Abstract
To examine the stability of repetitive-sequence (rep) PCR profiles, six species of bacteria were subcultured to blood agar plates and DNA was extracted from the cultures after 24, 48, and 72 h of incubation at 35°C. In addition, the same species were subcultured to fresh blood plates daily and DNA was extracted from the cultures after growth of 5, 10, and 15 subcultures, respectively. rep PCR analysis demonstrated that all rep PCR fingerprints from a single species were identical.
Methods for molecular typing of bacteria are powerful tools that can be used to determine whether isolates recovered from different patients or the environment are related and, in so doing, provide evidence for a common source of transmission of infection. Molecular typing can also be used to establish colonization of an individual by the same organism over time or to determine the relatedness of strains of bacteria possessing different antimicrobial resistance profiles.
Epidemiological analysis of bacterial isolates has gained popularity over the past three decades, and many distinct approaches have been used for this purpose, including phenotypic and genotypic typing schemes. The limitations of phenotypic analysis have led to the progressive development of genotypic strategies, including analysis of plasmid content and plasmid restriction patterns (6), random amplified polymorphic DNA analysis (8), repetitive-sequence (rep) PCR (2, 13, 14), restriction fragment length polymorphism (RFLP) and pulsed-field gel electrophoresis (PFGE) (4), and multilocus sequence analysis of bacterial DNA (9). While RFLP/PFGE is considered the current “gold standard” for typing bacteria, the process is somewhat time consuming and technically demanding, limiting the laboratory's ability to process large numbers of organisms simultaneously.
Repetitive DNA sequences of approximately 30 to 400 bp have been found interspersed throughout a variety of prokaryotic (and eukaryotic) genomes (10). In bacteria, the most common repeat sequences are highly conserved across species. Some of these sequences, such as the repetitive extragenic palindrome sequence, are thought to play a role in gene regulation (7). Unlike the repetitive sequence analysis used in forensic medicine, the method developed for assessment of bacterial relatedness (termed rep PCR) examines the distance between repeat sequences by using primers directed outwardly from these sequences. If repetitive sequences are within a distance that can be spanned by Taq polymerase extension, products of various molecular sizes (depending upon the distance between repeats) are obtained. The amplified products are separated by gel electrophoresis; the gel image is then scanned and the banding patterns are analyzed visually or by using gel comparison software.
Because repetitive sequences are interspersed throughout the genome, rep PCR is potentially capable of surveying the entire genome and has been shown to have similar or better strain differentiation power compared to other methods (12). rep PCR is rapid by comparison to RFLP/PFGE, has been shown to be reproducible, and requires few additional resources other than those already available in most molecular laboratories.
Although molecular typing using rep PCR has become relatively popular, some assumptions regarding the performance of this method have been made that have not yet been substantiated. For example, molecular typing analyses have generally relied on DNA extracted from overnight or 24-h subcultures of the strains being examined. Presumably, this practice was adopted to exclude confounding factors that might alter rep PCR banding patterns due to increasing culture age and/or clonal expansion during growth of multiple subcultures. However, the practice of using overnight cultures for analysis might be unnecessary. To challenge this ritual, we evaluated six representative organisms for changes in rep PCR patterns with increasing culture age and upon repeated subculture.
Strains.
The organisms that were evaluated during this study consisted of Pseudomonas aeruginosa (ATCC 27853), Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), Enterococcus faecalis (ATCC 29212), a previously described strain of Staphylococcus epidermidis (3), and a clinical isolate of Acinetobacter baumannii. All strains were initially subcultured onto blood agar plates and incubated at 35°C. To evaluate changes in rep PCR pattern with culture age, DNA was extracted from these cultures after 24, 48, and 72 h of incubation without subculture; we also grew sequential daily subcultures for a total of 15 days and extracted DNA from overnight growth after production of 5, 10, and 15 subcultures, respectively. For each extraction, several colonies were combined to provide the starting material.
DNA preparation.
DNA extraction of gram-negative bacteria was performed with QIAamp DNA Mini kits (QIAGEN, Inc., Valencia, Calif.) according to manufacturer's instructions. For gram-positive bacteria, cells were first treated with ice-cold 70% ethanol followed by lysozyme (5) prior to extraction with the QIAamp Mini kit.
PCR.
rep PCR was performed as previously described (2) using primers directed against the enterobacterial repetitive intergenic consensus sequence (ERIC) repeat element. An exception was made with E. faecalis, for which amplification was performed using the RW3A primer as previously described for S. aureus by Del Vecchio et al. (1). All amplifications were performed using random amplified polymorphic DNA-to-go PCR beads (Amersham-Pharmacia Biotech, Inc., Piscataway, N.J.) in a final volume of 25 μl, including 100 ng of purified genomic DNA and 125 pmol of each ERIC primer or 75 pmol of the RW3A primer for E. faecalis.
Electrophoresis and pattern analysis.
PCR products were electrophoresed in a 1.5% agarose gel (Type 1-A; Sigma Chemical Co., St. Louis, Mo.), stained in 5 μg of ethidium bromide/ml, and visualized using a Gel Doc 1000 combined with Molecular Analyst software (Bio-Rad Laboratories, Hercules, Calif.). Banding patterns were compared using the unweighted pair group method with average linkages as previously described (11). An 85% relatedness cutoff was used to define homology groups.
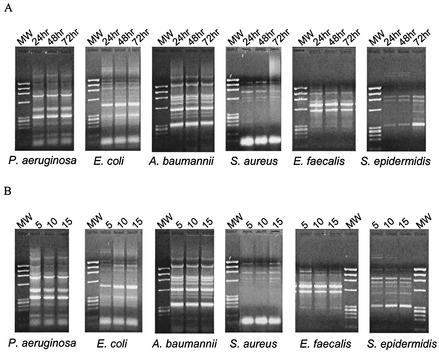
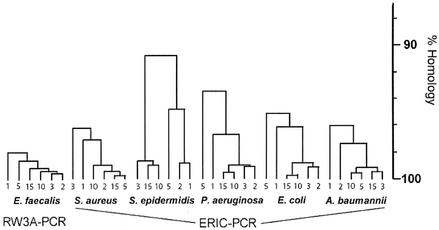
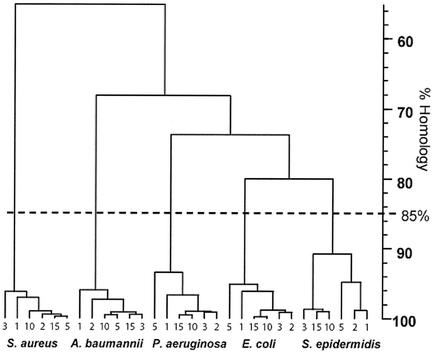
Figure 1 shows the rep PCR patterns obtained from test strains with increasing culture age (Fig. 1A) or following growth of sequential subcultures (Fig. 1B). Macroscopically, the banding patterns appear very similar; this is confirmed in the individual dendrograms generated by the gel comparison software, as illustrated in Fig. 2, which shows that even after 72 h of incubation or growth of 15 sequential daily subcultures, all banding profiles from a single strain show greater than 90% homology. To evaluate the ability of the software to differentiate between species, we also ran the analysis with the 30 patterns that had been generated with the ERIC primers. The complete dendrogram in Fig. 3 shows that the program did not find greater than 80% homology between the rep PCR banding profiles of any one species.
FIG. 1.
rep PCR banding patterns with increasing culture age (A) or daily sequential subcultures (B); 5, 10, and 15 refer to numbers of subcultures. MW, molecular weight.
FIG. 2.
Individual dendrograms generated by Molecular Analyst software. Culture ages are indicated as follows: 1, 24 h; 2, 48 h; and 3, 72 h. The numbers 5, 10, and 15 refer to numbers of subcultures.
FIG. 3.
Complete dendrogram of species amplified with ERIC primers. The dotted line indicates the empirically set cutoff of 85% homology used to indicate identity.
The central issue for the epidemiological analysis of bacteria recovered from putative nosocomial or community outbreaks is that of employing methods that provide stable markers of identity that are reproducible over time and between laboratories. One of the key factors in molecular typing, therefore, is the quality of the nucleic acid extracts used for the analysis. Although rep PCR is rapidly becoming a popular method for molecular typing of bacteria, certain assumptions have been made regarding the parameters of this method. One of these assumptions is that bacterial genomic DNA for analysis should be obtained from overnight or 24-h cultures to prevent the possibility of altered banding patterns arising from aging cultures and genomic deterioration or clonal selection during multiple subcultures. The results of this investigation show that the banding patterns of the study strains of bacteria maintained a >90% relatedness level after 72 h of continuous incubation or after growth of 15 daily sequential subcultures. These findings were true for both gram-positive and -negative bacteria and for amplification using either ERIC or RW3A repeat sequence primers. Taken together, we interpret this as indicating that 3-day-old cultures are acceptable for rep PCR analysis and that there is no need to prepare fresh subcultures of test strains to perform epidemiological relatedness studies. In addition, it is possible to use isolates that have been subcultured multiple times without fear of banding profile deterioration. A possible drawback is that we combined several colonies for the analysis; if changes in rep PCR patterns were to occur at low frequency, they would be difficult to detect unless a large number of individual colonies were analyzed or until the new rep PCR type became a significant percentage of the population. Additionally, for broad interpretation of these results, it would be advisable to verify them in a wider range of clinical isolates and also in variably permissive strains that contain a phage or plasmid. One sidelight of our study is that we did not find greater than 80% homology between any of the species examined, which serves as an internal control of the method and also demonstrates the potential use of rep PCR for species identification as previously proposed (7, 12, 13).
REFERENCES
- 1.Del Vecchio, V. G., J. M. Petroziello, M. J. Gress, F. K. McCleskey, G. P. Melcher, H. K. Crouch, and J. R. Lupski. 1995. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J. Clin. Microbiol. 33:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunne, W. M., Jr., and S. Maisch. 1995. Epidemiological investigation of infections due to Alcaligenes species in children and patients with cystic fibrosis: use of repetitive-element-sequence polymerase chain reaction. Clin. Infect. Dis. 20:836-841. [DOI] [PubMed] [Google Scholar]
- 3.Dunne, W. M., Jr., H. Qureshi, H. Pervez, and D. A. Nafziger. 2001. Staphylococcus epidermidis with intermediate resistance to vancomycin: elusive phenotype or laboratory artifact? Clin. Infect. Dis. 33:135-137. [DOI] [PubMed] [Google Scholar]
- 4.Goering, R. V. 1993. Molecular epidemiology of nosocomial infection: analysis of chromosomal restriction fragment patterns by pulsed-field gel electrophoresis. Infect. Control Hosp. Epidemiol. 14:595-600. [DOI] [PubMed] [Google Scholar]
- 5.Kalia, A., A. Rattan, and P. Chopra. 1999. A method for extraction of high-quality and high-quantity genomic DNA generally applicable to pathogenic bacteria. Anal. Biochem. 275:1-5. [DOI] [PubMed] [Google Scholar]
- 6.Liu, P. Y., Y. J. Lau, B. S. Hu, J. M. Shyr, Z. Y. Shi, W. S. Tsai, Y. H. Lin, and C. Y. Tseng. 1995. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J. Clin. Microbiol. 33:1779-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupski, J. R., and G. M. Weinstock. 1992. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J. Bacteriol. 174:4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maslow, J. N., M. E. Mulligan, and R. D. Arbeit. 1993. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin. Infect. Dis. 17:153-164. [DOI] [PubMed] [Google Scholar]
- 11.van Belkum, A., M. Sluijuter, R. de Groot, H. Verbrugh, and P. W. Hermans. 1996. Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 34:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Versalovic, J., F. J. deBruijn, and J. R. Lupski. 1997. Rep-PCR based DNA fingerprinting of bacterial genomes, p. 437-454. In F. J. deBruijn, J. R. Lupski, and G. M. Weinstock (ed.), Bacterial genomes: physical structure and analysis. Chapman & Hall, New York, N.Y.
- 13.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woods, C. R., J. Versalovic, T. Koeuth, and J. R. Lupski. 1993. Whole-cell repetitive element sequence-based polymerase chain reaction allows rapid assessment of clonal relationships of bacterial isolates. J. Clin. Microbiol. 31:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]