Abstract
Plant viral vectors allow expression of heterologous proteins at high yields, but so far, they have been unable to express heterooligomeric proteins efficiently. We describe here a rapid and indefinitely scalable process for high-level expression of functional full-size mAbs of the IgG class in plants. The process relies on synchronous coinfection and coreplication of two viral vectors, each expressing a separate antibody chain. The two vectors are derived from two different plant viruses that were found to be noncompeting. Unlike vectors derived from the same virus, noncompeting vectors effectively coexpress the heavy and light chains in the same cell throughout the plant body, resulting in yields of up to 0.5 g of assembled mAbs per kg of fresh-leaf biomass. This technology allows production of gram quantities of mAbs for research purposes in just several days, and the same protocol can be used on an industrial scale in situations requiring rapid response, such as pandemic or terrorism events.
Keywords: monoclonal antibody, potato virus X, tobacco mosaic virus
Although the ability of plants to express full-size human antibodies was discovered 17 years ago (1–3), the idea of industrial-scale antibody production in plants has been abandoned by most companies, mostly because of limitations of existing expression protocols. Stably transformed (transgenic) plants are able to express correctly folded and functional antibodies of both the IgG and IgA classes, but yields are generally very low (usually in the range of 1–40 μg/g of fresh biomass); in addition, the time necessary to generate the first grams of research antibody material is very long, requiring >2 years (4–8).
Transient expression systems, on the other hand, allow production of research quantities of antibody material much faster. However, the early versions of transfection systems, such as Agrobacterium-mediated transient expression or viral vector-mediated expression, cannot provide for high-level coexpression of two or several polypeptides necessary for the assembly of heterooligomeric proteins, in particular IgG antibodies (9–12).
Recently, we have developed a scalable transient expression technology (magnifection) that is based on replication of viral vectors delivered to multiple parts of a plant body (systemic delivery) by Agrobacterium (13). Such a technology is in essence an en masse infiltration of whole, mature plants with a diluted agrobacteria suspension carrying T-DNAs encoding viral replicons. The magnifection process allows expression of various proteins, but, until now, it has been used to express only single-polypeptide proteins or homooligomers (14). Attempts to express two or more different polypeptides from one viral replicon failed because of drastically reduced expression levels obtained with bicistronic constructs (unpublished results).
Therefore, we decided to explore expression protocols that involve two or more viral replicons. We report here a general solution for coexpression of high amounts of two heterologous polypeptides by using two different viral vectors, each expressing a separate polypeptide. The vectors described here are built on the backbones of two noncompeting viruses: tobacco mosaic virus (TMV) and potato virus X (PVX). This expression technology leads to yields of assembled full-size monoclonal antibody at levels as high as 0.5 g of mAb per kg of fresh leaf biomass (one to two orders of magnitude higher than other transient expression systems). The molecules produced are fully functional, and the first gram of material can be produced in <2 weeks after infiltration. Because the ratio of heavy chain (HC) to light chain (LC) expression and other expression/processing parameters have not been fully optimized, the protocol likely has the potential for further yield improvement.
Results
Two TMV-Based Replicons Expressing Different Genes Segregate Early During Cell-to-Cell Movement.
Expression of heterooligomeric proteins requires expression of two (or more) different proteins within one cell. One approach to achieving this goal would consist of using two TMV-based viral vectors, each one expressing a different protein subunit. To test this strategy, viral vectors expressing GFP and red fluorescent protein from Discosoma (DsRED) were inoculated into Nicotiana benthamiana leaves by using Agrobacterium-mediated delivery (constructs shown in Fig. 1). Six days postinfiltration (dpi), infiltrated leaf areas exhibited a mosaic of either GFP- or DsRED-expressing sectors, suggesting early segregation and spatial separation of distinct TMV populations and, therefore, a lack of coexpression (Fig. 2A). Confirming this observation, only 4–5% of protoplasts prepared from infected leaf tissue coexpressed both reporter genes (Fig. 2 C and D).
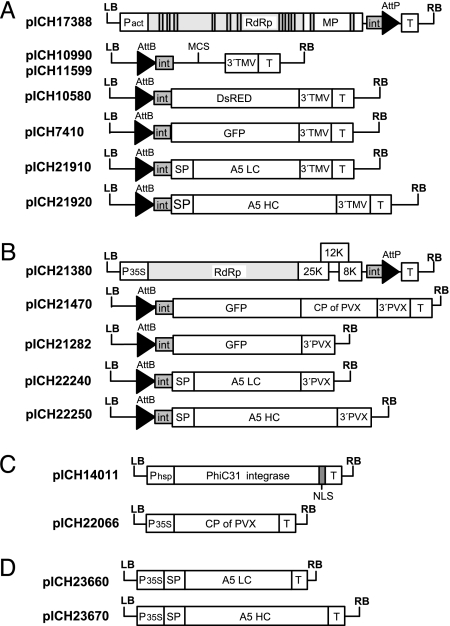
Fig. 1.
Plasmid constructs. (A) TMV-based provectors: 5′ module pICH17388; 3′ module cloning vectors pICH10990 and pICH11599 (differ in the structure of the multicloning site); and 3′ modules for expression of DsRED (pICH10580), GFP (pICH7410), A5 LC (pICH21910), and A5 HC (pICH21920). (B) PVX-based provectors: 5′ module pICH21380; and 3′ modules for expression of GFP [coat protein (CP)-containing vector pICH21470, CP-less vector pICH21282], A5 LC (pICH22240), and A5 HC (pICH22250). (C) Integrase module pICH14011 and construct for expression of the PVX CP (pICH22066). (D) Constructs for expression of the A5 LC (pICH23660) and the A5 HC (pICH23670) under control of the 35S promoter. LB and RB, binary left and right borders, respectively; Pact, Arabidopsis actin 2 promoter; P35S, 35S promoter; Phsp, Arabidopsis heat shock protein hsp 81.1 promoter; T, nos terminator; RdRp, RNA-dependent RNA polymerase; MP, movement protein; int, intron; AttP and AttB, recombination sites; MCS, multicloning site; 3′TMV and 3′PVX, 3′ untranslated regions of TMV and PVX, respectively; SP, signal peptide; NLS, nuclear localization signal.
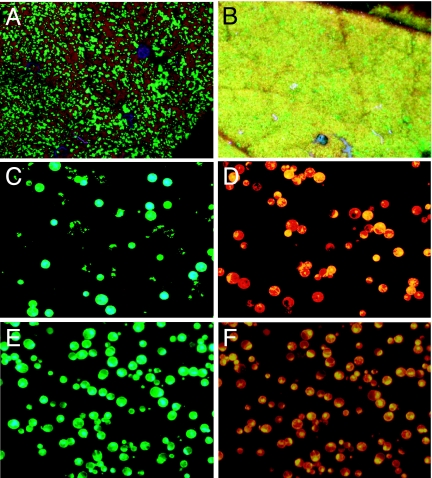
Fig. 2.
Coexpression of GFP and DsRED in N. benthamiana leaves by using viral vectors (6 dpi). Leaf sectors infected with a mixture of two TMV constructs expressing GFP or DsRED (A) or coinfiltrated with a PVX vector expressing GFP and a TMV vector expressing DsRED (B) are viewed under the UV light of a hand-held lamp. Protoplasts prepared from leaf areas coinfiltrated with TMV constructs (C and D) or PVX and TMV constructs (E and F) are viewed under blue (C and E) or red (D and F) light.
TMV and PVX Are Able to Replicate Within the Same Cell with High Efficiency.
To test whether viral vectors derived from TMV and PVX could be used to coexpress two proteins of interest, N. benthamiana leaves were coinoculated with a mixture of agrobacteria carrying a DsRED-containing TMV vector or a GFP-containing PVX vector. At 6 dpi, the infiltrated areas displayed a uniform pattern of yellow fluorescence, indicating that both genes were coexpressed in a majority of cells (Fig. 2B). Observation of mesophyll protoplasts prepared from the infiltrated areas revealed that GFP and DsRED were coexpressed in 95% of the cells (Fig. 2 D and F).
We also made PVX vectors lacking CP, and we found that, as with TMV vectors, removal of the CP leads to higher level of expression of the protein of interest in the infiltrated leaf (results not shown). Because PVX vectors lacking CP are unable to move from cell to cell (15, 16), they were inoculated at a high density of agrobacterial cells (a 10−1 dilution relative to a saturated overnight culture) to transfect a majority of plant cells in the infiltrated area. Moreover, it is known that TMV MP can complement cell-to-cell movement of PVX viruses lacking CP (17), and therefore, in mixed infections, MP expressed from TMV is expected to provide cell-to-cell movement ability to the PVX construct. Observation of mesophyll protoplasts prepared from infiltrated areas revealed that coinfection of TMV with CP-less PVX provided coexpression of GFP and DsRED in only 67% of the cells. To increase this value, viral vectors were coinfiltrated with a construct expressing PVX CP from the 35S promoter (pICH22066 construct). With this combination, coexpression of TMV and PVX increased to ≈82%. This combination, which provides a high level of expression of both genes and high level of coexpression, was used for the rest of this study.
Coexpression of IgG HC and LC by Using TMV and PVX Vectors.
The noncompeting vector technology was then applied for expression of immunoglobulins, which are natural tetramers composed of two identical HCs and two identical LCs. The human tumor-specific mAb A5, which belongs to the IgG1 subclass, was chosen as a first candidate. The HC- and LC-coding sequences (including their native human endoplasmic reticulum-targeting signal peptides) were subcloned into 3′ TMV and 3′ PVX provector constructs. The resulting constructs were agroinfiltrated into N. benthamiana leaves, together with the corresponding 5′ provector modules, a source of recombinase, and a construct for expression of PVX CP (a total of six constructs, including two 5′ provector modules and two 3′ provector modules). Two different combinations were tested: TMV-HC + PVX-LC and PVX-HC + TMV-LC. As a control, HC and LC were expressed separately by using TMV vectors. In all cases, slight toxicity symptoms appeared in infected leaves at 5–6 dpi and progressed further. Toxicity symptoms were stronger for the combination PVX-HC + TMV-LC.
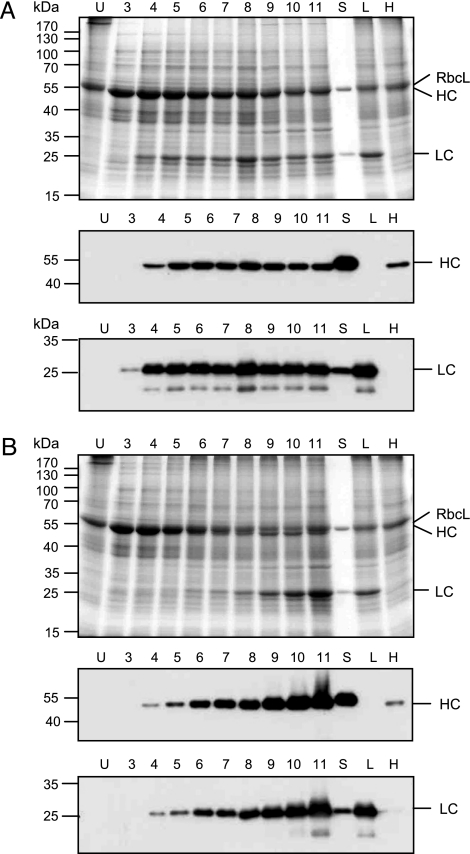
Expression of the HC and LC was analyzed by SDS/PAGE run under reducing conditions followed by Coomassie blue staining or Western blotting probed with HC- and LC-specific antibodies. Accumulation of the HCs and LCs was analyzed from 3 to 11 dpi. For the PVX-HC + TMV-LC combination, accumulation of the LC reached a maximum at 4 dpi and remained stable until 11 dpi (Fig. 3A). The level of HC increased until 5–6 dpi and remained relatively stable until 11 dpi. For the reverse combination, TMV-HC + PVX-LC, accumulation of the LC was slower, but it increased continuously until 10–11 dpi (Fig. 3B). Expression of the HC showed similar kinetics. At 11 dpi, and for both combinations, tissue in expressing areas displayed significant necrotic symptoms. However, the level of expression of both chains remained high, suggesting their relative stability. For both combinations, the LC accumulated at a substantially higher level compared with the HC. The difference was stronger for the PVX-HC + TMV-LC combination.
Fig. 3.
Accumulation of A5 HCs and LCs in N. benthamiana leaves coinfected with TMV and PVX provectors. (A) The LC is expressed with TMV and the HC with PVX. (B) The LC is expressed with PVX and the HC with TMV. Proteins are separated in 12% polyacrylamide gels run under reducing conditions. (B Top) Coomassie blue staining. (B Middle) Western blot probed with HRP-conjugated goat anti-human IgG (γ-chain-specific) antibodies (Sigma). (B Bottom) Western blot probed with HRP-conjugated rabbit anti-human IgG (λ-chain-specific) antibodies (Sigma). U, uninfected tissue; 3–11, dpi; S, A5 standard; L, LC expressed alone with TMV provectors (6 dpi); H, HC expressed alone with TMV provectors (6 dpi); RbcL, large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase.
HCs and LCs Coexpressed with TMV and PVX Vectors Form Assembled Antibodies with Specific Antigen-Binding Activity.
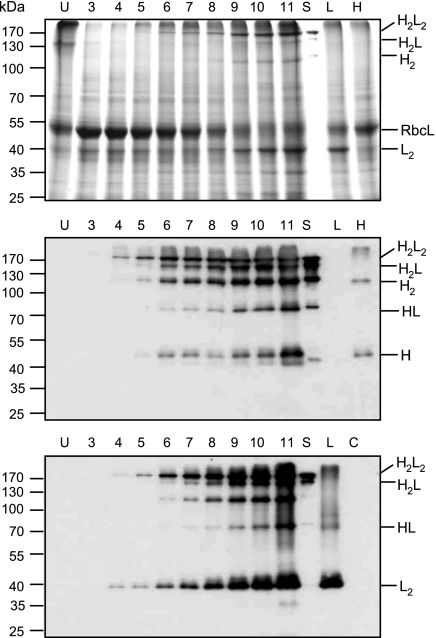
To test whether the HC and LC assembled into functional monoclonal antibodies, crude protein extracts from the combination TMV-HC + PVX-LC were separated on SDS/PAGE under nonreducing conditions and stained with Coomassie blue or analyzed by Western blotting. Probing Western blots with γ-chain-specific antibodies revealed molecular forms present both in N. benthamiana protein extracts prepared from coexpressing tissue and in the control mAb A5 produced in CHO cells (Fig. 4 Middle). In contrast, and as expected, the pattern was different in extracts in which the HC was expressed alone. Two bands of ≈50 kDa and 110 kDa most likely correspond to monomeric and dimeric forms of the HC, respectively. In addition to the dimeric form of the HC, another product of 110 kDa is detected with both anti-γ- and anti-λ-chain antibodies when the HC and LC are coexpressed. This product may be produced by proteolytic cleavage of the fully assembled mAb, as has been reported for other antibodies expressed in plants (18). Probing with anti-λ-chain antibodies revealed a complex pattern as well (Fig. 4 Bottom). The two higher molecular mass forms (at ≈170 kDa) were present in plant extracts coexpressing the HCs and LCs and in the control mAb, but they were not found when the LC was expressed alone. These two bands were also detected by using anti-γ-chain antibodies, and they most likely represent assembled IgG molecules, heterotetramer H2L2 and heterotrimer H2L. Of these forms, the heterotetramer is the most intense band visible after Coomassie blue staining. In addition, an abundant band of ≈40 kDa was detected in plant tissue coexpressing the HCs and LCs or expressing the LC alone. This band disappears after treatment with 2-mercaptoethanol, and it most likely corresponds to a dimeric form of the LC. Abundant dimerization of the LC might be a consequence of the excess amount of LC relative to the HC, and it suggests that the available pool of HC may be a limiting factor for the formation of IgG heterodimers. A less abundant form of ≈70 kDa detected with both λ- and γ-chain-specific antibodies could be interpreted as a HL complex based on size.
Fig. 4.
Accumulation of assembled mAb A5 in N. benthamiana leaves coinfected with HC-expressing TMV provectors and LC-expressing PVX provectors. Proteins were separated in 10% polyacrylamide gels under nonreducing conditions and stained with Coomassie blue (Top) or blotted to nylon membrane. The Western blots were probed with goat anti-human IgG (γ-chain-specific) antibodies (Middle) and rabbit anti-human IgG (λ-chain-specific) antibodies (Bottom). U, uninfected tissue; 3–11 dpi; S, A5 standard; L, LC expressed alone with TMV provectors (6 dpi); H, HC expressed alone with TMV provectors (6 dpi); H2L2, IgG heterotetramer containing two HCs and two LCs; H2L, heterotrimer containing two HCs and one LC; H2, HC homodimer; RbcL, large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase; HL, heterodimer containing one HC and one LC; L2, LC homodimer; H, HC monomeric form.
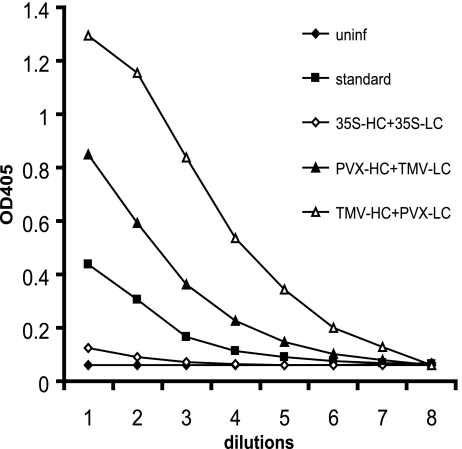
Next, plant material was analyzed by using antigen-specific ELISA for estimation of the specific antigen-binding activity of expressed mAbs. Microtiter plates were coated with the antigen peptide, and HRP-labeled anti-human IgG (whole-molecule) antibodies were used for detection of bound A5 mAbs. The absorbance values for N. benthamiana tissue coexpressing A5 chains were significantly higher compared with uninfected leaves (Fig. 5), confirming the efficient assembly of Ig molecules with specific antigen-binding activity.
Fig. 5.
Accumulation of mAb A5 in N. benthamiana leaves after coinfiltration with 35S-promoter constructs or viral vectors, analyzed by using antigen-specific ELISA. Serial 2-fold dilutions of crude extracts from leaves coinfected with viral provectors (PVX-HC + TMV-LC, 8 dpi; TMV-HC + PVX-LC, 10 dpi) and 35S-promoter constructs (35S-LC + 35S-HC, 2 dpi) were compared with serial dilutions of the mAb A5 standard expressed in CHO cells and crude protein extracts from uninfected tissue (uninf) used as a negative control.
Noncompeting Viral Vectors Provide a Higher Level of Expression than Standard Transient Expression Cassettes.
We compared the expression level of mAb A5 obtained by using noncompeting viral vectors or standard expression cassettes consisting of a coding sequence under the control of a constitutive promoter. N. benthamiana leaves were coinfected with a mixture of agrobacterial cultures carrying constructs pICH23660 (35S promoter-LC) and pICH23670 (35S promoter-HC). Expression levels of the HCs and LCs, as well as of the assembled mAbs, were monitored from 1 to 6 dpi by Western blotting. Expression was maximal at 2 dpi (data not shown).
The yield of assembled recombinant mAb A5 obtained with mixed viral infection or with standard promoter constructs was determined by using antigen-specific ELISA. The antibody concentration was calculated by comparing titration curves built for the plant samples with a curve prepared for the control mAb A5 expressed in Chinese hamster ovary (CHO) cells, which was used as quantification standard (Fig. 5). The yield obtained by coexpression of promoter constructs was 10–15 μg/g of fresh weight, depending on the experiment. The yield obtained by using viral constructs with the combination PVX-HC + TMV-LC was between 200 and 300 μg/g of fresh weight depending on the experiment. The highest yield was obtained by using the combination TMV-HC + PVX-LC with values ≈500 μg/g of fresh weight.
Purification of Functional A5 Antibodies from Plant Tissue.
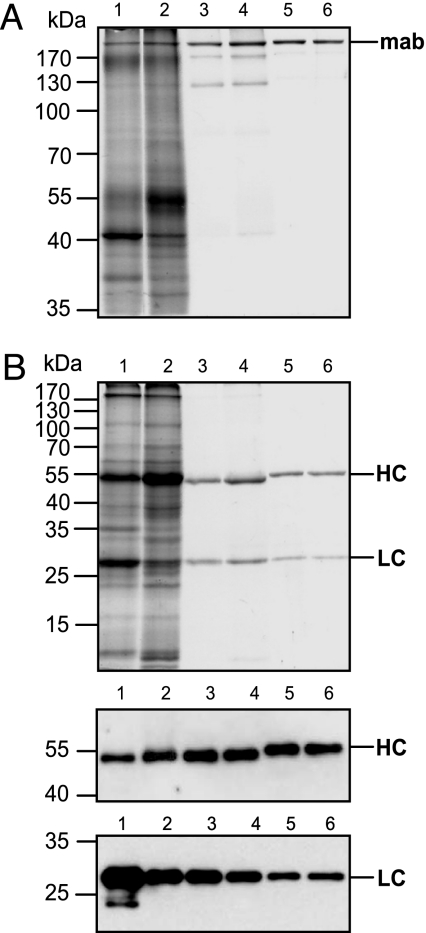
The A5 antibody was purified from N. benthamiana leaves coinfected with TMV and PVX vectors by using protein A magnetic beads. Purified mAb A5 migrates under nonreducing conditions as a major band of ≈170 kDa, corresponding to the H2L2 heterotetramer, and some minor bands. An identical pattern was also obtained with the standard antibody produced in CHO cells (Fig. 6A). Under reducing conditions mAb A5 disassociates into HCs and LCs that migrate at ≈50 and 25 kDa in the same stoichiometric ratio as the standard mAb (Fig. 6B). The HC of the plant-derived antibody migrates slightly faster than that of the control antibody. This result is as expected because the plant-expressed antibody was specifically engineered to be nonglycosylated. Antigen-specific ELISA confirmed the specific antigen-binding activity of isolated mAb A5 with a titration curve similar to the standard (data not shown).
Fig. 6.
Purification of the A5 mAb by using protein A magnetic beads. (A) Plant-derived and standard mAbs migrating in a 10% polyacrylamide gel under nonreducing conditions. (B) Plant-derived and standard mAbs resolved in a 12% polyacrylamide gel under reducing conditions. (B Top) PAGE with Coomassie blue staining. (B Middle) Western blot probed with γ-chain-specific antibodies. (B Bottom) Western blot probed with λ-chain-specific antibodies. Lane 1, PVX-HC + TMV-LC, crude extract; lane 2, TMV-HC + PVX-LC, crude extract; lane 3, PVX-HC + TMV-LC, purified mAb; lane 4, TMV-HC + PVX-LC, purified mAb; lanes 5 and 6, A5 standard.
Discussion
Until now, plant viral vectors have not been able to express heterooligomeric proteins efficiently. The efficacy of viral replication usually decreases as gene insert length increases; a viral vector containing both antibody chains in a single construct would be quite large (>2.4 kb) and therefore less efficient for replication. In addition, as shown in this study, if both polypeptide chains are expressed from two different vectors built around the same virus backbone, the amount of cells coexpressing both polypeptides becomes extremely small. Similar observations have been reported for other viruses belonging to several genera, including potyviruses (19), potexviruses such as PVX (19, 20), and the alfamovirus avian myeloblastosis virus in the Bromoviridae family (21).
Although two related viral vectors/viruses are obviously not able to replicate within the same cell efficiently, it has long been known that some viruses belonging to different groups can coexist in mixed populations (22). It has also been shown that, as for PVX and TMV in this work, other combinations of viruses such as PVX and PVY are able to coinfect the same cells efficiently (19). It is therefore likely that viral vectors built on other noncompeting virus combinations could be used for expression of heterooligomeric proteins.
The most astonishing result of this study is the level of coexistence and coexpression provided by the noncompeting viral vectors. It is interesting that the level of expression provided by one of the two vectors within the same cell is almost independent of the expression level controlled by the other, indicating that the expression levels did not reach the stage when the material/energy resources of the cell become a bottleneck. Why noncompeting vectors are able to replicate efficiently in the same cell and move efficiently from cell to cell is unclear. One possibility may be that noncompeting viruses interact with different sets of host proteins required for replication/movement.
The expression levels achieved in our experiments are by no means fully optimized, as evidenced by the presence of the other oligomeric products composed of either LC or HC or both. Expression levels of heterologous recombinant proteins achieved by magnifection technology were as high as 5 g of recombinant protein per kg of fresh-leaf biomass (13, 23). In contrast, the levels of expression of fully assembled IgG antibodies observed in this study are ≈0.5 g/kg, or 1/10 as high. This yield is nevertheless 10–100 times greater than the yields obtained with other transient expression systems. In this work, only two expression combinations were tested: (i) the LC expressed from the TMV-based vector and the HC expressed from the PVX-based vector, and (ii) vice versa. The best yield was achieved when the HC was expressed from the TMV-based vector. Because expression levels achieved with viral vectors depend on gene insert size (among other parameters) and because the optimal ratio of expression levels of the two chains needs to be determined empirically (see ref. 24), additional fine-tuning of the expression levels provided by each of the vectors should further improve the yield of assembled functional mAbs.
Another way of further increasing the expression could be to coexpress chaperones known to be involved in antibody folding/assembly (25). Given the extremely high expression of the two polypeptide chains (because of expression from viral vectors) and given the fact that expression of both chains requires multiple host protein factors for proper processing (targeting to the endoplasmic reticulum, folding, glycosylation), it seems likely that some of the components of the processing machinery may become limiting and that boosting their expression might result in an increased yield of mature antibody.
In addition to the results reported here, we have successfully used the protocol described in this paper to express six additional antibodies, representing glycosylated and aglycosylated, chimeric and humanized mAbs of the IgG1 or IgG2 classes (unpublished data). The protocol therefore appears to be a broadly applicable solution for expression of heterooligomers containing two different polypeptide chains.
In conclusion, the technology proposed here allows for a very rapid and versatile assembly of full IgG mAbs, and thus it represents a universal platform for rapid research and development as well as continuous and unlimited industrial scale-up. This technology could be especially suited for situations requiring very rapid large-scale manufacturing of antibodies or other protein complexes such as virus-like particles, for example, in cases of pandemic or terrorist events.
Materials and Methods
Plasmid Constructs.
To make the TMV-based 5′ module pICH17388, a 5,007-bp SphI–EcoRI fragment consisting of the RdRp- and MP-coding sequences with an analogous 6,436-bp fragment (but containing 11 introns) from pICH17272 (13) was substituted into pICH-NOP (23) (Fig. 1). A 684-bp BsaI–BamHI fragment containing the DsRED-coding sequence was inserted into the TMV-based 3′ module cloning vector pICH10990, generating the pICH10580 construct. Similarly, a 717-bp fragment containing the GFP-coding sequence was placed into a 3′ provector module, resulting in the pICH7410 construct. Synthetic genes for the human mAb A5 LCs and HCs were used for subcloning into viral vectors. A 730-bp NcoI–NotI fragment, containing the LC-coding sequence, was blunted at the NotI site and ligated into the TMV-based 3′ module cloning vector pICH11599 digested with NcoI–XbaI and blunted at the XbaI site, resulting in pICH21910. The same strategy was used to subclone a 1,421-bp NcoI–NotI fragment containing the A5 HC-coding sequence into pICH11599, resulting in construct pICH21920. A PVX-based 5′ module (pICH21380) was made by cloning a fragment consisting of the 35S promoter PVX RdRp and triple gene block sequences (subcloned from construct pA3151, obtained from Y. Dorokhov, Moscow State University), which is derived from pPVX201, described in ref. 26, followed by a fragment containing intron sequences (an AttP recombination site), the nopaline synthase (nos) terminator (from pICH17388) in a binary vector. PVX-based 3′ modules for GFP expression, either containing CP (pICH21470) or lacking CP (pICH21282), were made from pICH7410 by replacing the TMV 3′ untranslated region with the 3′ end of PVX (the CP-coding sequence and 3′ untranslated region of PVX for pICH21470 or the PVX 3′ untranslated region alone for pICH21282). Similarly, the TMV-based 3′ provector module pICH21920 was converted into a PVX-based 3′ provector construct (pICH22250) by replacing the TMV 3′ untranslated region with the 3′ untranslated region of PVX. This replacement was achieved by ligation of the 575-bp HindIII–NdeI fragment of pICH21282 blunted at the HindIII site with an SalI–NdeI fragment of pICH21920 blunted at the SalI site. A 723-bp NcoI–EcoRI fragment containing the coding sequence of the A5 LC was subcloned into pICH22250 digested with NcoI–EcoRI, resulting in pICH22240. A construct for the expression of A5 LC under the control of the 35S promoter, pICH23660, was made in several cloning steps. The final construct contained, in sequential order, the 1,337-bp 35S promoter, the 62-bp 5′ untranslated leader of TMV (ω), the 711-bp A5 LC-coding sequence including the N-terminal endoplasmic reticulum-targeting presequence, and the 274-bp nopaline synthase terminator. A corresponding construct for the expression of A5 HC, pICH23670, was made by insertion of the 1,404-bp NcoI–PacI fragment containing the A5 HC-coding sequence into pICH23660 digested with NcoI–PacI. The pICH14011 construct was made on the basis of the pICH10881 plasmid (23) by replacing the Arabidopsis actin 2 promoter with the promoter of the hsp81.1 gene from the same plant (GenBank accession no. AJ010947).
Agroinfiltration Procedure.
Equal volumes of overnight-grown Agrobacterium cultures, OD600 ≈1.8, were mixed and sedimented at 6,000 × g for 3 min. The pellet was resuspended in a solution containing 10 mM Mes (pH 5.5) and 10 mM MgSO4, resulting in a 10−1 dilution for each individual culture. Leaves of 6- to 7-week-old greenhouse-grown N. benthamiana plants were infiltrated by using a syringe without a needle. Alternatively, the agroinfiltration was performed with unsedimented overnight cultures directly diluted in the solution described above.
SDS/PAGE and Western Blotting.
Samples (100 mg) of N. benthamiana leaves infiltrated with Agrobacterium were ground in liquid nitrogen with 300 μl of protein extraction buffer (0.1 M Tris/150 mM NaCl/0.1% Tween 20, pH 8.0). Crude leaf extracts were resolved on 10% (nonreducing conditions) or 12% (reducing conditions) polyacrylamide gels by using the buffer system of Laemmli (27) followed by Coomassie brilliant blue G-250 staining with PageBlue protein staining solution (Fermentas, Vilnius, Lithuania). For Western blot analysis, proteins from SDS/PAGE were electrophoretically transferred to Hybond P membrane (GE Healthcare, Munich, Germany) by using blotting buffer [25 mM Tris/192 mM glycine/20% (vol/vol) methanol, pH 8.3]. Membranes were blocked with a 5% (wt/vol) solution of skim milk powder in TBST (50 mM Tris/100 mM NaCl/0.05% Tween 20, pH 7.4) and probed with goat anti-human λ-LC HRP-conjugated antibodies (Sigma, Dorset, U.K.) or goat anti-human IgG γ-chain specific HRP-conjugated antibodies (Sigma) diluted in a 2.5% (wt/vol) solution of skim milk powder in TBS (1:10,000 and 1:5,000, respectively). Bound antibodies were detected by using the ECL detection reagent (GE Healthcare).
Mesophyll Protoplasts Isolation.
N. benthamiana leaf areas inoculated with Agrobacterium were sliced into 2- to 3-mm strips, which were then macerated for 10–16 h at 25°C in a solution containing 0.4% cellulase Onozuka R-10 (Duchefa Biochemie, Haarlem, The Netherlands), 0.4% Macerozyme R-10 (Duchefa), 0.5 M sucrose, 7 mM CaCl2, and 90 mM glycine. Released protoplasts were filtered through a nylon mesh with 50-μm diameter pores, and then they were resuspended in W5 medium (28).
Monitoring of GFP and DsRED Fluorescence.
Leaves expressing GFP and DsRED were observed under UV illumination generated by a hand-carried UV B-100AP lamp (UVP, Upland, CA). Protoplasts were viewed with a DM IL microscope (Leica, Vienna, Austria) with GFP plant fluorescence filter sets (excitation filter, 470/40 nm; barrier filter, 525/50 nm) or DsRED filter sets (excitation filter, 546/12 nm; barrier filter, LP 560 nm).
ELISA.
Ninety-six-well Nunc-Immuno MaxiSorp surface plates (Nalge Nunc, Rochester, NY) were coated with synthetic peptide antigen coupled to BSA (1 μg/ml; Sigma–Aldrich, St. Louis, MO) diluted in 50 mM sodium carbonate buffer (pH 10.0). The wells were blocked with 250 μl of TBST containing 5% skim milk powder. Leaf tissue (100 mg) was homogenized in 600 μl of protein-extraction buffer. Plates were loaded with 100 μl of serial 2-fold dilutions of crude plant extracts and standard mAb A5. After incubation with HRP-conjugated rabbit anti-human IgG (whole-molecule) antibodies (Sigma), plates were treated with 100 μl of ABTS [2,2′-azinobis-(3-ethylbenzthiazoline-6-sulfonate) (Calbiochem, San Diego, CA)] as a peroxidase substrate. The color reaction was stopped with 100 μl of 0.625 M oxalic acid. Absorbance was measured at 405 nm by using a universal microplate reader EL800 (Bio-Tek Instruments, Winooski, VT). Calculation of the data was performed by using Excel 2000 software (Microsoft, Redmond, WA).
Isolation of Plant-Derived Recombinant mAbs.
Total soluble proteins were extracted from agroinoculated N. benthamiana leaf areas with a buffer containing 100 mM sodium phosphate, 150 mM NaCl, and 0.05% Tween 20 (pH 8.0). Small-scale isolation of mAb A5 from crude protein extracts was performed by using one-step affinity purification with protein A magnetic beads (New England Biolabs, Beverly, MA) according to the instructions of the manufacturer.
Abbreviations
- CP
coat protein
- dpi
days postinfiltration
- DsRED
red fluorescent protein from Discosoma
- HC
heavy chain
- LC
light chain
- MP
movement protein
- PVX
potato virus X
- RdRp
RNA-dependent RNA polymerase
- TMV
tobacco mosaic virus.
Footnotes
Conflict of interest statement: No conflicts declared.
See Commentary on page 14645.
References
- 1.Hiatt A, Cafferkey R, Bowdish K. Nature. 1989;342:76–78. doi: 10.1038/342076a0. [DOI] [PubMed] [Google Scholar]
- 2.Duering K, Hippe S, Kreuzaler F, Schell J. Plant Mol Biol. 1990;15:281–293. doi: 10.1007/BF00036914. [DOI] [PubMed] [Google Scholar]
- 3.Ma JK, Hiatt A, Hein M, Vine ND, Wang F, Stabila P, van Dolleweerd C, Mostov K, Lehner T. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- 4.Schillberg S, Fischer R, Emans N. Cell Mol Life Sci. 2003;60:433–445. doi: 10.1007/s000180300037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoger E, Sack M, Nicholson L, Fischer R, Christou P. Curr Pharm Des. 2005;11:2439–2457. doi: 10.2174/1381612054367535. [DOI] [PubMed] [Google Scholar]
- 6.Klimyuk V, Marillonnet S, Knaeblein J, McCaman M, Gleba Y. In: Modern Biopharmaceuticals. Knaeblein J, editor. Germany: Wiley, Weinheim; 2005. pp. 893–917. [Google Scholar]
- 7.Ma JK-C, Drake PMW, Chargelegue D, Obregon P, Prada A. Vaccine. 2005;23:1814–1818. doi: 10.1016/j.vaccine.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Ko K, Steplewski Z, Glogowska M, Koprowski H. Proc Natl Acad Sci USA. 2005;102:7026–7030. doi: 10.1073/pnas.0502533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verch T, Yusibov V, Koprowski H. J Immunol Methods. 1998;220:69–75. doi: 10.1016/s0022-1759(98)00149-5. [DOI] [PubMed] [Google Scholar]
- 10.Vaquero C, Sack M, Chandler J, Drossard J, Schuster F, Monecke M, Schillberg S, Fischer R. Proc Natl Acad Sci USA. 1999;96:11128–11133. doi: 10.1073/pnas.96.20.11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathuria S, Sriraman R, Nath R, Sack M, Pal R, Artsaenko O, Talwar GP, Fischer R, Finnern R. Hum Reprod. 2002;17:2054–2061. doi: 10.1093/humrep/17.8.2054. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez M, Ramirez NI, Ayala M, Freyre F, Perez L, Triguero A, Mateo C, Selman-Housein G, Gavilondo JV, Pujol M. Biotechnol Bioeng. 2005;89:188–194. doi: 10.1002/bit.20333. [DOI] [PubMed] [Google Scholar]
- 13.Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, Gleba Y. Nat Biotechnol. 2005;23:718–723. doi: 10.1038/nbt1094. [DOI] [PubMed] [Google Scholar]
- 14.Gleba Y, Klimyuk V, Marillonnet S. Vaccine. 2005;23:2042–2048. doi: 10.1016/j.vaccine.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Forster RL, Beck DL, Guilford PJ, Voot DM, Van Dolleweerd CJ, Andersen MT. Virology. 1992;191:480–484. doi: 10.1016/0042-6822(92)90215-b. [DOI] [PubMed] [Google Scholar]
- 16.Cruz SS, Roberts AG, Prior DA, Chapman S, Oparka KJ. Plant Cell. 1998;10:495–510. doi: 10.1105/tpc.10.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedorkin O, Solovyev A, Yelina N, Zamyatnin A, Jr, Zinovkin R, Makinen K, Schiemann J, Morozov SYu. J Gen Virol. 2001;82:449–458. doi: 10.1099/0022-1317-82-2-449. [DOI] [PubMed] [Google Scholar]
- 18.Peterson E, Owens SM, Henry RL. AAPS J. 2006;8:E383–E390. doi: 10.1007/BF02854909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietrich C, Maiss E. J Gen Virol. 2003;84:2871–2876. doi: 10.1099/vir.0.19245-0. [DOI] [PubMed] [Google Scholar]
- 20.Diveki Z, Salanki K, Balazs E. Biochimie. 2002;84:997–1002. doi: 10.1016/s0300-9084(02)00007-x. [DOI] [PubMed] [Google Scholar]
- 21.Hull R, Plaskitt A. Virology. 1970;42:773–776. doi: 10.1016/0042-6822(70)90322-3. [DOI] [PubMed] [Google Scholar]
- 22.Goodman RM, Ross AF. Virology. 1974;58:16–24. doi: 10.1016/0042-6822(74)90137-8. [DOI] [PubMed] [Google Scholar]
- 23.Marillonnet S, Giritch A, Gils M, Kandzia R, Klimyuk V, Gleba Y. Proc Natl Acad Sci USA. 2004;101:6852–6857. doi: 10.1073/pnas.0400149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlatter S, Stansfield SH, Dinnis DM, Racher AJ, Birch JR, James DC. Biotechnol Prog. 2005;21:122–133. doi: 10.1021/bp049780w. [DOI] [PubMed] [Google Scholar]
- 25.Nuttall J, Vine N, Hadlington JL, Drake P, Frigerio L, Ma JK. Eur J Biochem. 2002;269:6042–6051. doi: 10.1046/j.1432-1033.2002.03302.x. [DOI] [PubMed] [Google Scholar]
- 26.Baulcombe DC, Chapman S, Santa Cruz S. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Medgyesy P, Menczel L, Maliga P. Mol Gen Genet. 1980;179:693–696. [Google Scholar]