M. intracellulare ATCC 35762 were also extracted from Mycobacterium Growth Indicator Tube (MGIT) broth on the day when positive fluorescence was detected. One hundred microliters of positive MGIT broth was pipetted into a 1.5-ml screw-cap polypropylene tube, and the tube was boiled for 20 min to obtain the crude lysate. For the sensitivity study, purified DNAs from M. tuberculosis H37Rv, M. avium ATCC 25291, and M. intracellulare ATCC 13950 were isolated with Isoplant (Nippon Gene, Toyama, Japan) according to the manufacturer's instructions. DNAs were quantified by using a DNA DipStick kit (Invitrogen Co., Carlsbad, Calif.) as recommended by the manufacturer. The number of genomic copies per LAMP mixture and the number of genomic copies per Amplicor reaction solution were calculated by assuming a molecular size of 4.4 Mbp for each of these three species.
TABLE 1.
Specificity of LAMP primers for identification of mycobacterial species
| Strain | Amplification by LAMP primera:
|
|||
|---|---|---|---|---|
| MTB | MAV | MIN | Muniv | |
| Mycobacteria | ||||
| M. tuberculosis H37Rv | + | − | − | + |
| M. tuberculosis H37Ra | + | − | − | + |
| M. tuberculosis clinical isolate 1 | + | − | − | + |
| M. tuberculosis clinical isolate 2 | + | − | − | + |
| M. bovis BCG Tokyo | + | − | − | + |
| M. avium ATCC 15769b | − | + | − | + |
| M. avium ATCC 25291 | − | + | − | + |
| M. avium ATCC 35718 | − | + | − | + |
| M. avium clinical isolate 1 | − | + | − | + |
| M. avium clinical isolate 2 | − | + | − | + |
| M. intracellulare ATCC 13950 | − | − | + | + |
| M. intracellulare ATCC 35847 | − | − | + | + |
| M. intracellulare ATCC 35762 | − | − | + | + |
| M. intracellulare clinical isolate 1 | − | − | + | + |
| M. intracellulare clinical isolate 2 | − | − | + | + |
| M. kansasii ATCC 12478 | − | − | − | + |
| M. marinum ATCC 927 | − | − | − | + |
| M. simiae ATCC 25275 | − | − | − | + |
| M. scrofulaceum ATCC 19981 | − | − | − | + |
| M. gordonae ATCC 14470 | − | − | − | + |
| M. lentiflavum clinical isolate | − | − | − | + |
| M. szulgai ATCC 35799 | − | − | − | + |
| M. xenopi ATCC 19250 | − | − | − | + |
| M. gastri ATCC 15754 | − | − | − | + |
| M. nonchromogenicum ATCC 19530 | − | − | − | + |
| M. triviale ATCC 23292 | − | − | − | + |
| M. terrae clinical isolate | − | − | − | + |
| M. fortuitum ATCC 6841 | − | − | − | + |
| M. chelonae ATCC 19237 | − | − | − | + |
| M. abscessus ATCC 19977 | − | − | − | + |
| M. smegmatis ATCC 14468 | − | − | − | + |
| M. flavescens ATCC 14474 | − | − | − | + |
| M. phlei ATCC 11758 | − | − | − | + |
| M. diernhoferi ATCC 19340 | − | − | − | + |
| M. vaccae ATCC 15483 | − | − | − | + |
| Nonmycobacterial species | ||||
| Corynebacterium flavescens NBRC 14136c | − | − | − | + |
| Corynebacterium xerosis NBRC 12684 | − | − | − | + |
| Nocardia farcinica NBRC 15532 | − | − | − | + |
| Rhodococcus equi NBRC 3730 | − | − | − | + |
| Streptococcus salivarius NBRC 13956 | − | − | − | − |
| Klebsiella pneumoniae NBRC 3318 | − | − | − | − |
| Pseudomonas aeruginosa NBRC 3080 | − | − | − | − |
+, amplification was found after 35 min of incubation; −, amplification was not found after 60 min of incubation.
Source, American Type Culture Collection.
Source, Natoinal Institute of Technology and Evaluation Biological Resource Center.
Clinical sputum specimens.
Sixty-six sputum specimens from 58 patients with suspected mycobacteriosis were obtained from the Kobe City General Hospital and Kobe City Public Health Office. After decontamination by N-acetylcysteine-NaOH treatment and subsequent concentration by centrifugation, DNA was extracted from these specimens with a respiratory specimen preparation kit (Roche Diagnostics GmbH). Fifty microliters of the DNA lysate (200 μl) was used for Amplicor tests (Roche Diagnostics GmbH), which were performed according to the manufacturer's guidelines, and 2.75 μl of the DNA lysate was used for the LAMP reaction. The sputum specimens were also subjected to acid-fast staining and culture. All of the clinical isolates were identified by using either Accu-Probe (Gen-Probe, Inc., San Diego, Calif.) or DDH Mycobacteria “Kyokuto” (Kyokuto Pharmaceutical Ind., Co., Ltd., Tokyo, Japan).
DNA oligonucleotides.
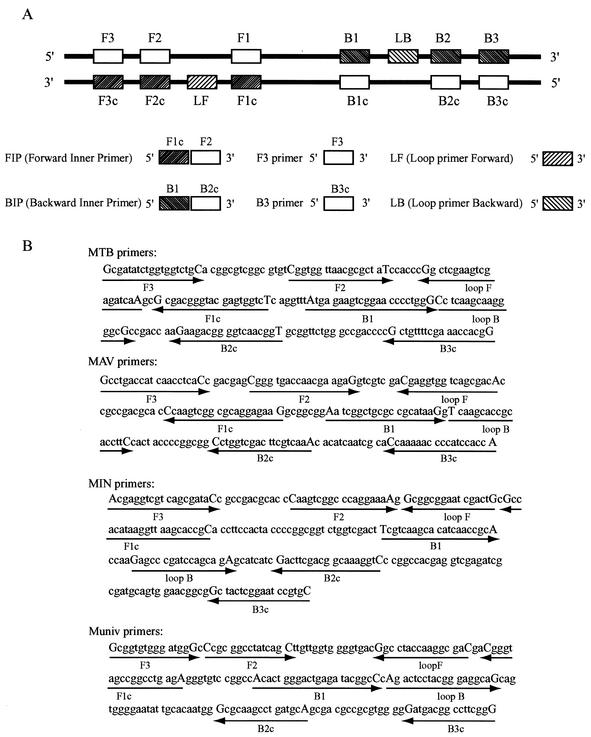
We used six primers: outer primers (F3 and B3), a forward inner primer (FIP), a backward inner primer (BIP), and loop primers (loop F and loop B). They recognize eight distinct regions on the target DNA (Fig. 1A). FIP consists of a complementary sequence of F1 and a sense sequence of F2. BIP consists of a sense sequence of B1 and a complementary sequence of B2. Species-specific primers MTB for M. tuberculosis complex, MAV for M. avium, and MIN for M. intracellulare were designed to target the gyrB gene sequences (Fig. 1B). The genus universal primers, Muniv, for the genus Mycobacterium, are specific for universally conserved mycobacterial 16S ribosomal DNA (rDNA) sequences (Fig. 1B). All of the sequence data for designing the primers were obtained from the Identification and Classification of Bacteria (ICB) gyrB database for gyrB (8) and from the ribosomal differentiation of medical microoganisms (RIDOM) database for 16S rDNA (6, 7).
FIG. 1.
(A) Schematic representation of primers used in this study. Construction of the inner primers FIP and BIP is shown. F1c and B2c, complementary sequences of F1 and B2, respectively. (B) Nucleotide sequences of gyrB and 16S rDNA used for designing the primers. Recognition sequences of the primers are shown between capital letters. A right arrow indicates that a sense sequence is used for the primer. A left arrow indicates that a complementary sequence is used for the primer.
LAMP reaction.
The LAMP reaction was performed with a Loopamp DNA amplification kit (Eiken Chemical Co., Ltd., Tochigi, Japan). A reaction mixture (12.5 μl) containing 1.6 μM each inner primer (FIP and BIP), 0.2 μM each outer primer (F3 and B3), 0.8 μM each loop primer (F and B), 2× reaction mix (6.25 μl), Bst DNA polymerase (0.5 μl), and the specified amounts of DNA lysates (1.0 μl for a solid-medium isolate and 2.75 μl for a liquid-medium isolate or sputum specimen) was incubated at 63°C for 35 min (for solid-medium isolates) or 60 min (for liquid-medium isolates or sputum samples) and was heated at more than 80°C for 2 min to terminate the reaction.
Analysis of LAMP products.
LAMP amplicons in the reaction tube were directly detected with the naked eye by adding 1.0 μl of 1/10-diluted original SYBR Green I (Molecular Probes Inc.) to the tube and observing the color of the solution. The solution turned green in the presence of a LAMP amplicon, while it remained orange with no amplification. For further confirmation, some of the amplified products were also detected by agarose gel electrophoresis. Five-microliter aliquots of LAMP products and 2-μl aliquots of products digested with restriction enzymes (BsaI for MTB amplicons, HaeIII for MAV amplicons, and Tth111I for MIN amplicons) were electrophoresed in 3% agarose gels, followed by staining with ethidium bromide. The sensitivities of electrophoresis and SYBR Green I inspection with the naked eye were compared by using serially diluted LAMP products.
RESULTS
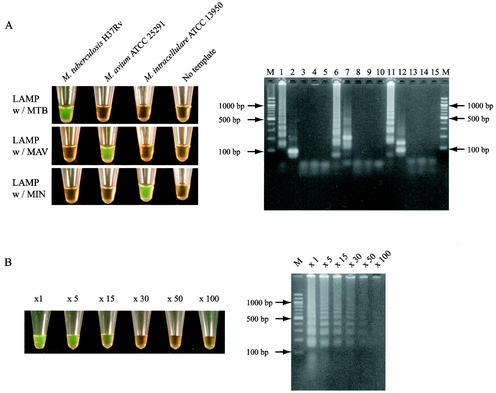
A successful LAMP reaction with species-specific primers produced many bands of different sizes (Fig. 2A). When the sample tube did not contain target DNA, no amplification was seen. To confirm that the amplification products had corresponding DNA structures, the amplified products were digested with restriction enzymes and the sizes of the fragments were analyzed by electrophoresis. BsaI cuts between F1c and B1 for MTB amplicons, HaeIII cuts B2 for MAV amplicons, and Tth111I cuts B1c for MIN amplicons. The sizes of the fragments generated after digestion were in good agreement with the sizes predicted theoretically from the expected DNA structures: 88 and 89 bp by BsaI digestion, 200 bp by HaeIII digestion, and 90 and 142 bp by Tth111I digestion (Fig. 2A). A LAMP reaction mixture which contained amplified fragments turned green after the addition of SYBR Green I, whereas a solution with no amplicons retained the original orange color of SYBR Green I (Fig. 2A). Thus, the results of the LAMP reactions can simply be judged by the naked eye. The sensitivities of inspection by the naked eye and detection by electrophoresis were compared using variously diluted LAMP products (Fig. 2B). The results demonstrated that electrophoresis is slightly more sensitive (detecting 30- to 50-fold-diluted products) than inspection of color change by the naked eye (detecting 15-fold-diluted products).
FIG. 2.
(A) Visual inspection and electrophoretic analysis of LAMP amplified products. Lane M, 100-bp ladder used as a size marker; lanes 1, 3, and 4, LAMP carried out with MTB primers in the presence of genomic DNA from M. tuberculosis, M. avium, and M. intracellulare, respectively; lane 2, LAMP product from lane 1 after digestion with BsaI; lanes 5, 10, and 15, LAMP carried out in the absence of template DNA with MTB, MAV, and MIN primers, respectively; lanes 6, 8, and 9, LAMP carried out with MAV primers in the presence of genomic DNA from M. avium, M. tuberculosis, and M. intracellulare, respectively; lane 7, LAMP product from lane 6 after digestion with HaeIII; lanes 11, 13, and 14, LAMP carried out with MIN primers in the presence of genomic DNA from M. intracellulare, M. tuberculosis, and M. avium, respectively; lane 12, LAMP product from lane 11 after digestion with Tth111I. (B) Sensitivities of visual inspection and electrophoretic analysis of LAMP amplified products. The number above each tube or lane represents the dilution of the LAMP product: ×1, no dilution; ×5, 5-fold dilution; ×15, 15-fold dilution; ×30, 30-fold dilution; ×50, 50-fold dilution; ×100, 100-fold dilution.
To evaluate the species specificities of the newly designed primers, we tested 35 mycobacterial strains and 7 nonmycobacterial species which were grown on solid media (Table 1). Significant amplification of the DNAs isolated from the targeted organisms was observed after a 35-min incubation. In contrast, nontargeted strains were not amplified even after 60 min of incubation. Universal primers for the genus Mycobacterium were also used to confirm extraction of the proper DNAs. The primers amplified all of the mycobacteria after 35 min of incubation. Besides the mycobacteria, four actinomycetes were also amplified. When the method was applied to liquid-medium cultures of M. tuberculosis H37Rv, M. tuberculosis H37Ra, M. bovis BCG Tokyo, M. avium ATCC 15769, M. avium ATCC 25291, M. avium ATCC 35718, M. intracellulare ATCC 13950, M. intracellulare ATCC 35847, and M. intracellulare ATCC 35762, consistent results for solid-medium cultures were obtained with the 60-min incubation.
The LAMP reaction with a 60-min incubation and visual inspection has a sensitivity equivalent to that of the Amplicor test (Table 2). Both methods showed a detection limit of 5 to 50 genomes per test for the three mycobacterial species. The sensitivity of the LAMP assay was 10 to 100 times lower when the reaction time was shortened to 35 min.
TABLE 2.
Sensitivities of LAMP and Amplicor assays for M. tuberculosis complex, M. avium, and M. intracellulare
| Assay | Resulta for:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
M. tuberculosis complex
|
M. avium
|
M. intracellulare
|
||||||||||||||||
| 10,000 copies | 1,000 copies | 100 copies | 50 copies | 5 copies | 0.5 copy | 10,000 copies | 1,000 copies | 100 copies | 50 copies | 5 copies | 0.5 copy | 10,000 copies | 1,000 copies | 100 copies | 50 copies | 5 copies | 0.5 copy | |
| Amplicor | + | + | + | + | ± | − | + | + | + | + | ± | − | + | + | + | + | ± | − |
| LAMP, 60 min | + | + | + | + | ± | − | + | + | + | + | − | − | + | + | + | + | ± | − |
| LAMP, 35 min | + | + | ± | − | − | − | + | + | − | − | − | − | + | + | − | − | − | − |
+, triplicate assay showed all positive; ±, triplicate assay showed both positive and negative; −, triplicate assay showed all negative.
Residual DNA lysates prepared for Amplicor testing from 66 sputum specimens were tested in the LAMP assay (Table 3). Five Amplicor-positive samples (samples 19 to 21 for M. tuberculosis complex, sample 25 for M. intracellulare, and sample 33 for M. avium) were negative by the LAMP assay. Other results were identical for these two assays. Eight samples negative by both the Amplicor and LAMP tests (samples 26, 32, and 36 to 40) were culture positive. Among these, five samples were identified as either M. abscessus (samples 37. 38a, and 38b) or M. kansasii (samples 39 and 40), which are not targeted by these two assays. Four culture-negative samples (samples 13, 14, 21, and 30b) were positive by Amplicor, and three of these (samples 13, 14, and 30b) were positive by LAMP. When the samples positive for M. tuberculosis complex, M. avium, and M. intracellulare by any one of these three methods were summed up, 8 of 41 positive samples showed false-negative results by LAMP. These samples were retested with the MTB primer set after 50 copies of M. tuberculosis DNA were added to each reaction mixture. All samples gave positive results. This indicates that the possible reason for these false-negative results is a sensitivity issue and is not related to inhibition of amplification. Both the Amplicor and LAMP methods detected all of the smear-positive samples, which were identified as M. tuberculosis complex, M. avium, or M. intracellulare.
TABLE 3.
Comparison of LAMP assay with Amplicor for direct identification of mycobacteria in sputum specimens
| Specimen no.a | Smear resultb | Species identification of culture isolates | Result with the following assay and primer:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Amplicorb
|
LAMPb
|
||||||||
| MTB | MAV | MIN | IC | MTB | MAV | MIN | |||
| 1 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 2 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 3 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 4 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 5 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 6 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 7 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 8 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 9 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 10 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 11 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 12 | Pos | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 13 | Pos | Culture negative | Pos | — | — | Pos | Pos | — | — |
| 14 | Pos | Culture negative | Pos | — | — | Pos | Pos | — | — |
| 15a | — | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 15b | — | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 15c | — | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 16 | — | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 17 | — | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 18 | — | M. tuberculosis complex | Pos | — | — | Pos | Pos | — | — |
| 19 | — | M. tuberculosis complex | Pos | — | — | Pos | — | — | — |
| 20 | — | M. tuberculosis complex | Pos | — | — | Pos | — | — | — |
| 21 | — | Culture negative | Pos | — | — | Pos | — | — | — |
| 22a | Pos | M. intracellulare | — | — | Pos | Pos | — | — | Pos |
| 22b | Pos | M. intracellulare | — | — | Pos | Pos | — | — | Pos |
| 23a | Pos | M. intracellulare | — | — | Pos | Pos | — | — | Pos |
| 23b | Pos | M. intracellulare | — | — | Pos | Pos | — | — | Pos |
| 24 | — | M. intracellulare | — | — | Pos | Pos | — | — | Pos |
| 25 | — | M. intracellulare | — | — | Pos | Pos | — | — | — |
| 26 | — | M. intracellulare | — | — | — | Pos | — | — | — |
| 27 | Pos | M. avium | — | Pos | — | Pos | — | Pos | — |
| 28 | Pos | M. avium | — | Pos | — | Pos | — | Pos | — |
| 29 | Pos | M. avium | — | Pos | — | Pos | — | Pos | — |
| 30a | Pos | M. avium | — | Pos | — | Pos | — | Pos | — |
| 30b | Pos | Culture negative | — | Pos | — | Pos | — | Pos | — |
| 31 | Pos | M. avium | — | Pos | — | Pos | — | Pos | — |
| 32 | — | M. avium | — | — | — | Pos | — | — | — |
| 33 | — | M. avium | — | Pos | — | Pos | — | — | — |
| 34 | — | M. avium | — | Pos | — | Pos | — | Pos | — |
| 35 | — | M. avium | — | Pos | — | Pos | — | Pos | — |
| 36 | — | M. avium | — | — | — | Pos | — | — | — |
| 37 | Pos | M. abscessus | — | — | — | Pos | — | — | — |
| 38a | Pos | M. abscessus | — | — | — | Pos | — | — | — |
| 38b | — | M. abscessus | — | — | — | Pos | — | — | — |
| 39 | Pos | M. kansasii | — | — | — | Pos | — | — | — |
| 40 | — | M. kansasii | — | — | — | Pos | — | — | — |
| 41a | — | Culture negative | — | — | — | Pos | — | — | — |
| 41b | — | Culture negative | — | — | — | Pos | — | — | — |
| 42 | — | Culture negative | — | — | — | Pos | — | — | — |
| 43 | — | Culture negative | — | — | — | Pos | — | — | — |
| 44a | — | Culture negative | — | — | — | Pos | — | — | — |
| 44b | — | Culture negative | — | — | — | Pos | — | — | — |
| 45 | — | Culture negative | — | — | — | Pos | — | — | — |
| 46 | — | Culture negative | — | — | — | Pos | — | — | — |
| 47 | — | Culture negative | — | — | — | Pos | — | — | — |
| 48 | — | Culture negative | — | — | — | Pos | — | — | — |
| 49 | — | Culture negative | — | — | — | Pos | — | — | — |
| 50 | — | Culture negative | — | — | — | Pos | — | — | — |
| 51 | — | Culture negative | — | — | — | Pos | — | — | — |
| 52 | — | Culture negative | — | — | — | Pos | — | — | — |
| 53 | — | Culture negative | — | — | — | Pos | — | — | — |
| 54 | — | Culture negative | — | — | — | Pos | — | — | — |
| 55 | — | Culture negative | — | — | — | Pos | — | — | — |
| 56 | — | Culture negative | — | — | — | Pos | — | — | — |
| 57 | — | Culture negative | — | — | — | Pos | — | — | — |
| 58 | — | Culture negative | — | — | — | Pos | — | — | — |
The same number with different lowercase letters represents different sputum specimens obtained from the same patient.
Pos, positive; —, negative.
DISCUSSION
We used a novel nucleic acid amplification method, LAMP, for detection of M. tuberculosis complex, M. avium, and M. intracellulare either as culture isolates or in sputum specimens. The mycobacterial species can be identified in 35 min from a solid-medium culture and in 60 min from a liquid-medium culture or sputum specimen after DNA extraction. The longer incubation time required for liquid than for solid media would be due to the small number of cells in positive MGIT broth. After extended cultivation of positive MGIT broth for a couple of days, a 35-min LAMP reaction showed positive results with the targeted organisms. The LAMP operation is quite simple. It starts with the mixing of the buffer, primers, DNA lysates, and DNA polymerase in a tube; then the mixture is incubated at 63°C for a certain period. There is no necessity for heat denaturation of the template DNAs. The only equipment needed for the LAMP reaction is a regular laboratory water bath or a heat block that furnishes a constant temperature of 63°C. Visual judgment eliminates the need for any laborious and time-consuming postamplification operations such as hybridization and electrophoresis as well as the need for special equipment.
During the past decade, various nucleic acid amplification-based methods such as the PCR-based Roche Amplicor system (2, 3), the rRNA amplification-based Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test system (1, 19, 24), ligase chain reaction (12), the Q-beta replicase amplified assay (22), the nucleic acid sequence-based amplification assay (23), and strand displacement amplification (4, 11, 18) have been developed to address the need for rapid and sensitive diagnosis of M. tuberculosis and other mycobacterial infections. These methods require either precision instruments for the amplification or elaborate methods for detection of the amplified products, which are the major obstacles to wide use of these methods in relatively small scale clinical laboratories such as those in private clinics. In this regard, the LAMP-based assay developed in this study has the advantages of rapid reaction, simple operation, and easy detection.
One of the most attractive characteristics of LAMP is the visual judgment of nucleic acid amplification. This can be achieved due to the high specificity and high amplification efficiency of LAMP. Mori et al. (13) originally reported that the LAMP amplicons can be detected by confirming the presence of magnesium pyrophosphate, a white precipitate generated as a by-product during the reaction. Although this is a quite simple approach, detecting a small amount of the white precipitate by the naked eye is not always easy; therefore, the detection limit is apparently inferior to that of electrophoresis. To increase the rate of recognition by the naked eye, we added SYBR Green I to the reaction solution. By this approach, the detection limit of LAMP could be improved so as to approach that of electrophoresis (Fig. 2B).
The sensitivity study using purified DNA indicated that the LAMP-based assay has a detection limit equivalent to that of Amplicor (Table 2). When we compared the results obtained from 66 sputum specimens, 5 samples were positive by the Amplicor test but negative by LAMP (Table 3). Four of the five samples were culture positive. None of the LAMP-positive samples were negative by Amplicor. The existence of amplification inhibitors in false-negative samples was disproved by spiking samples with 50 copies of M. tuberculosis DNA. These results indicate that the sensitivity of the LAMP assay on sputum samples is slightly lower than that of Amplicor. The discrepancy between the results obtained with the purified DNA and the sputum can be explained by the different sample sizes used in these two assays. The Amplicor uses 50 μl of the DNA lysates, while the LAMP assay uses only 2.75 μl. In this study, we used the Amplicor protocol for DNA extraction from sputum specimens. When a more-compatible DNA extraction method for the LAMP assay is developed, it will increase the rate of detection of mycobacteria in clinical specimens. Amplicor contains an internal amplification control (IAC) (20) designed to detect inhibition in the processed samples. The lack of IAC in our LAMP assay would pose a problem for its use in routine diagnosis. For a larger-scale multicenter study of the LAMP-based assay in the future, we are currently constructing an IAC that will be amplified simultaneously in a separate tube with the target sample.
Identification of the species of mycobacterial isolates is another critical requirement for clinical laboratories. The conventional biochemical tests for identification of mycobacterial species are time-consuming because of the slow growth of mycobacteria on culture media. The LAMP-based assay can identify M. tuberculosis complex, M. avium, and M. intracellulare from a solid-medium culture in 60 min: 20 min for DNA extraction, 35 min for the LAMP reaction, and 1 min for detection. When the incubation time was shortened to 35 min, the sensitivity of the LAMP assay decreased (Table 2); however, it was still high enough for species identification of solid-medium culture isolates. We also confirmed that this assay with the 60-min incubation time can be used with MGIT broth on the day on which positive fluorescence is detected. While the LAMP assay is more advantageous than all of the currently available DNA probe methods (5, 21) in its simple operation and rapid reaction, developing a panel of species-specific primers that covers all of the clinically critical mycobacterial species is our next direction in developing this promising method for wider clinical use.
In conclusion, the LAMP-based assay developed in this study allows rapid and accurate identification of M. tuberculosis complex, M. avium, and M. intracellulare in both culture isolates and sputum specimens. Due to its easy operation without sophisticated equipment, it will be simple enough to use in small-scale hospitals, primary care facilities, and clinical laboratories in developing countries if the remaining issues such as sample preparation, nucleic acid extraction, and cross-contamination controls are addressed.
REFERENCES
- 1.Abe, C., K. Hirano, M. Wada, Y. Kazumi, M. Takahashi, Y. Fukasawa, T. Yoshimura, C. Miyagi, and S. Goto. 1993. Detection of Mycobacterium tuberculosis in clinical specimens by polymerase chain reaction and Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test. J. Clin. Microbiol. 31:3270-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beavis, K. G., M. B. Lichty, D. L. Jungkind, and O. Giger. 1995. Evaluation of Amplicor PCR for direct detection of Mycobacterium tuberculosis from sputum specimens. J. Clin. Microbiol. 33:2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpentier, E., B. Drouillard, M. Dailloux, D. Moinard, E. Vallee, B. Dutilh, J. Maugein, E. Bergogne-Berezin, and B. Carbonnelle. 1995. Diagnosis of tuberculosis by Amplicor Mycobacterium tuberculosis test: a multicenter study. J. Clin. Microbiol. 33:3106-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Down, J. A., M. A. O'Connell, M. S. Dey, A. H. Walters, D. R. Howard, M. C. Little, W. E. Keating, P. Zwadyk, Jr., P. D. Haaland, D. A. McLaurin III, and G. Cole. 1996. Detection of Mycobacterium tuberculosis in respiratory specimens by strand displacement amplification of DNA. J. Clin. Microbiol. 34:860-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez, R., and B. A. Hanna. 1987. Evaluation of Gen-Probe DNA hybridization systems for the identification of Mycobacterium tuberculosis and Mycobacterium avium-intracellulare. Diagn. Microbiol. Infect. Dis. 8:69-77. [DOI] [PubMed] [Google Scholar]
- 6.Harmsen, D., J. Rothganger, M. Frosch, and J. Albert. 2002. RIDOM: ribosomal differentiation of medical micro-organisms database. Nucleic Acids Res. 30:416-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harmsen, D., J. Rothganger, C. Singer, J. Albert, and M. Frosch. 1999. Intuitive hypertext-based molecular identification of micro-organisms. Lancet 353:291. [DOI] [PubMed] [Google Scholar]
- 8.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachnik, J., B. Ackermann, A. Bohrssen, S. Maass, C. Diephaus, A. Puncken, M. Stermann, and F. C. Bange. 2002. Rapid-cycle PCR and fluorimetry for detection of mycobacteria. J. Clin. Microbiol. 40:3364-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindbrathen, A., P. Gaustad, B. Hovig, and T. Tonjum. 1997. Direct detection of Mycobacterium tuberculosis complex in clinical samples from patients in Norway by ligase chain reaction. J. Clin. Microbiol. 35:3248-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Little, M. C., J. Andrews, R. Moore, S. Bustos, L. Jones, C. Embres, G. Durmowicz, J. Harris, D. Berger, K. Yanson, C. Rostkowski, D. Yursis, J. Price, T. Fort, A. Walters, M. Collis, O. Llorin, J. Wood, F. Failing, C. O'Keefe, B. Scrivens, B. Pope, T. Hansen, K. Marino, K. Williams, et al. 1999. Strand displacement amplification and homogeneous real-time detection incorporated in a second-generation DNA probe system, BDProbeTecET. Clin. Chem. 45:777-784. [PubMed] [Google Scholar]
- 12.Moore, D. F., and J. I. Curry. 1998. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by ligase chain reaction. J. Clin. Microbiol. 36:1028-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289:150-154. [DOI] [PubMed] [Google Scholar]
- 14.Nagamine, K., T. Hase, and T. Notomi. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16:223-229. [DOI] [PubMed] [Google Scholar]
- 15.Nagamine, K., Y. Kuzuhara, and T. Notomi. 2002. Isolation of single-stranded DNA from loop-mediated isothermal amplification products. Biochem. Biophys. Res. Commun. 290:1195-1198. [DOI] [PubMed] [Google Scholar]
- 16.Nagamine, K., K. Watanabe, K. Ohtsuka, T. Hase, and T. Notomi. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47:1742-1743. [PubMed] [Google Scholar]
- 17.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfyffer, G. E., P. Funke-Kissling, E. Rundler, and R. Weber. 1999. Performance characteristics of the BDProbeTec system for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J. Clin. Microbiol. 37:137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfyffer, G. E., P. Kissling, E. M. Jahn, H. M. Welscher, M. Salfinger, and R. Weber. 1996. Diagnostic performance of amplified Mycobacterium tuberculosis direct test with cerebrospinal fluid, other nonrespiratory, and respiratory specimens. J. Clin. Microbiol. 34:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenstraus, M., Z. Wang, S. Y. Chang, D. DeBonville, and J. P. Spadoro. 1998. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J. Clin. Microbiol. 36:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanguinetti, M., B. Posteraro, F. Ardito, S. Zanetti, A. Cingolani, L. Sechi, A. De Luca, L. Ortona, and G. Fadda. 1998. Routine use of PCR-reverse cross-blot hybridization assay for rapid identification of Mycobacterium species growing in liquid media. J. Clin. Microbiol. 36:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, J. H., G. Radcliffe, S. Rigby, D. Mahan, D. J. Lane, and J. D. Klinger. 1997. Performance of an automated Q-beta replicase amplification assay for Mycobacterium tuberculosis in a clinical trial. J. Clin. Microbiol. 35:1484-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Vliet, G. M., R. A. Schukkink, B. van Gemen, P. Schepers, and P. R. Klatser. 1993. Nucleic acid sequence-based amplification (NASBA) for the identification of mycobacteria. J. Gen. Microbiol. 139:2423-2429. [DOI] [PubMed] [Google Scholar]
- 24.Vlaspolder, F., P. Singer, and C. Roggeveen. 1995. Diagnostic value of an amplification method (Gen-Probe) compared with that of culture for diagnosis of tuberculosis. J. Clin. Microbiol. 33:2699-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeboah-Manu, D., M. D. Yates, and S. M. Wilson. 2001. Application of a simple multiplex PCR to aid in routine work of the mycobacterium reference laboratory. J. Clin. Microbiol. 39:4166-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]