Abstract
Background
Transformation-associated recombination (TAR) cloning in yeast is a unique method for selective isolation of large chromosomal fragments or entire genes from complex genomes. The technique involves homologous recombination, during yeast spheroplast transformation, between genomic DNA and a TAR vector that has short (~ 60 bp) 5' and 3' gene targeting sequences (hooks).
Result
TAR cloning requires that the cloned DNA fragment carry at least one autonomously replicating sequence (ARS) that can function as the origin of replication in yeast, which prevents wide application of the method. In this paper, we describe a novel TAR cloning system that allows isolation of genomic regions lacking yeast ARS-like sequences. ARS is inserted into the TAR vector along with URA3 as a counter-selectable marker. The hooks are placed between the TATA box and the transcription initiation site of URA3. Insertion of any sequence between hooks results in inactivation of URA3 expression. That inactivation confers resistance to 5-fluoroorotic acid, allowing selection of TAR cloning events against background vector recircularization events.
Conclusion
The new system greatly expands the area of application of TAR cloning by allowing isolation of any chromosomal region from eukaryotic and prokaryotic genomes regardless of the presence of autonomously replicating sequences.
Keywords: transformation-associated recombination cloning, gene isolation, counter-selection
Background
Draft sequences of human, mouse, fruit fly, and nematode genomes have recently been released, and the sequencing of several other complex genomes is underway. The next challenge – sequence completion and verification – requires the closing of the gaps between contigs, isolation and characterization of centromeric and telomeric regions that are poorly represented in libraries, and verification of contig assembly. These goals could be greatly simplified by a cloning strategy that would allow selective isolation of any genomic region. Driven by the needs of functional genomics, a novel approach for DNA cloning by homologous recombination in Escherichia coli has been recently developed [1,2]. While the new approach greatly simplify cloning protocols, peculiarities of E. coli transformation do not allow to apply recombinogenic engineering to large size of DNA fragments.
We developed an alternative cloning procedure based on transformation-associated recombination (TAR) in the yeast Saccharomyces cerevisiae [3,4]. TAR cloning allows to selectively isolate genomic fragments up to 300 kb. The following two general TAR cloning approaches were developed and exploited [5,6]. (i) When DNA sequence information is available from both the 3' and 5' flanking regions of the gene of interest, the gene is isolated using a vector with two short unique sequences that flank the gene. The hooks can be as small as 60 bp [7]. (ii) When DNA sequence information is available only as 3' or 5' flanking regions, a modified version of TAR cloning, called radial TAR cloning, is used. In radial TAR cloning, one hook is a unique sequence from the chromosomal region of interest and the other is a repeated sequence that occurs frequently and randomly throughout the genomic DNA (i.e., Alu repeats in human DNA and B1 repeats in mouse DNA). The radial cloning method involves isolation of a set of nested overlapping fragments that extend from the gene-specific targeting hook to upstream or downstream Alu (or B1) positions in the gene of interest. TAR cloning has been used successfully to isolate over a dozen single copy genes from human and mouse genomes [[6-13] and NK, unpublished data].
TAR cloning requires that the cloned DNA fragment carry at least one autonomously replicating sequence (ARS) that can function as the origin of replication in yeast. Sequences that function as ARSs in yeast occur on average every 20 to 40 kb in all the eukaryotic genomes thus far examined [14,15], which suggests that most mammalian chromosomal regions can be readily isolated by TAR-cloning with a vector that lacks an ARS. The ARS frequency in chromosomal regions that consist of multiple repetitive elements, however, such as the centromere and telomere, may be reduced, meaning that these regions may not be isolatable by the original TAR method.
This report describes a general TAR cloning system for isolation of genomic regions regardless of the presence of ARS elements. In this new system, the TAR vector contains an ARS and a counter-selectable marker. Negative genetic selection eliminates the background due to vector recircularization caused by end-joining during yeast transformation. This novel method provides a powerful tool for structural and functional analysis of eukaryotic and prokaryotic genomes.
Results
Vectors with a counter-selectable marker at the end of a targeting hook are not efficient for gene isolation from complex genomes
Isolation of ARS-less genomic regions by TAR cloning requires incorporation of yeast origin of replication into the vector. It is well documented, however, that linearized ARS/CEN vectors circularize with high efficiency via illegitimate end-joining during yeast transformation [[16] and references therein]. To eliminate background resulting from re-circularization of the vector during TAR cloning, we investigated two different strategies based on use of a counter-selectable marker.
The first strategy exploited a vector with a counter-selectable marker placed at the end of one of the targeting hooks. This strategy is similar to one widely used for gene disruption in mice [17] and was recently successful in isolating chromosomal fragments from a prokaryotic genome [18]. To apply the strategy, we constructed a TAR vector which, when linearized, had the structure URA3-Hook1-CEN6-ARSH4-HIS3-Hook2. Hook1 and Hook2 represent two targeting sequences, while URA3 is a marker that can be selected for or against in yeast. Circularization of the vector does not prevent URA3 expression and, therefore, the clones containing the vector without the DNA insert can not grow on media containing 5-FOA. If the TAR vector recombines with a genomic DNA fragment, the URA3 gene is lost and the resulting YAC clone becomes resistant to 5-FOA.
We used the selection system described above to isolate the Tg.AC transgene from the mouse genome. The Tg.AC transgenic mouse carries approximately 40 copies of the transgene integrated into a unique site on chromosome 11 [[11] and references therein]. Each copy includes the v-Ha-ras gene and a simian virus 40 (SV40) polyadenylation signal and is under control of a zeta-globin promoter. The TAR vector used in this experiment, pARS-SV211-URA, contained a 211-bp transgene-specific hook and a B1 repeat as the second hook (see Methods). The vector was linearized between the B1 targeting hook and URA3 and co-transformed with genomic DNA into yeast spheroplasts. Transformants were selected on histidine-minus synthetic medium containing 5-FOA. No transgene-positive clones were found among the 1,000 transformants analyzed. Because the standard TAR cloning technique (i.e., one that uses a vector lacking ARS and a counterselectable marker) yields about 1%-2% transgene-positive transformants [7,15], this result demonstrates that the specific targeting hook recombines much less efficiently when it is protected by a ~ 1.1 kb nonhomologous URA3 sequence. To study in greater detail how the presence of nonhomologous DNA at the end of one of the hooks reduces gene capture efficiency, we constructed a set of TAR vectors with different lengths of nonhomologous DNA next to the hook. Each vector contained a 60-bp transgene-specific hook and a common B1 repeat as the second hook. Figure 1 summarizes the results of radial TAR cloning of the transgene with those vectors. As can be seen, the yield of gene-positive clones progressively declined as the length of the nonhomologous DNA increased. Even 37 bp of nonhomologous DNA decreased TAR cloning efficiency ~ 5 times. These results demonstrate that TAR cloning vectors with a counter-selectable marker at the end of one of the hooks are inefficient for cloning the genes from complex mammalian genomes.
Figure 1.
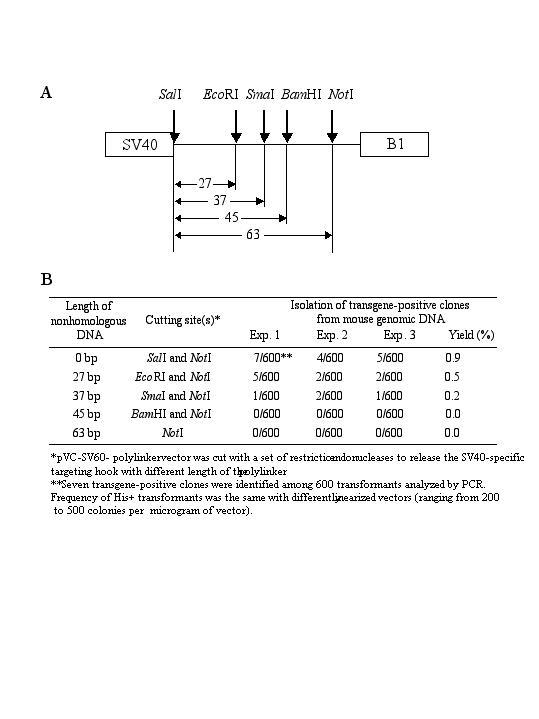
Effect on gene capture of length of nonhomologous sequences placed at the end of the hook. The diagram shows only one of the targeting hooks of the pVC-SV60-polylinker TAR vector. The vector was cut with a set of endonucleases to release the SV40-specific targeting hook with a different length polylinker (A). Yield of transgene-positive clones obtained with different TAR constructs (B).
Rationale for TAR cloning of genomic regions lacking ARS sequences
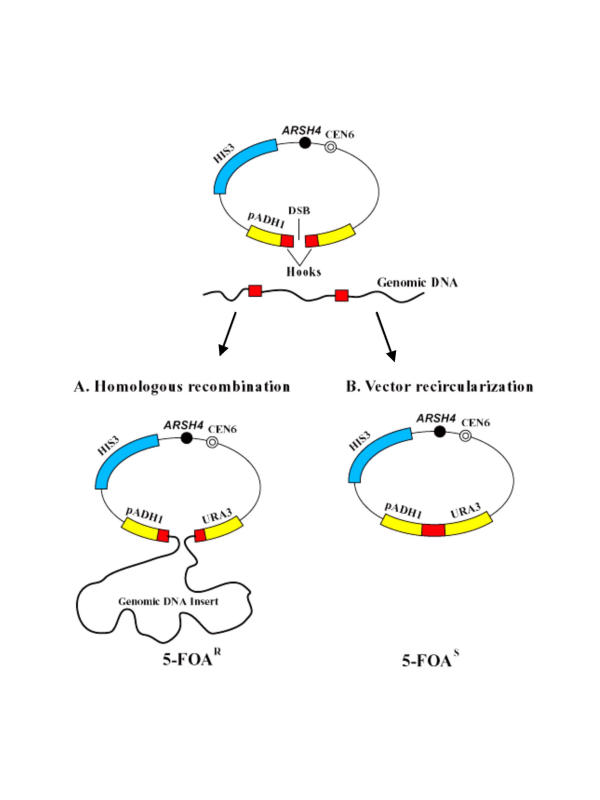
To reduce the background caused by recircularization of ARS-containing vectors, we developed a different gene capture strategy (Figure 2). As in the first approach, the TAR vector contained a yeast centromere (CEN6), a yeast ARS element (ARSH4), a positive-selectable marker (HIS3), and a negative-selectable marker (URA3). In this approach, however, the targeting hooks are placed between the promoter and the open reading frame of the URA3 gene. The promoter derived from the S. pombe ADH1 gene exhibits specific spacing requirements for its function such that the permissive distance between the TATA element and the transcription initiation site is at most 130 bp [19,20]. A greater distance results in transcription initiation at an alternative site and inactivation of URA3 expression. The specific spacing requirements allow selection against the recircularized vector. Figure 2 shows the outcomes of two different events involving such a TAR vector. In Figure 2A, the TAR vector recombines with a genomic DNA fragment via the targeting hooks, which results in separation of the TATA box from the transcription initiation site in the URA3 gene by the DNA insert. The resulting YAC does not express URA3 and, therefore, the clone can be selected by medium containg 5-FOA. Figure 2B shows the results of nonhomologous end-joining leading to re-circularization of the vector. In this case, the distance between the TATA element and the transcription initiation site is not changed, and URA3 is expressed normally, rendering the clone sensitive to 5-FOA.
Figure 2.
TAR cloning of genomic regions lacking ARS sequences. The TAR vector carries a yeast centromere (CEN6), a yeast positive selectable marker (HIS3), a negative-selectable marker (URA3), and two gene-specific targeting hooks. URA3 is a hybrid gene containing the open reading frame of the S. cerevisiae URA3 gene and the promoter of the S. pombe ADH1 gene. The promoter tolerates the insertion of up to a 130-bp sequence between the TATA box and the transcription initiation site. Further increase of the distance between the TATA box and the transcription initiation site inactivates URA3 expression. The reason the targeting hooks inserted into the ADH1 promoter do not disrupt URA3 expression is that their combined length is less than 130 bp. This configuration allows selection of TAR cloning events against the vector re-circularization. (A) Homologous recombination between the gene-specific targeting hooks and the genomic fragment containing the gene of interest leads to insertion of the genomic fragment between the TATA box and the transcription initiation site. Such clones can be selected by their ability to grow on media containing 5-FOA, which abolishes URA3 expression. (B) Nonhomologous end-joining forms a circular vector. The URA3 marker is normally expressed in these clones, so they do not grow on media containing 5-FOA. (R = resistant, S = sensitive to 5-FOA, DBS = double strand break.)
Isolation of specific chromosomal regions with a TAR vector containing ARS and a counter-selectable marker
We used the TAR cloning strategy described above to isolate the v-Ha-rasTg.Ac transgene region from mouse DNA. The vector pARS-TGAC contained 60 bp of SV40 as a transgene-specific targeting hook and 50 bp of a common mouse repeat B1 as a second hook. Both selected targeting sequences do not contain an ATG initiation codon that is out-of-frame with the vector URA3, which avoids the translational incompetence of URA3 mRNA and resistance to 5-FOA (see Methods). The targeting hooks were placed in the promoter region of URA3 as shown in Figure 2. Genomic DNA and the linearized TAR cloning vector were combined with yeast spheroplasts and the His+Ura- transformants were selected on medium containing 5-FOA to identify clones with genomic DNA inserts. Yield of the transformants varied from 100 to 300 colonies/ug vector, 2 ug genomic DNA, and 2 × 108 spheroplasts (Table 3). Eight transgene-positive YAC clones were identified among 950 His+ 5-FOA-resistant transformants screened by PCR for the presence of the zeta-globin promoter sequence (Table 3). Among 8 YAC clones characterized by CHEF electrophoresis, size of inserts varied from 50 to 200 kb. Approximately the same yield of gene-positive clones had been previously observed during TAR cloning of this genomic region with vectors lacking ARS and a counter-selectable marker [7,15]. No transgene-positive clones were found when 5-FOA selection was omitted (Table 3), indicating that the new TAR cloning method is highly selective and efficient for isolation of multicopy genes.
Table 3.
Isolation of the mouse transgene Tg.AC and the human HPRT gene with ARS-containing TAR vectors
| Targeted region | Experiment | Mean No. of transformants per ug vector (range) | No. of gene-positive clones/No. of clones analyzed (%) |
| Tg.AC mouse transgene | 1 | 86 (60–120) | 3/250 (1.2) |
| 2 | 319 (242–380) | 2/400 (0.5) | |
| 3 | 99 (52–166) | 3/300 (1.0) | |
| Control* | 1,910 (1,132–8,600) | 0/3000 (0.0) | |
| Human HPRT | 1 | 72 (52–108) | 1/300 (0.33) |
| 2 | 24 (10–60) | 1/150 (0.67) | |
| 3 | 22 (8–38) | 1/175 (0.57) |
*Control TAR cloning experiment was carried out with the same TAR vector but transformants were selected on SD medium lacking 5-FOA
We also tested whether TAR cloning with an ARS-containing vector and negative selection can be used to isolate a single copy gene from a complex genome, such as HPRT from the human genome. We designed a TAR cloning vector pARS-HPRT carrying a 56-bp 5' HPRT-specific targeting hook and a 60-bp 3' HPRT-specific hook and inserted them into the promoter region of URA3 (see Methods). His+Ura- transformants were selected on medium containing 5-FOA to identify recombinant clones. Transformant yields ranged from 20 to 70 colonies (Table 3). A low yield is predicted for a TAR vector containing two unique targeting hooks. We screened 625 randomly picked transformants by PCR for the presence of HPRT, and 3 (~ 0.5%) of them carried all the HPRT exon sequences (Table 3). All gene-positive clones contained circular YACs with ~ 40 kb DNA inserts (data not shown). Such a high yield of HPRT-positive clones has been previously observed with a similar vector lacking the ARS element [15]. Our results indicate that the novel TAR cloning system is highly efficient, highly selective, and sufficiently sensitive to isolate a single copy gene from the large and complex genomes.
Selective isolation of centromeric regions
The new TAR cloning system is useful for isolation of heterochromatic regions enriched by repeats and presumably poor in ARS elements, such as a mammalian centromere. Human centromeres contain primarily alpha (or alphoid) satellite DNA. Alphoid DNA consists of 171-bp repeating units organized into higher order repeats. The length of centromeric alphoid arrays of different human chromosomes varies from 0.2 to 5 Mb.
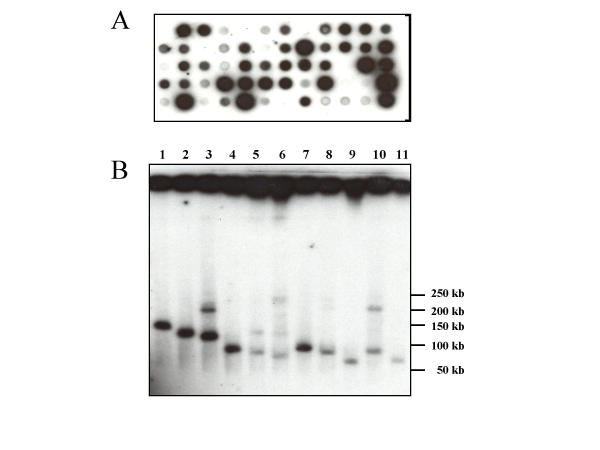
To apply the new selection strategy for isolation of large blocks of alphoid DNA, we created a TAR vector pARS-ALF with the structure SAT-pADH1-CEN6-ARSH4-HIS3-URA3-SAT, where SAT represents two 40-bp fragments from the alphoid DNA consensus sequence (see Methods). As in the constructs described above, the targeting hooks were inserted into the URA3 promoter region. TAR cloning of human centromeric DNA was carried out as described above. The average yield of His+Ura- transformants was 250. To detect the colonies with alphoid DNA inserts, randomly selected transformants were tested by dot-hybridization with an alphoid DNA-specific probe. The probe has no homology to the targeting sequences in the TAR vector (see Methods). Roughly 50% of the clones reacted with the probe, indicating high selectivity of TAR cloning (Fig. 3A and Table 4). Twenty alphoid DNA-positive clones were further characterized. The size of YAC inserts was determined by clamped homogeneous electrical field gel electrophoresis and visualized with an alphoid DNA probe. The size of the YACs varied from 50 kb to 210 kb (Fig. 3B). Some isolates had more than one band, suggesting that some satellite sequences may be unstable in yeast cells, as has been previously reported [21]. Alternatively, the transformants may have acquired more than one YAC. The relatively large sizes of the inserts suggest that genomic alphoid DNA fragments recombine with targeting hooks in the vector by sequences located at DNA fragment ends, rather than by internal sequences.
Figure 3.
Characterization of alphoid DNA YACs obtained by an ARS-containing TAR vector. (A). Detection of alphoid DNA-positive clones by colony hybridization (B). CHEF analysis of circular YAC isolates containing alphoid DNA arrays. Chromosomal size DNA was isolated from transformants, expose to a low dose of gamma-rays, separated by CHEF and blot hybridized with alphoid DNA probe. Strong signals at the positions of the starting wells correspond to large circular DNA molecules. The leading bands correspond to molecules linearized by the radiation.
Table 4.
Selective cloning of alphoid DNA arrays from human genome with a ARS-containing TAR vector
| Transformant selection medium | Genomic DNA in transformation mix | Number of transformants per ug vector (range) | Yield of clones with alphoid DNA arrays (%) |
| SD-His-FoA* | no | 5 (2–7) | N/A |
| SD-His- | no | 2,095 (1,930–2,270) | N/A |
| SD-His-FoA | yes | 245 (188–380) | 50.0 |
| SD-His- | yes | 9,225 (8,560–9,620) | 3.5 |
Transformants were selected on either synthetic (SD) histidine minus or histidine minus plates containing 5-fluoroorotate (5-FOA) to select clones that had lost the URA3 marker. 5-FOA was excluded from top agar because of its toxic effect on yeast spheroplasts regeneration.
Because the human genome contains approximately 250,000 copies of alphoid DNA monomer, it is possible that alphoid DNA could be cloned into an ARS-containing vector even without counter-selection. To test this conjecture, we performed TAR cloning as above but selected His+, rather than the His+Ura-, transformants. As expected, the number of transformants was roughly one order of magnitude higher than that obtained with counter-selection (Table 4). Dot-hybridization of transformants showed that approximately 3% of the clones were alphoid DNA-positive. Thus, although alphoid DNA arrays represent a significant fraction of the genome, TAR cloning of those regions is inefficient without counter-selection. Similar results were obtained during cloning of genomic fragments using the Alu-containing vector pARS-Alu90 (data not shown).
In conclusion, the results demonstrate that our novel selection system is useful for isolation of large centromeric regions that are poorly clonable in bacterial artificial chromosome vectors [22].
Discussion
Over a decade ago, Furter-Graves and Hall [20] demonstrated that the S. pombe ADH1 gene promoter is tolerant to insertion of up to 125 base pairs between the TATA box and the transcription start site. That observation led to the development of a system for investigating the expansion of trinucleotide repeats in yeast in which the promoter is combined with the URA3 gene, a widely used counter-selectable marker [19,23]. In our work, we took advantage of the specific spacing requirements of the ADH1 promoter to develop a novel TAR cloning procedure that allows isolation of genomic regions that lack ARS elements. A prerequisite for development of the system was an estimate of the minimal size of targeting hooks required for TAR cloning. That size (60 bp for each hook) exactly matches the maximal size of an insert tolerated with the ADH1 promoter. The use of the system allowed us to include a yeast origin of replication into the TAR vector and to select TAR cloning events against vector recircularization.
The new system greatly expands the applicability of TAR cloning. It eliminates the limitation of the previously described TAR cloning protocols by eliminating the need for an ARS sequence in the region to be cloned. Now, any region of a complex genome can be selectively isolated in yeast. Because TAR cloning produces multiple gene isolates, it allows the isolation of both parental alleles of the gene which can then be used for haplotype analysis [24]. Recently we found that up to 15% DNA divergence does not preclude efficient gene isolation by TAR [25]. Such tolerance to DNA divergence allows the isolation of highly polymorphic alleles and gene and homologues with the same TAR vector. Moreover, the new system provides an alternative approach to selectively clone large fragments of bacterial genomes [18] that are poor in sequences that can function as ARSs in yeast [14].
The new TAR cloning system has several potential applications for the Human Genome Project. The goal of the current phase of the Project is to close gaps and improve the quality of the data, which is to be accomplished by collecting additional sequence data. With TAR cloning, the desired regions could be isolated from the genome directly, rather than through construction and screening of new bacterial artificial chromosome libraries. Recently we successfully applied this approach to close the gaps in the draft of human genome sequence [26]. Characterization of the TAR isolates has shown that the gap sequences to be toxic to E. coli, which may explain why those regions were not found in bacterial artificial chromosome libraries. Verification of contig assembly includes characterization of multiple, independently isolated chromosome-specific clones. TAR cloning may be the simplest way to achieve that goal. We previously demonstrated that Alu-containing TAR cloning vectors that lack ARS elements are efficient for selective isolation of human DNA from human-rodent monochromosomal hybrid cell lines [27]. Our new counter-selectable system allows construction of representative chromosome-specific libraries. One of such library for chromosome 19 has been constructed in our laboratory and is now being used to improve the accuracy of chromosome 19 draft sequences.
Among the other advantages of the new TAR cloning system is its ability to isolate centromeric and telomeric regions, which have been generally excluded as potential targets by the Human Genome Project because of the difficulty of cloning large blocks of repetitive DNA. When centromeric DNA is digested by a restriction endonuclease, the resulting fragments are usually either too small or too large for efficient cloning in bacterial artificial chromosomes [22]. In recent work we reported isolation of human centromeric DNA using a TAR vector lacking ARS element [28]. While the isolation was selective, a standard TAR cloning technique does not allow to recover a significant fraction of alphoid DNA arrays that does not contain ARS. In our present study, using the new TAR cloning strategy clones of human alphoid DNA up to ~ 210 kb in length were isolated with high selectivity. Most of the TAR clones contained exclusively alphoid DNA, which in some cases was organized in higher-order repeat arrays (data not shown). Approximately 15% of the isolates contained alphoid DNA arrays interrupted with nonalphoid DNA sequences, including Alu and LINE repeats. Those clones can be used as starting material for construction of physical maps of human centromeres as well as for functional analysis of centromeres.
Methods
Yeast strains, transformation, and selection of gene-positive clones
The highly transformable S. cerevisiae strain VL6-48N (MATalpha, his3-Δ200, trp1-Δ1, ura3-Δ1, lys2, ade2-101, met14), which has HIS3 and URA3 deletions, was used for transformation. The strain is a derivative of VL6-48 with the ura3-52 gene replaced by the KanMX cassette [15]. Spheroplasts were generated as described previously [29]. Agarose plugs (60 ul) containing approximately 2 ug of high molecular weight DNA were prepared from normal human fibroblasts MRC-5 (ATCC) or from liver cells of the Tg.AC mouse [11]. Linearized TAR cloning vector (1 ug) was added to the plugs, treated with agarase, and mixed with yeast spheroplasts. Optimum conditions for TAR cloning have been recently described [30]. Transformants were selected on synthetic histidine-minus plates containing 5-fluoroorotic acid (5-FOA) to select clones that no longer express URA3 [31]. Because of its toxic effect on yeast spheroplasts, 5-FOA was not included in the top agar.
Construction of TAR vectors
Table 1 lists the TAR cloning vectors used in this study. All those vectors were created with the Bluescript-based yeast-E. coli shuttle vector pRS313 (HIS3-CEN6-ARSH4)[32]. The cassette containing the URA3 gene driven by the Shizosaccharomyces pombe ADH1 promoter was derived from the pBL94 plasmid [19]. In the original cassette, the ADH1 promoter and URA3 were separated by a 35-bp adaptor containing an out-of-frame ATG initiation codon. In our construct, we deleted this adaptor to prevent use of the start codon for translation; that would result in nonexpression of the functional URA3 gene and, therefore, an inability to grow in the absence of uracil. The deletion of the adaptor also provided an opportunity to incorporate longer DNA fragments between the TATA box and the transcription initiation site without inactivation of URA3. Construction of the modified ADH1-URA3 cassette included PCR amplification of the ADH1 promoter and the URA3 gene from the pBL94 plasmid using two pairs of primers, ADH-F plus ADH-R and URA-ARS-F plus URA-ARS-R (Table 2). The first pair amplifies the ADH1 promoter as an ApaI-XhoI fragment; the second amplifies the URA3 gene as a ClaI-BamHI fragment. The PCR fragments were inserted into a pRS313 polylinker [32] to generate the basic ARS-containing TAR vector designated as pARS-VN. In this vector, the TATA box and the initiation transcription site in URA3 are separated by XhoI-ClaI sites, which were used to clone targeting hooks.
Table 1.
TAR vectors used in this study
| Plasmid | ARS presence | Hook 1 | Hook 2 | Linearization site |
| pARS-TGAC | + | 60 bp SV40 | 50 bp B1 | SalI |
| pARS-HPRT | + | 56 bp 5' HPRT | 60 bp 3' HPRT | SalI |
| pARS-ALF | + | 40 bp alphoid DNA | 40 bp alphoid DNA | MluI |
| pARS-Alu90 | + | 45 bp Alu | 45 bp Alu | SmaI |
| pARS-Alu110 | + | 45 bp Alu | 65 bp Alu | SmaI |
| pARS-Alu120 | + | 45 bp Alu | 75 bp Alu | SmaI |
| pARS-Alu130 | + | 65 bp Alu | 65 bp Alu | SmaI |
| pARS-Alu150 | + | 75 bp Alu | 75 bp Alu | SmaI |
| pARS-Alu190 | + | 115 bp Alu | 75 bp Alu | SmaI |
| pARS-SV211-URA | + | 211 bp SV40 | 130 bp B1 | BamHI |
| pVC-SV211 | - | 211 bp SV40 | 130 bp B1 | BamHI |
| pVC-SV60 | - | 60 bp SV40 | 130 bp B1 | SalI |
| pVC-SV60-polylinker | - | 60 bp SV40 | 130 bp B1 | SalI, EcoRI, BamHI, SmaI, NotI |
Table 2.
Primers used in this study
| Primer | Sequence | Restriction site |
| Group A* | ||
| ADH-F | 5'-atgcgggcccCCTACAACAACTAAGAAAATGGC-3' | Apa1 |
| ADH-R | 5'-atgcatgcctcgagCCACTATTTATACCATGGGAGG-3' | Xho1 |
| URA-ARS-F | 5'-atgcatcgatGTCGAAAGTTACATATAAGGAACGT-3' | Cla1 |
| URA-ARS-R | 5'-atgcggatccCAGTTTTTTAGTTTTGCTGGCCGC-3' | BamH1 |
| alu45-F | 5'-atgcatcgatGTCCCAGCTACTTGGGAGGC-3' | Cla1 |
| alu45-R | 5'-atgcatgcctcgagGGAGTGCAGTGGCGGGATCT-3' | Xho1 |
| alu65-F | 5'-atgcatcgatCGTAGTGGCGGGCGCCTGTA-3' | Cla1 |
| alu65-R | 5'-atgcatgcctcgagTCTCGCTCTGTCGCCCAGGC-3' | Xho1 |
| alu75-F | 5'-atgcatcgatATTAGCCGGGCGTAGTGGCG-3' | Cla1 |
| alu75-R | 5'-atgcatgcctcgagGAGACGGAGTCTCGCTCTGT-3' | Xho1 |
| alu190-F | 5'-atgcatcgatCTAAAACGGTGAAACCCCGT-3' | Cla1 |
| SV-ARS-F | 5'-atgcatcgatTTATACACTTAAAAATTTTA-3' | Cla1 |
| SV-ARS-R | 5'-atgcatgcctcgagCTGCCTCCCAAGTGCTGGGATTAAAGGCGTGCGCCACCtTGCCCG-3' | Xho1 |
| HP-ARS-F1 | 5'-atgcatgcctcgagCCTCGGCTTCTTCTGGGA-3' | Xho1 |
| HP-ARS-R1 | 5' atgcatgcgtcgacGTTTGCAGGCTCACTAGGTAG 3' | SalI |
| HP-ARS-F2 | 5' atgcatgcgtcgacGTAGGATATGCCCTTGACTAT 3' | SalI |
| HP-ARS-R2 | 5'-atgcatcgatGAAGCAATTACTTACATTCAA-3' | Cla1 |
| ALF-F | 5'-atgcatcgatAAGAGTGTTTCAAAACTGCT-3' | Cla1 |
| ALF-R | 5'-atgcatgcctcgagCATTCTCAGAAACTTCTTTGTGAaGTTTGC-3' | Xho1 |
| Group B* | ||
| ALU-DET-F | 5'-GGCCGGGCGCGGTGGCTCACGCCTGTAA-3' | |
| ALU-DET-R | 5'-TGTATTTTTAGTAGAGACGGG-3' | |
| Y-SAT-F | 5'-GAATCTGCAAGTGGATATTTG-3' | |
| Y-SAT-R | 5'-TCTGTGAGATGAATGCACACA-3' |
Upper case letters indicate homologous regions. Lower case letters indicate nonhomoloqous sequences. Bold letters indicate endonuclease restriction sites used for cloning PCR products. Underlined lower case letters indicate nucleotide substitutions introduced into the primer to eliminate an out-of-frame ATG initiation codon in the targeting hook. *Group A includes primers used for plasmid construction; group B includes primers used for Alu- and Sat-probes.
To determine the distance permitted between the TATA box and the transcription initiation site in pARS-VN, we cloned different sized Alu targeting hooks (45/45 bp, 45/65 bp, 45/75 bp, 65/65 bp, 75/75 bp, and 115/75 bp) into vector XhoI-ClaI sites. The Alu hooks were PCR-amplified from the 3' end of a 290-bp Alu consensus sequence [33] with pairs of alu-F, alu-R primers (Table 2). Each pair of Alu targeting hooks is separated by a SmaI site present in the Alu consensus sequence. Six constructs – pARS-Alu90, pARS-Alu110, pARS-Alu120, pARS-Alu130, pARS-Alu150, and pARS-Alu190 – with different sized Alu sequences (from 90 bp to 190 bp) were tested for URA3 expression after transformation into VL6-48N. Expression of URA3 (determined by the sensitivity of His+ transformants to 5-FOA) was abolished with 150-bp and 190-bp Alu inserts. Based on those results, we concluded that targeting sequences (hooks) with a combined length of up to 130 bp cloned into the ADH1 promoter region would allow selection for insertion events during TAR cloning.
We designed three pairs of targeting hooks to investigate the selectivity of the new TAR cloning system. The first pair contained 60 bp of the SV40-specific targeting sequence in the Tg.AC transgene [5,11] and 50 bp of the mouse B1 element. The hooks were PCR-amplified from the SV800 vector using specific primer pairs SV-ARS-F and SV-ARS-R (Table 2) and cloned into XhoI-ClaI sites of pARS-VN, yielding the vector pARS-TGAC. We mutated an out-of-frame ATG codon identified in a B1 template sequence during PCR amplification of the hook to avoid translational incompetence and resistance to 5-FOA. The second pair contained 56-bp and 60-bp specific sequences from the 5' and 3' ends of human HPRT. The hooks were amplified from HPRT yeast artificial chromosome (YAC) [6] with primer pairs HP-ARS-F and HP-ARS-R (Table 2) and cloned into the XhoI-ClaI sites of pARS-NK, yielding the vector pARS-HPRT. The third vector, pARS-ALF, was designed for cloning human alphoid DNA arrays. pARS-ALF contained two 40-bp fragments of alphoid DNA monomer that were PCR-amplified using the primer pair ALF-F and ALF-R from a 171-bp alphoid consensus sequence (X07685) [34]. None of the amplified targeting hooks contained an ATG codon that was out-of-frame in URA3. Vectors pARS-TGAC and pARS-HPRT were linearized by SalI, pARS-Alu by SmaI, and pARS-ALF by MluI, exposing the targeting sequences. The TAR vector pARS-SV211-URA was constructed by insertion of the S. cerevisiae URA3 gene between the targeting hooks of pVC-SV211 [7] and cloning of ARSH4 into the polylinker. URA3 was amplified as a ~ 1.1 kb fragment from pRS306 [32]. Before TAR cloning, the vector was linearized with BamHI to expose the B1 targeting sequence and the URA3 sequence placed at the end of the transgene-specific hook.
Three TAR vectors used in this study did not contain ARS elements. Two of them, pVC-SV211 and pVC-SV60, contain a 130-bp mouse B1 element as a common hook and, respectively, 211 bp and 60 bp of the trangene-specific targeting hooks. These vectors have been described [7]. We constructed an additional vector to investigate how the presence of nonhomologous DNA at the end of a hook affects the efficiency of gene capture. The pVC-SV60-polylinker plasmid contains the same hooks as pVC-SV60 [7], but the hooks are separated by approximately 70 bp in the pBluscript polylinker. This configuration allowed us to linearize the vector by a different combination of enzymes, producing a set of TAR constructs with different lengths of nonhomologous DNA at the end of the specific hook (Fig. 1).
Detection of positive clones among yeast transformants
We used two pairs of primers to identify transformants containing a human HPRT sequence. IN1R plus IN1L amply a 516-bp sequence in intron 1, and 46L plus 47R amplify a 575-bp sequence in exon 2, along with flanking introns [15]. We used a pair of primers specific to the zeta-globin promoter region to screen PCR transformants for the Tg.AC transgene sequence [7]. These primers generate a 419-bp PCR product that is diagnostic for recombination between the TAR vector and genomic Tg.AC transgene sequences. Yeast genomic DNA was isolated from transformants and PCR-amplified as described previously [29].
Detection of recombinant clones obtained during TAR cloning with Alu- and alphoid DNA-containing vectors was carried out by colony hybridization. For this purpose, we designed two specific probes – a 56-bp alphoid DNA fragment (Sat-probe) and a 124-bp Alu fragment (Alu-probe). Because those sequences are not present in the TAR constructs, they are diagnostic for recombinant clones. The probes were labeled for hybridization by PCR using Alu and alphoid DNA monomers as templates and repeat-specific primers (Table 2).
Characterization of YAC clones
Chromosome-size DNA was prepared from yeast transformants carrying circular YACs, separated by clamped homogeneous electrical field gel electrophoresis (CHEF), transferred onto a nylon membrane, and hybridized with an alphoid DNA probe or Alu-probe (see above). The size of circular YACs was estimated by melting the agarose plugs with yeast genomic DNA prior to CHEF analysis. This treatment linearized approximately 20% of the YACs in the 100–400 kb size range. Southern blot analysis of YAC DNA was performed with 32P labeled probes and the protocol described by Church and Gilbert [35].
Abbreviations
ARS, autonomously replicating sequence; 5-FOA, 5-fluoroorotic acid; TAR, transformation-associated recombination; YAC, yeast artificial chromosome.
Acknowledgments
Acknowledgments
We thank Robert Lahue for the pBL94 plasmid and Miriam Bloom (SciWrite Biomedical Writing & Editing Services) for professional editing. This research was partially supported by the Biological and Environmental Research Program (BER), US Department of Energy, Interagency Agreement No. DE-AI02-01ER63079.
Contributor Information
Vladimir N Noskov, Email: noskovv@mail.nih.gov.
Natalay Kouprina, Email: kouprinn@mail.nih.gov.
Sun-Hee Leem, Email: leems@mail.nih.gov.
Ilia Ouspenski, Email: ouspenski@mail.nih.gov.
J Carl Barrett, Email: Barrett@mail.nih.gov.
Vladimir Larionov, Email: larionov@mail.nih.gov.
References
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/78475. [DOI] [PubMed] [Google Scholar]
- Larionov V., Kouprina N, Graves J, Chen XN, Korenberg JR, Resnick MA. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc Natl Acad Sci USA. 1996;93:491–496. doi: 10.1073/pnas.93.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Graves J, Resnick MA. Highly selective isolation of human DNAs from rodent-human hybrid cells as circular yeast artificial chromosomes by transformation-associated recombination cloning. Proc Natl Acad Sci USA. 1996;93:13925–13930. doi: 10.1073/pnas.93.24.13925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V, Kouprina N, Solomon G, Barrett JC, Resnick MA. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc Nat Acad Sci USA. 1997;94:7384–7387. doi: 10.1073/pnas.94.14.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Annab L, Graves J, Afshari C, Barret JC, Resnick MA, Larionov V. Functional copies of a human gene can be directly isolated by transformation-associated recombination cloning with a small 3' end target sequence. Proc Natl Acad Sci USA. 1998;95:4469–4474. doi: 10.1073/pnas.95.8.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov V, Koriabine M, Solomon G, Randolph M, Barrett JC, Leem SH, Stubbs L, Kouprina N, Larionov V. Defining the minimal length of sequence homology required for selective gene isolation by TAR cloning. Nucleic Acids Res. 2001;29:E32. doi: 10.1093/nar/29.6.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancilla M, Tainton K, Barry A, Larionov V, Kouprina N, Resnick M, Du Sart D, Choo A. Direct cloning of human 10q25 neocentromere DNA transformation-associated recombination (TAR) in yeast. Genomics. 1998;47:399–404. doi: 10.1006/geno.1997.5129. [DOI] [PubMed] [Google Scholar]
- Annab L, Kouprina N, Solomon G, Cable L, Hill D, Barrett JC, Larionov V, Afshari C. Isolation of a functional copy of the human BRCA1 gene by transformation-associated recombination in yeast. Gene. 2000;250:201–208. doi: 10.1016/S0378-1119(00)00180-3. [DOI] [PubMed] [Google Scholar]
- Kim J, Noskov VN, Lu X, Bergmann A, Ren X, Warth T, Richardson P, Kouprina N, Stubbs L. Discovery of a novel paternally expressed ubiquitin-specific processing protease gene through comparative analysis of a imprinted region of mouse chromosome 7 and human chromosome 19q13.4. Genome Res. 2000;10:1138–1147. doi: 10.1101/gr.10.8.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humble M, Kouprina N, Noskov V, Graves J, Garner E, Tennant R, Resnick MA, Larionov V, Cannon RE. Radial TAR cloning from the Tg.AC transgene. Genomics. 2000;70:292–299. doi: 10.1006/geno.2000.6384. [DOI] [PubMed] [Google Scholar]
- Leem SH, Londono-Vallejo JA, Kim JH, Bui H, Tubacher E, Solomon G, Park JE, Horikawa I, Kouprina N, Barrett JC, Larionov V. Cloning of the human telomerase gene: Complete genomic sequence and analysis of tandem repeats polymorphisms in intronic regions. Oncogene. 2002;21:769–777. doi: 10.1038/sj.onc.1205122. [DOI] [PubMed] [Google Scholar]
- Nihei N, Kouprina N, Larionov V, Oshima J, Martin GM, Ichikawa T, Barrett JC. Functional Evidence for a Metastasis Suppressor Gene for Rat Prostate Cancer within a 60 kb Region on Human Chromosome 8p21-p12. Cancer Research. 2002;62:367–370. [PubMed] [Google Scholar]
- Stinchomb DT, Thomas M, Kelly I, Selker , Davis RW. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc Natl Acad Sci USA. 1980;77:4559–4563. doi: 10.1073/pnas.77.8.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov V, Kouprina N, Leem SH, Koriabine M, Barrett JC, Larionov V. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 2002;30:e8. doi: 10.1093/nar/30.2.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Resnick MA. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat Res. 2000;451:71–89. doi: 10.1016/S0027-5107(00)00041-5. [DOI] [PubMed] [Google Scholar]
- Mansour S, Thomas K, Capecchi M. Disruption of the proto oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;409:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, Zhou Y, Kaul R, Clendenning JB, Olson MV. Genetic variation at the O-antigen biosysthetic locus in Pseudomonas aeruginosa. J Bacteriol. 2002;184:3614–3622. doi: 10.1128/JB.184.13.3614-3622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furter-Graves E, Hall BD. DNA sequence elements required for transcription initiation of the Shizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol Gen Genet. 1990;223:407–417. doi: 10.1007/BF00264447. [DOI] [PubMed] [Google Scholar]
- Neil DL, Villasante A, Fisher RB, Vetrie D, Cox B, Tyler-Smith C. Structural instability of human tandemly repeated DNA sequences cloned in yeast artificial chromosome vectors. Nucleic Acids Res. 1990;18:142–148. doi: 10.1093/nar/18.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong P. A bacterial artificial chromosome library for sequencing of complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfsmeier ML, Dixon MJ, Pessoa-Brandao L, Pelletier R, Miret JJ, Lahue RS. Cis-elements governing trinucleotide repeats in Saccharomyces cerevisiae. Genetics. 2001;157:1569–1579. doi: 10.1093/genetics/157.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Leem SH, Sunwoo Y, Kouprina N. Separation of long-range human TERT gene haplotypes by transformation-associated recombination cloning in yeast. Oncogene. 2003;22:2452–2456. doi: 10.1038/sj.onc.1206316. [DOI] [PubMed] [Google Scholar]
- Noskov VN, Leem SH, Solomon G, Mullokandov M, Chae JI, Yoon YH, Shin YS, Kouprina N, Larionov V. A Novel strategy for analysis of gene homologues and segmental genome duplications. J Mol Evol. 2003;56:1–9. doi: 10.1007/s00239-002-2442-x. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Leem SH, Solomon G, Ly A, Koriabine M, Otstot J, Pak E, Dutra A, Zha S, Barrett JC, Larionov Segments missing from the draft human genome sequence can be isolated by TAR cloning in yeast. EMBO Reports. 2003;4:253–262. doi: 10.1038/sj.embor.embor766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Campbell M, Graves J, Campbell E, Meincke L, Tesmer J, Grady D, Doggett N, Moyzis R, Deaven L, Larionov V. Construction of human chromosome 16- and 5-specific YAC/BAC libraries by in vivo recombination in yeast (TAR cloning) Genomics. 1998;53:21–28. doi: 10.1006/geno.1998.5469. [DOI] [PubMed] [Google Scholar]
- Kouprina N, Ebersole T, Koriabine M, Pak E, Rogozin I, Katoh M, Oshimura M, Ogi K, Peredelchuk M, Solomon S, Brown W, Barrett JC, Larionov V. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 2003;31:922–934. doi: 10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N, Larionov V. Selective isolation of mammalian genes by TAR cloning. Current Protocols in Human Genetics. 1999;1:5.17.1–5.17.21. doi: 10.1002/0471142905.hg0517s49. [DOI] [PubMed] [Google Scholar]
- Leem SH, Noskov VN, Park JE, Kim SI, Larionov V, Kouprina N. Optimum conditions for selective isolation of genes from complex genomes by transformation-associated recombination cloning. Nucleic Acids Res. 2003;31:e29. doi: 10.1093/nar/gng029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selectable agent in yeast molecular genetics. Methods Enzymol. 1987;154:64–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Alegria-Hartman M, Deininger PL. A consensus Alu repeat probe for physical mapping. Genet Anal Tech Appl. 1994;11:34–38. doi: 10.1016/1050-3862(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Vessel B, Choo KH. Human alpha-satellite DNA-consensus sequence and conserved regions. Nucleic Acids Res. 1987;15:6751–6752. doi: 10.1093/nar/15.16.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]