Abstract
Corticotropin-releasing factor (CRF) receptor subtypes 1 and 2 have been implicated in rodent models of anxiety, but much less is known about the CRF system and social behavior. Both corticosterone and central CRF receptors modulate pair bonding in the monogamous prairie vole. Using receptor autoradiography, we mapped CRFR1 and CRFR2 in the brains of two monogamous vole species, the prairie vole and pine vole, and two promiscuous vole species, the meadow vole and montane vole. We found markedly different patterns of brain CRFR1 and CRFR2 binding among the four species, including species differences in the olfactory bulb, nucleus accumbens, lateral septum, hippocampus, laterodorsal thalamus, cingulate cortex, superior colliculus, and dorsal raphe. Interestingly, we also observed striking sex differences in voles: CRFR2 binding was higher in the encapsulated bed nucleus of the stria terminalis in males than females for all four vole species. These results suggest possible sites of action for CRF-induced facilitation of pair bond formation in prairie voles, as well as potential sex differences in the CRF modulation of pair bonding. Further examination of CRF receptors in vole species may reveal a novel role for CRF in social behavior. Ultimately, our results identify several brain regions with conserved CRF receptor patterns across rodent and primate species, in contrast to several brain regions with phylogenetically plastic CRF receptor patterns, and have interesting implications for the evolution of CRF receptor patterns and behavior.
Indexing terms: corticotropin-releasing factor, corticotrophin-releasing hormone, nucleus accumbens, lateral septum, hippocampus, bed nucleus of the stria terminalis
Microtine rodents offer a well-established animal model for the comparative study of social behaviors (Insel and Young, 2001; Young and Wang, 2004). Prairie voles (Microtus ochrogaster) and pine voles (Microtus pinetorum), which exhibit monogamous social structure in nature, show species-typical social behaviors such as pair bonding, affiliation, and biparental care (Getz et al., 1981; Gruder-Adams and Getz, 1985). In contrast, congener meadow voles (Microtus pennsylvanicus) and montane voles (Microtus montanus) show a strikingly opposite social structure, demonstrating behaviors such as promiscuity, social isolation, and minimal paternal care of offspring (Salo et al., 1993; Shapiro and Dewsbury, 1990). Genetic differences that have evolved between prairie, pine, meadow, and montane voles are thought to contribute to species differences in social organization and brain receptors; these species differences in social behavior have been linked to the brain distribution of the neuropeptide receptors for oxytocin and vasopressin (Insel and Shapiro, 1992; Insel et al., 1994; Lim et al., 2004b; Young et al., 1999). Much of the research in the last decade has focused very closely on these neurohypophysial peptides and pair bonding in monogamous prairie voles. However, more recent research has uncovered another candidate neuropeptide, the corticotropin-releasing factor (CRF) system, which also appears to regulate pair bonding in monogamous prairie voles (DeVries et al., 2002).
There is extensive literature linking CRF to stress, anxiety, and the regulation of the hypothalamic-pituitary-adrenal (HPA) axis. However, there are relatively few studies implicating the CRF system in social behavior. One study using prairie voles found that exogenous corticosterone administered to male prairie voles facilitated pair bond formation (DeVries et al., 1996). This effect was mimicked by swim stress, which had been previously shown to activate the HPA axis in voles, prior to pairing (DeVries et al., 1995). Adrenalectomy blocked pair bond formation, and this effect was rescued by corticosterone replacement (DeVries et al., 1996). Interestingly, the opposite phenomenon was observed in female prairie voles, where exogenous corticosterone actually inhibited partner preference formation (DeVries et al., 1996). Since CRF released from the paraventricular nucleus of the hypothalamus causes the release of ACTH from the anterior pituitary, and in turn corticosterone from the adrenal cortex, these data suggest that CRF release may play a role in stress-induced activation of the HPA axis during pair bond formation. However, a later study by the same group found that CRF administered intracerebroventricularly (i.c.v.), at doses too low to facilitate anxiety, were sufficient to facilitate partner preference in male prairie voles (DeVries et al., 2002). Central administration of alpha-helical CRF, an antagonist that binds to both CRFR1 and CRFR2, blocked partner preference in males (DeVries et al., 2002). These data suggest that CRF may play a role in pair bond formation via anxiety-independent mechanisms and through the involvement of centrally acting brain receptors.
Since the CRF system has been shown to be critical for the regulation of pair bonding, one might predict that neural circuits for this peptide system would differ between monogamous and promiscuous species. In addition, because corticosterone exerts opposite effects on pair bond formation in male and female prairie voles, one might predict that sex differences could potentially exist at the level of central CRF receptors in the brain. Given that corticosterone can exert canonical feedback at both hypothalamic as well as suprahypothalamic brain regions, it is possible that corticosterone could affect CRF release centrally within the brain and, thus, depending on gender-specific CRF receptor expression, activate different neural circuits in male versus female pair bond formation (McEwen et al., 1968; Sapolsky et al., 1990). To test these hypotheses, we first mapped the distributions of CRFR1 and CRFR2 in male and female prairie and meadow voles using receptor autoradiography. As an additional axis of comparison, we mapped CRFR1 and CRFR2 brain distribution in a different species pair, the monogamous pine vole and the promiscuous montane vole. We show evidence for both species and sex differences in several brain regions that could be related to the differences in their social structure across the four species. Based on a semiquantitative comparison of CRF receptor distributions found in the four vole species to rat, mouse, and rhesus macaque distributions found in previous studies (Aguilera et al., 1987; De Souza et al., 1985; Potter et al., 1994; Primus et al., 1997; Rominger et al., 1998; Sanchez et al., 1999; Steckler and Holsboer, 1999; Van Pett et al., 2000), we discuss putative brain CRF systems that appear to be 1) evolutionarily conserved across species, 2) plastic between species, and 3) potentially associated with monogamous social organization.
MATERIALS AND METHODS
Subjects
Animals were adult male and female prairie voles (70–100 days of age) from our laboratory breeding colony that were originally derived from field-captured voles in Illinois, USA. Adult male and female meadow voles of the same age were derived from stock obtained from the laboratory breeding colony of Dr. Zuoxin Wang at Florida State University. Adult male and female pine voles were obtained from the laboratory breeding colony of Dr. John Vandenbergh at North Carolina State University. Montane vole subjects were adult males and females from our laboratory breeding colony originally derived from stock obtained from the National Institutes of Health. After weaning at 21 days of age, subjects were housed in same-sex sibling pairs or trios and water and Purina rabbit chow provided ad libitum. All cages were maintained on a 14:10 light:dark cycle with the temperature at 20°C. Thirty-two animals were used in the generation of data for these experiments (n = 8 per species of vole, and four males and four females per species). All experiments were approved by the institutional guidelines set by the animal care and use committee of Emory University and conformed to the guidelines set by the NIH.
CRF receptor autoradiography
Due to the lack of high-affinity iodinated ligands selective for the CRFR2 subtype, we performed receptor autoradiography with [125I-Tyr0]-sauvagine, a ligand with high affinity for both CRFR1 (Kd = 0.2–0.4 nM) and CRFR2 (Kd = 0.1-0.3 nM) (Grigoriadis et al., 1996; Primus et al., 1997). An incubation period of 2 hours was shown to be optimal for rhesus macaque brain tissue (Sanchez et al., 1999). To identify CRFR2 receptor binding sites, we combined [125I-Tyr0]-sauvagine with an excess of unlabeled CP-154,526, a selective CRFR1 small molecule antagonist (Schulz et al., 1996). To identify CRFR1 receptor binding sites, we used subtraction of optical density readings of total CRF receptor binding minus specific CRFR2 receptor binding. The subtraction technique is further described in the Data Analysis section (below).
Animals were deeply anesthetized with isoflurane (10 drops isoflurane placed onto a cotton ball within a chamber measuring 27 cubic inches), rapidly decapitated, their brains removed, and snap-frozen in dry ice. Brains were sliced on a cryostat at 20 μm, sections thaw-mounted onto Superfrost Plus slides (Fisher, Pittsburgh, PA), and stored at −80° C until use. A 1:7 series of sections were collected from the olfactory bulbs through the hindbrain. Three adjacent sets of sections were processed for CRF total binding (Fig. 1A,B), CRFR2 autoradiography (Fig. 1C,D), and CRF receptor nonspecific binding (Fig. 1E,F), similar to previously described protocols (Lim et al., 2004a; Sanchez et al., 1999; Skelton et al., 2000).
Fig. 1.
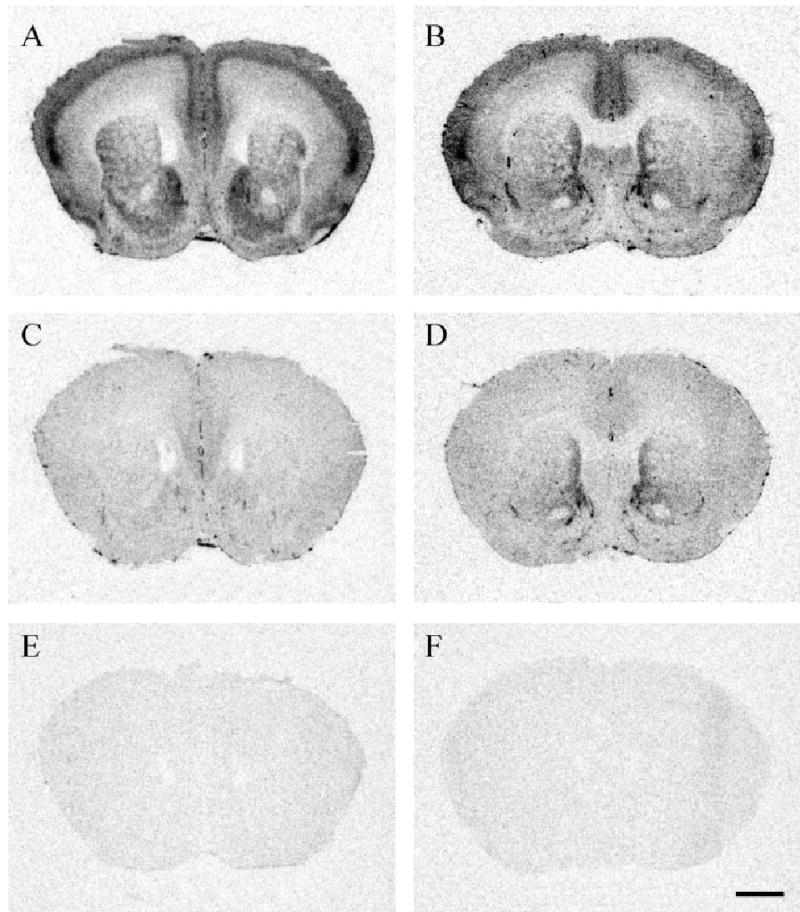
Validation of CRF receptor autoradiography technique. A: Meadow vole brain section incubated with [125I-Tyr0]-sauvagine, which labels both CRFR1 and CRFR2. B: Prairie vole brain section incubated with [125I-Tyr0]-sauvagine. C: Meadow vole brain section incubated with [125I-Tyr0]-sauvagine plus the CRFR1 selective inhibitor CP-154,526, which ultimately labels only CRFR2. D: Prairie vole brain section labeling for CRFR2. E: Meadow vole brain section incubated with [125I-Tyr0]-sauvagine plus cold sauvagine, ultimately showing the nonspecific binding. F: Prairie vole brain section showing nonspecific binding. Scale bar = 1 mm in F (applies to A–F).
Slides were thawed at room temperature until completely dried and lightly fixed for 2 minutes in a 0.1% paraformaldehyde-PBS solution (pH 7.4). Slides were then rinsed twice in 50 mM Tris base (pH 7.4) solution for 10 minutes each, then incubated in tracer for 2 hours. The tracer buffer consisted of a 50 mM Tris base, 10 mM MgCl, 0.1% bovine serum albumin, 0.05% bacitracin, plus 0.2 nM [125I-Tyr0]-sauvagine (PerkinElmer/NEN, Boston, MA). This tracer binds both CRFR1 and CRFR2. Slides were then rinsed with 50 mM Tris base plus 10 mM MgCl (pH 7.4) for 4 × 5 minutes, plus 30 minutes with stirring with a magnetic bar on a stir plate. Slides were then dipped in deionized H2O, blown dry with cool air, and apposed to Kodak MR film for 85 hours with [125I] microscale standards (PerkinElmer/NEN). Representative brain sections are shown in Figure 1A,B.
CRFR2 autoradiography
An adjacent set of slides were processed for CRFR2 autoradiography. CRFR2 binding was measured by incubating [125I-Tyr0]-sauvagine, which binds to both CRFR1 and CRFR2, with unlabeled CP-154,526-1 (butyl-[2,5-dimethyl-7-(2,4,6-trimethylphenyl)-7H-pyrrolo[2,3-d]-pyrimidin-4-yl]-ethylamine), a selective CRFR1 antagonist which was synthesized at Emory University and kindly provided by Michael J. Owens, Ph.D. This CP-154,526 compound has a high affinity for CRFR1 sites, with Ki of 2.7 nM, but has low affinity for CRFR2 sites (Schulz et al., 1996). The concentration of CP-154,526 used in this study (1 μM) would compete with [125I-Tyr0]-sauvagine for CRFR1 binding sites, but not CRFR2 binding sites, because the Ki for inhibition of binding by [125I-Tyr0]-sauvagine to CRFR2 is greater than 10 μM (Schulz et al., 1996).
Slides were thawed at room temperature until completely dried and lightly fixed for 2 minutes in a 0.1% paraformaldehyde-PBS solution (pH 7.4). Slides were then rinsed twice in 50 mM Tris base (pH 7.4) solution for 10 minutes each, then incubated in tracer for 2 hours. The tracer buffer consisted of a 50 mM Tris base, 10 mM MgCl, 0.1% bovine serum albumin, 0.05% bacitracin, plus 0.2 nM [125I-Tyr0]-sauvagine (PerkinElmer/NEN) and 1 μM CP-154,526. Slides were then rinsed with 50 mM Tris base plus 10 mM MgCl (pH 7.4) for 4 × 5 minutes, plus 30 minutes with stirring with a magnetic bar on a stir plate. Slides were then dipped in deionized H2O, blown dry with cool air, and apposed to Kodak MR film for 85 hours with [125I] microscale standards (PerkinElmer/NEN). This protocol has been successfully employed for both rat and monkey brain tissue (Sanchez et al., 1999; Skelton et al., 2000). Representative brain sections are shown in Figure 1C,D.
CRF receptor nonspecific binding
A third set of adjacent slides were processed for nonspecific binding for both the CRFR1 and CRFR2. The tracer buffer contained 0.2 nM [125I-Tyr0]-sauvagine plus 1 mM cold sauvagine (American Peptide, Sunnyvale, CA). Slides were apposed to Kodak MR film for 85 hours. This protocol for nonspecific binding has been successfully employed for both rat and monkey brain tissue (Sanchez et al., 1999; Wigger et al., 2004). Representative brain sections are shown in Figure 1E,F.
Acetylcholinesterase (AChE) stain
Following CRFR1 autoradiography and film development, slides were counterstained for acetylcholinesterase to better delineate the brain regions for image analysis. We modified the traditional AChE protocol to amplify AChE signal since the tissue had already undergone receptor autoradiography (Lim et al., 2004a). Briefly, slides were incubated for 5 hours in an enzymatic solution containing 0.0072% ethopropazine, 0.075% glycine, 0.05% cupric sulfate, 0.12% acetyl thiocholine iodide, 0.68% sodium acetate, pH 5.0. Slides were then rinsed in ddH2O, developed for 30 minutes in a solution composed of 0.38% sodium sulfide at pH 7.8, rinsed again in dH2O, and then exposed to a silver intensification solution (1% silver nitrate) for 10 minutes. Slides were rinsed in dH2O, air-dried, dehydrated in ascending ethanols, cleared with Clearing Agent (Electron Microscopy Sciences, Fort Washington, PA), and coverslipped with DPX (Electron Microscopy Sciences). Images were compared with AChE-stained brain sections in rat and mouse brain atlases (Paxinos and Franklin, 2001; Paxinos and Watson, 1998).
Data analysis
Total CRF receptor binding, CRFR2 binding, and nonspecific binding were quantified using AIS 6.0 software (Imaging Research, Ontario, Canada). AChE-stained sections were used to delineate the borders of brain regions of interest before sampling. Measurements were taken bilaterally and averaged for each brain region across two or three sections. All optical density readings were automatically converted into decompositions per minute per milligram tissue (DPM/mg) based on a known set of [125I] microscale standard values included on each film. Brain regions were quantified by an experimenter blind to the species and sex of the individuals.
Specific CRFR2 binding values were obtained by subtracting nonspecific binding values from [125I-Tyr0]-sauvagine binding in the presence of 1 μM CP-154,526. Specific CRFR1 binding values were calculated by subtracting nonspecific binding from the total [125I-Tyr0]-sauvagine, and then subtracting CRFR2 binding. Only values above two standard deviations from the nonspecific binding values were considered detectable.
Statistics
Due to the large number of regions sampled, brain regions were regrouped into anatomically or functionally similar groups which were comprised of 7–10 subregions. The regional groupings are detailed in Tables 1–4. Each group was analyzed using a two-between, one-within subject repeated measures ANOVA, where species and sex were the between-subject factors, and brain region the within-subject factor. Significant interactions were followed by tests for simple effects (univariate t-tests) as previously described (Nair et al., 1999; Stevens, 1996). The alpha level was set at 0.05 divided by 8 (0.00625), to control for multiple comparisons. Therefore, a P-value less than or equal to 0.05 after this modified Bonferroni adjustment was considered significant.
TABLE 1.
Comparison of Prairie and Meadow Voles: CRFR1 Binding Using 125-I-Sauvagine and CRFR2 Subtraction1
| Region | Abbreviation | Meadow Female Mean ± SEM | Meadow Male Mean ± SEM | Prairie Female Mean ± SEM | Prairie Male Mean ± SEM |
|---|---|---|---|---|---|
| Cortical | |||||
| Prefrontal | PFCtx | 3748 ± 394 | 3472 ± 624 | 5021 ± 625 | 3434 ± 304 |
| Insular | IFCtx | 6041 ± 533 | 5204 ± 1150 | 6989 ± 819 | 4681 ± 446 |
| Orbitofrontal | OFCtx | 4432 ± 474 | 3867 ± 594 | 5193 ± 1022 | 2689 ± 193 |
| Cingulate rostral | CgCtx1 | 4530 ± 333 | 3856 ± 639 | 4229 ± 435 | 3083 ± 256 |
| Cingulate middle | CgCtx2 | 4313 ± 367 | 2721 ± 94 | 3912 ± 416 | 2475 ± 221 |
| Cingulate caudal | CgCtx3 | 4022 ± 167 | 2576 ± 83 | 3688 ± 240 | 2653 ± 105 |
| Retrosplenial | RSCtx | 2523 ± 245 | 1684 ± 129 | 1838 ± 357 | 1684 ± 212 |
| Septal-hippocampal | |||||
| Lateral septum | LS | ||||
| dorsal rostral | rLSD | 2386 ± 683 | 1018 ± 345 | 4697 ± 1004 | 2295 ± 376 |
| intermediata rostral | rLSI | 1525 ± 470 | 801 ± 168 | 1998 ± 678 | 1599 ± 586 |
| dorsal caudal | cLSD | 2336 ± 803 | 1839 ± 313 | 6173 ± 1267 | 3309 ± 188 |
| intermediata caudal | cLSI | 1817 ± 642 | 1208 ± 636 | 1816 ± 695 | 1157 ± 593 |
| Hippocampus | HC | ||||
| CA1 | CA1 | 717 ± 241 | ND | 1024 ± 108 | 642 ± 174 |
| CA2 | CA2 | 779 ± 245 | ND | 811 ± 116 | ND |
| CA3 | CA3 | 923 ± 113 | 708 ± 242 | 929 ± 240 | 773 ± 205 |
| dentate gyrus | DG | 611 ± 296 | 580 ± 228 | 723 ± 201 | 655 ± 292 |
| Extended amygdata | |||||
| Bed nucleus of the stria terminals | BnST | ||||
| medial | mBnST | 1349 ± 140 | 939 ± 132 | 1249 ± 209 | 600 ± 152 |
| lateral | IBnST | 1039 ± 55 | ND | 860 ± 113 | 677 ± 130 |
| caudal | cBnST | 705 ± 528 | 672 ± 532 | 957 ± 322 | 693 ± 281 |
| Amygdala | Amyg | ||||
| central | CeA | 667 ± 192 | ND | 879 ± 216 | 697 ± 275 |
| basolateral rostral | rBLA | 1558 ± 157 | 1002 ± 257 | 1126 ± 227 | 1143 ± 422 |
| medial rostral | rMeA | 2095 ± 382 | 1771 ± 243 | 1935 ± 254 | 1430 ± 322 |
| cortical rostral | rCoA | 2778 ± 442 | 3010 ± 374 | 3043 ± 196 | 2382 ± 415 |
| basolateral caudal | cBLA | 1358 ± 211 | 1392 ± 256 | 1413 ± 330 | 931 ± 273 |
| medial caudal | cMeA | 2335 ± 327 | 2778 ± 666 | 1443 ± 169 | 1306 ± 209 |
| cortical caudal | cCoA | 3958 ± 632 | 4127 ± 921 | 3360 ± 400 | 2472 ± 297 |
| Striatum and habenula | |||||
| Nucleus accumbens | NAcc | ||||
| shell | shNAcc** | 6260 ± 740 | 3882 ± 1248 | 1145 ± 218 | 1109 ± 56 |
| septal pole | spNAcc** | 1697 ± 164 | 1967 ± 256 | ND | 637 ± 409 |
| Caudate-putamen | CPu | ||||
| dorsal | CPuD | 1947 ± 184 | 1566 ± 430 | 1009 ± 277 | 790 ± 164 |
| ventral | CPuV** | 1905 ± 113 | 1758 ± 225 | 983 ± 234 | 836 ± 213 |
| Habenula | |||||
| medial | mHab** | 2172 ± 423 | 2078 ± 261 | 6926 ± 1182 | 5478 ± 1201 |
| lateral | lHab | 903 ± 294 | 545 ± 98 | 760 ± 139 | 553 ± 253 |
| Thalamus | |||||
| laterodorsal | ldThal | ND | ND | ND | ND |
| Olfactory and hindbrain | |||||
| Olfactory bulb | |||||
| external plexiform | OB** | 15474 ± 774 | 17132 ± 2022 | 9831 ± 1498 | 9447 ± 2536 |
| Superior colliculus | SColl* | 12600 ± 1766 | 10199 ± 2120 | 7297 ± 226 | 5638 ± 709 |
| Periaqueductal gray | PAG | 1512 ± 146 | 1196 ± 64 | 1362 ± 206 | 1147 ± 239 |
| Dorsal raphe | DR | 497 ± 281 | ND | 542 ± 501 | 847 ± 523 |
| Locus coeruleus | LC | 1621 ± 126 | 1483 ± 286 | 1835 ± 398 | 1536 ± 150 |
| Pontine dorsal tegmental nucleus | PDTg | 2063 ± 438 | 1763 ± 430 | 2057 ± 408 | 1433 ± 57 |
| Principal motor nucleus of 5 | Pr5 | 1509 ± 198 | 1060 ± 485 | 1173 ± 425 | 724 ± 306 |
| Cerebellum | |||||
| granular layer | CBg | 4611 ± 633 | 3348 ± 296 | 4650 ± 1025 | 3274 ± 355 |
Values represent mean DPM/mg tissue from n = 4 per group.
ND, not detected (<2 S.D. nonspecific binding);
P < 0.05 species effect;
P < 0.01 species effect;
P < 0.05 sex effect.
TABLE 4.
Comparison of Pine and Montane Voles: CRFR2 Binding Using 125-I-Sauvagine and CP-154,5261
| Region | Abbreviation | Meadow Female Mean ± SEM | Meadow Male Mean ± SEM | Prairie Female Mean ± SEM | Prairie Male Mean ± SEM |
|---|---|---|---|---|---|
| Cortical | |||||
| Prefrontal | PFCtx | 1390 ± 186 | 1033 ± 130 | 1913 ± 200 | 1567 ± 84 |
| Insular | IFCtx** | 1181 ± 99 | 987 ± 74 | 1667 ± 123 | 1490 ± 109 |
| Orbitofrontal | OFCtx | 1475 ± 130 | 1187 ± 127 | 2774 ± 530 | 1842 ± 143 |
| Cingulate rostral | CgCtx1* | 1351 ± 136 | 1151 ± 55 | 1751 ± 154 | 1699 ± 123 |
| Cingulate middle | CgCtx2** | 1286 ± 142 | 1118 ± 58 | 1772 ± 202 | 1820 ± 54 |
| Cingulate caudal | CgCtx3** | 1277 ± 150 | 1137 ± 91 | 1871 ± 179 | 2023 ± 169 |
| Retrosplenial | RSCtx | 1608 ± 200 | 1266 ± 57 | 1558 ± 150 | 1290 ± 94 |
| Septal-hippocampal | |||||
| Lateral septum | LS | ||||
| dorsal rostral | RLSD** | 9891 ± 2333 | 4869 ± 306 | 27167 ± 2369 | 30216 ± 4144 |
| intermediata rostral | RLSI** | 10054 ± 1655 | 7136 ± 654 | 23646 ± 2990 | 29935 ± 2470 |
| dorsal caudal | CLSD** | 8139 ± 1158 | 7593 ± 241 | 34474 ± 7817 | 32930 ± 3591 |
| intermediata caudal | CLSI* | 13629 ± 2659 | 8774 ± 453 | 24334 ± 5494 | 31306 ± 5716 |
| Hippocampus | HC | ||||
| CA1 | CA1* | 1216 ± 119 | 1123 ± 127 | 2422 ± 398 | 1750 ± 139 |
| CA2 | CA2 | 1360 ± 51 | 1088 ± 106 | 1784 ± 389 | 1492 ± 102 |
| CA3 | CA3** | 1110 ± 49 | 1022 ± 104 | 1596 ± 117 | 1613 ± 153 |
| dentate gyrus | DG | 1401 ± 50 | 1130 ± 27 | 1497 ± 133 | 1481 ± 95 |
| Extended amygdala | |||||
| Bed nucleus of the stria terminals | BnST | ||||
| medial | mBnST | 1945 ± 236 | 1603 ± 166 | 2508 ± 384 | 2675 ± 343 |
| lateral | lBnST | 1552 ± 426 | 1916 ± 247 | 2422 ± 313 | 1775 ± 239 |
| caudal | cBnST† | 2954 ± 1195 | 7088 ± 2378 | 5375 ± 1914 | 8536 ± 609 |
| Amygdala | Amyg | ||||
| central | CeA | 1043 ± 52 | 1017 ± 133 | 1169 ± 116 | 1143 ± 113 |
| basolateral rostral | rBLA | 2316 ± 393 | 1675 ± 452 | 1292 ± 172 | 1199 ± 148 |
| medial rostral | rMeA | 1442 ± 324 | 1438 ± 347 | 2264 ± 429 | 2927 ± 917 |
| cortical rostral | rCoA | 1470 ± 79 | 1242 ± 124 | 1504 ± 235 | 1702 ± 224 |
| basolateral caudal | cBLA | 1230 ± 101 | 1323 ± 107 | 1339 ± 181 | 1300 ± 93 |
| medial caudal | cMeA | 1168 ± 84 | 1318 ± 90 | 2237 ± 219 | 1485 ± 123 |
| cortical caudal | cCoA | 1593 ± 177 | 1393 ± 372 | 1455 ± 172 | 2222 ± 707 |
| Striatum and habenula | |||||
| Nucleus accumbens | NAcc | ||||
| shell | shNAcc | 1068 ± 54 | 1240 ± 321 | 1560 ± 166 | 1474 ± 160 |
| septal pole | spNAcc** | 1496 ± 193 | 949 ± 23 | 10608 ± 2223 | 13684 ± 4451 |
| Caudate-putamen | CPu | ||||
| dorsal | CPuD** | 820 ± 35 | 791 ± 45 | 1095 ± 122 | 982 ± 32 |
| ventral | CPuV* | 1071 ± 60 | 866 ± 55 | 1630 ± 177 | 1310 ± 158 |
| Habenula | |||||
| medial | mHab | 935 ± 40 | 814 ± 35 | 1129 ± 134 | 1037 ± 118 |
| lateral | lHab* | 1042 ± 53 | 965 ± 57 | 1325 ± 91 | 1167 ± 72 |
| Thalamus | |||||
| laterodorsal | ldThal** | 5308 ± 1117 | 7582 ± 1027 | 504 ± 39 | 1224 ± 125 |
| Olfactory and hindbrain | |||||
| Olfactory bulb | |||||
| external plexiform | OB* | 1587 ± 48 | 1611 ± 48 | 3023 ± 224 | 3778 ± 944 |
| Superior colliculus | SColl | 862 ± 146 | 1227 ± 157 | 1323 ± 267 | 1218 ± 35 |
| Periaqueductal gray | PAG | 984 ± 93 | 821 ± 68 | 1529 ± 178 | 1197 ± 129 |
| Dorsal raphe | DR** | 10575 ± 547 | 10593 ± 1377 | 19083 ± 2604 | 16370 ± 1686 |
| Locus coeruleus | LC | 827 ± 96 | 895 ± 41 | 1363 ± 190 | 1041 ± 86 |
| Pontine dorsal tegmental n. | PDTg | 863 ± 51 | 758 ± 90 | 1417 ± 159 | 1035 ± 92 |
| Principal motor nucleus of 5 | Pr5 | 4317 ± 706 | 3721 ± 531 | 4692 ± 606 | 5250 ± 291 |
| Cerebellum | |||||
| granular layer | CBg** | 932 ± 77 | 742 ± 97 | 1315 ± 145 | 1301 ± 102 |
Values represent mean DPM/mg tissue from n = 4 per group.
ND, not detected (<2 S.D. nonspecific binding);
P < 0.05 species effect;
P < 0.01 species effect;
P < 0.05 sex effect.
Photomicrograph production details
Digital images were obtained from film autoradiograms using a B-95 Northern Light light box (Imaging Research) and a SPOT camera (Diagnostic Instruments, Sterling Heights, MI) connected to a computer through the RT SPOT power supply. Digital images obtained from microscope slides were similarly taken using the SPOT camera setup. Images were then imported into Adobe PhotoShop 7.0 (Adobe Systems, San Jose, CA), cropped to the correct size, and minimally adjusted for brightness and contrast in order to clarify the scientific point of interest. All figure text, arrows, and scale bars were added using Adobe Illustrator 10.0.
RESULTS
Distribution of CRFR1 in meadow and prairie voles
Mean CRFR1 values (DPM/mg) from several brain regions in both male and female promiscuous meadow and monogamous prairie voles are shown in Table 1. Selected CRFR1 meadow and prairie vole species comparisons are shown in Figure 2.
Fig. 2.
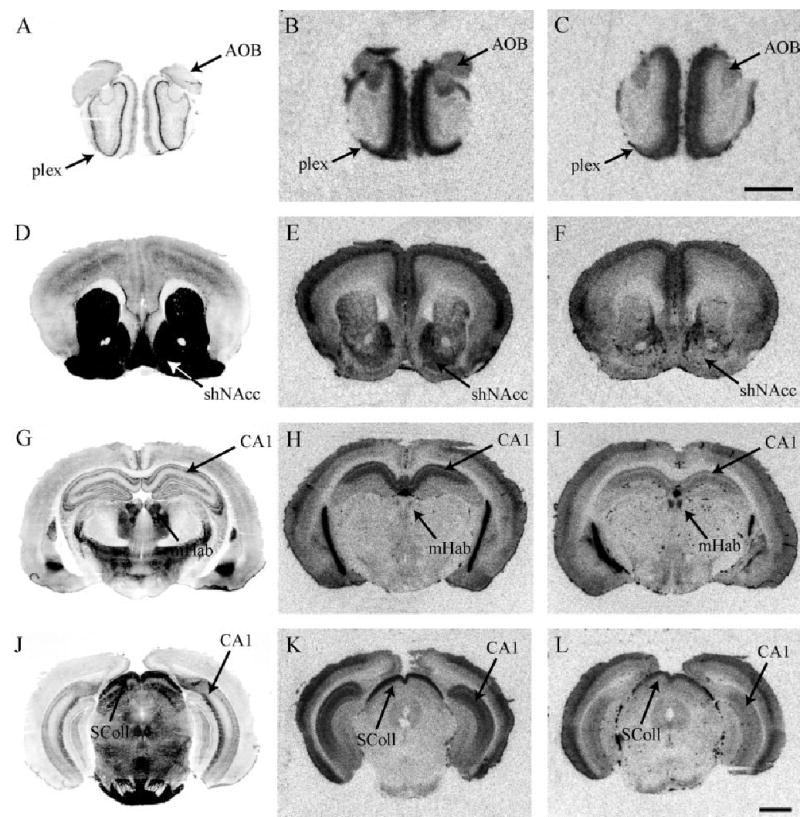
Total CRF receptor binding, highlighting differences in CRFR1 between promiscuous meadow and monogamous prairie vole species. Meadow vole (B) and prairie vole (C) brain sections through the olfactory bulb. Meadow vole (E) and prairie vole (F) brain sections through the nucleus accumbens. Note the strikingly dense CRFR1 binding in the shell of the NAcc in the meadow vole. Meadow vole (H) and prairie vole (I) brain sections showing the species difference in CRFR1 binding in the medial habenula nucleus. Note that the species difference in CRFR1 in the medial habenula is in the opposite direction as the species difference in the shell of the NAcc. Meadow vole (K) and prairie vole (L) hindbrain section highlighting the superior colliculus. Acetylcholinesterase-stained adjacent sections are shown to the left of each panel. Scale bars = 1 mm in C (applies to A,B,C), L (applies to D–L).
There were no significant species or sex differences in CRFR1 binding in the cortex, septal-hippocampal system, or extended amygdala. In cortical regions, prairie and meadow voles expressed high levels of CRFR1, as has been reported for rat, mouse, and nonhuman primate (Aguilera et al., 1987; De Souza et al., 1985; Potter et al., 1994; Primus et al., 1997; Sanchez et al., 1999; Van Pett et al., 2000). Prairie and meadow voles demonstrated only moderate CRFR1 binding in the septo-hippocampal system and a minimal level of binding in the BNST and amygdalar subregions. This is in contrast to other rodent and primate species which express higher levels of the receptor in these regions (Aguilera et al., 1987; De Souza et al., 1985; Potter et al., 1994; Primus et al., 1997; Sanchez et al., 1999; Van Pett et al., 2000).
Striatum and habenula
A significant species by brain region interaction (F = 25.1, P < 0.05) was found in the striatal and habenular regions, such that meadow voles showed significantly higher CRFR1 binding in the spNAcc, CPUv, and shNAcc than prairie voles (P < 0.01 for all regions) (Fig. 2E,F) (P < 0.01). In contrast, prairie voles showed approximately 3-fold higher density of CRFR1 binding in the mHab than meadow voles (P < 0.01) (Fig. 2H,I).
Olfactory and hindbrain
Both prairie and meadow voles showed CRFR1 labeling in these olfactory and hindbrain regions. A significant species by brain region interaction (F = 10.5, P < 0.01) followed by tests for simple effects revealed that prairie voles showed less labeling in both the OB (Fig. 2B,C) and SColl (Fig. 2K,L) as compared to meadow voles (P < 0.05).
While it may appear that prairie voles might have lower overall R1 binding or lower R1 affinity for [125I-Tyr0]-sauvagine than meadow voles, there were several brain regions where prairie voles have higher R1 binding density than meadow voles, such as in the medial habenula. Therefore, we doubt the differences that we observe are due to lower overall binding or affinity.
Distribution of CRFR2 in prairie and meadow voles
Mean values for CRFR2 binding (DPM/mg) from several brain regions in both male and female meadow and prairie voles are shown in Table 2. Selected CRFR2 species comparisons are shown in Figure 3.
TABLE 2.
Comparison of Prairie and Meadow Voles: CRFR2 Binding Using 125-I-Sauvagine and CP-154,5261
| Region | Abbreviation | Meadow Female Mean ± SEM | Meadow Male Mean ± SEM | Prairie Female Mean ± SEM | Prairie Male Mean ± SEM |
|---|---|---|---|---|---|
| Cortical | |||||
| Prefrontal | PFCtx | 3117 ± 83 | 3180 ± 112 | 2957 ± 78 | 2862 ± 123 |
| Insular | IFCtx | 2937 ± 79 | 2906 ± 74 | 2906 ± 57 | 2884 ± 121 |
| Orbitofrontal | OFCtx | 2845 ± 125 | 2805 ± 79 | 2809 ± 30 | 2828 ± 77 |
| Cingulate rostral | CgCtx1 | 3331 ± 150 | 3195 ± 153 | 3000 ± 67 | 2924 ± 66 |
| Cingulate middle | CgCtx2 | 3066 ± 121 | 2905 ± 206 | 2842 ± 81 | 2955 ± 81 |
| Cingulate caudal | CgCtx3 | 3007 ± 34 | 2918 ± 153 | 2832 ± 60 | 2858 ± 110 |
| Retrosplanial | RSCtx | 2846 ± 170 | 3305 ± 321 | 2660 ± 89 | 2768 ± 186 |
| Septal-hippocampal | |||||
| Lateral septum | LS | ||||
| dorsal rostral | rLSD | 5974 ± 475 | 6116 ± 572 | 4449 ± 298 | 4211 ± 579 |
| intermediata rostral | rLSl | 8306 ± 912 | 6662 ± 552 | 5830 ± 649 | 5583 ± 1138 |
| dorsal caudal | cLSD** | 6896 ± 604 | 6776 ± 602 | 4444 ± 182 | 4274 ± 485 |
| intermediata caudal | cLSI | 12374 ± 1601 | 9357 ± 747 | 7888 ± 1213 | 7635 ± 2000 |
| Hippocampus | HC | ||||
| CA1 | CA1** | 8534 ± 630 | 10404 ± 808 | 3598 ± 156 | 3978 ± 268 |
| CA2 | CA2** | 3727 ± 186 | 3763 ± 300 | 2830 ± 53 | 2832 ± 246 |
| CA3 | CA3** | 3332 ± 136 | 3339 ± 255 | 2656 ± 61 | 2735 ± 159 |
| dentate gyrus | DG | 4826 ± 727 | 4571 ± 494 | 2924 ± 88 | 2951 ± 166 |
| Extended amygdala | |||||
| Bed nucleus of the stria terminals | BnST | ||||
| medial | mBnST | 2638 ± 96 | 2650 ± 122 | 2437 ± 38 | 2559 ± 73 |
| lateral | lBnST | 2503 ± 45 | 2503 ± 98 | 2442 ± 36 | 2501 ± 84 |
| caudal | cBnST† | 4154 ± 302 | 14671 ± 861 | 2563 ± 109 | 6214 ± 541 |
| Amygdala | Amyg | ||||
| central | CeA | 2412 ± 78 | 2504 ± 105 | 2410 ± 91 | 2514 ± 84 |
| basolateral rostral | rBLA | 2846 ± 84 | 2916 ± 118 | 3379 ± 243 | 3050 ± 161 |
| medial rostral | rMeA | 2613 ± 40 | 2724 ± 121 | 2960 ± 171 | 3091 ± 197 |
| cortical rostral | rCoA | 2988 ± 107 | 3117 ± 122 | 2797 ± 129 | 2839 ± 68 |
| basolateral caudal | cBLA | 2817 ± 61 | 2679 ± 91 | 3154 ± 248 | 2982 ± 159 |
| medial caudal | cMeA | 2722 ± 105 | 3015 ± 197 | 3150 ± 155 | 3250 ± 257 |
| cortical caudal | cCoA | 3144 ± 104 | 3240 ± 194 | 2630 ± 57 | 2746 ± 133 |
| Striatum and habenula | |||||
| Nucleus accumbens | NAcc | ||||
| shell | shNAcc | 2823 ± 60 | 2804 ± 176 | 2922 ± 132 | 2822 ± 90 |
| septal pole | spNAcc** | 3258 ± 210 | 2969 ± 228 | 6981 ± 641 | 6185 ± 703 |
| Caudate-putamen | CPu | ||||
| dorsal | CPuD | 2503 ± 62 | 2381 ± 111 | 2594 ± 153 | 2700 ± 136 |
| ventral | CPuV** | 2812 ± 51 | 2572 ± 145 | 4738 ± 520 | 4740 ± 421 |
| Habenula | |||||
| medial | mHab | 2704 ± 20 | 2818 ± 131 | 2716 ± 47 | 2611 ± 131 |
| lateral | lHab | 2645 ± 40 | 2683 ± 145 | 2469 ± 67 | 2477 ± 97 |
| Thalamus | |||||
| laterodorsal | ldThal | ND | ND | ND | ND |
| Olfactory and hindbrain | |||||
| Olfactory bulb | |||||
| external plexiform | OB | 2909 ± 97 | 3040 ± 45 | 3060 ± 30 | 3053 ± 191 |
| Superior colliculus | SColl | 2938 ± 129 | 3116 ± 174 | 2725 ± 77 | 2799 ± 149 |
| Periaqueductal gray | PAG | 2482 ± 51 | 2548 ± 80 | 2407 ± 69 | 2408 ± 166 |
| Dorsal raphe | DR | 7047 ± 740 | 8852 ± 981 | 8694 ± 1672 | 7250 ± 1372 |
| Locus coeruleus | LC | 2843 ± 138 | 2591 ± 100 | 2989 ± 189 | 2591 ± 181 |
| Pontine dorsal tegmental nucleus | PDTg | 2451 ± 32 | 2473 ± 106 | 3154 ± 203 | 2950 ± 223 |
| Principal motor nucleus of 5 | Pr5 | 3791 ± 160 | 3499 ± 237 | 4959 ± 470 | 5433 ± 952 |
| Cerebellum | |||||
| granular layer | CBg | 2689 ± 78 | 2751 ± 147 | 2807 ± 125 | 2631 ± 127 |
Values represent mean DPM/mg tissue from n = 4 per group.
ND, not detected (<2 S.D. nonspecific binding);
P < 0.05 species effect;
P < 0.01 species effect;
P < 0.05 sex effect.
Fig. 3.
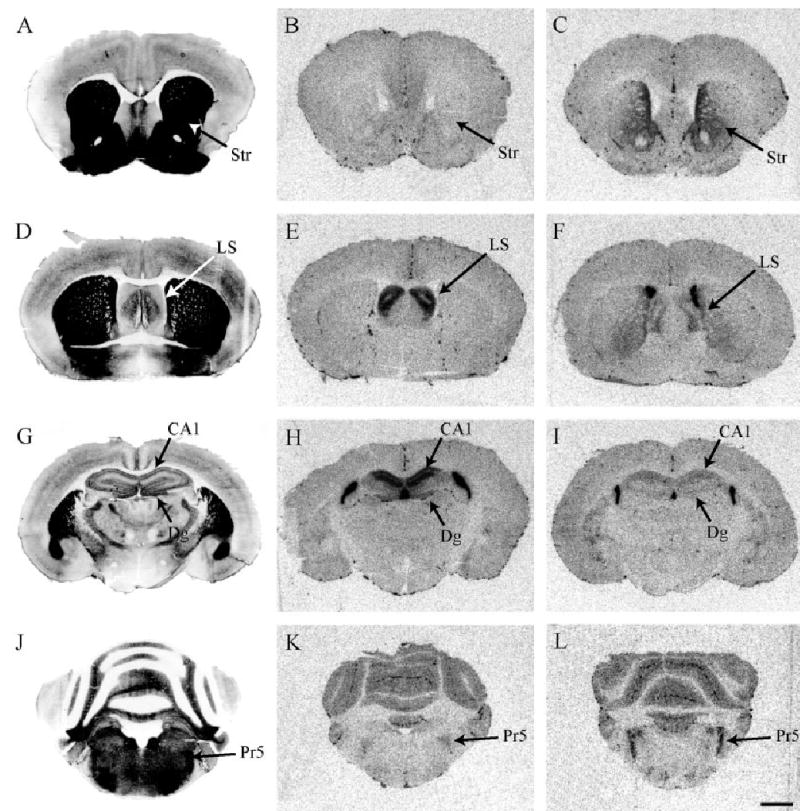
CRFR2 binding in promiscuous meadow and monogamous prairie vole species. Meadow vole (B) and prairie vole (C) sections through the striatum. Meadow vole (E) and prairie vole (F) brain sections through the lateral septum. Note that the species difference in CRFR2 in the lateral septum is in the opposite direction as the species difference in the striatum. Meadow vole (H) and prairie vole (I) brain sections through the hippocampus. Note the dense labeling in the CA1 fields. Meadow vole (K) and prairie vole (L) hindbrain sections through the principal nucleus of five. Acetylcholinesterase-stained adjacent sections are shown to the left of each panel. Scale bar = 1 mm in L (applies to A–L).
No species or sex differences were found in the cortex, extended amygdala, olfactory, and hindbrain nuclei. There was a nonsignificant trend for higher binding in the principal motor nucleus of 5 in prairie versus meadow voles (Fig. 3K,L). Among the hindbrain nuclei, the dorsal raphe had the highest levels of binding, while in cortical, extended amygdalar, olfactory, and other hindbrain nuclei, CRFR2 binding was minimal in both species. These findings are generally consistent with other rodent and nonhuman primate studies, although these species tend to show much higher CRFR2 binding in the extended amygdala (Potter et al., 1994; Sanchez et al., 1999; Van Pett et al., 2000).
Septal-hippocampal regions
As reported for other rodent species (but not in primate) (Potter et al., 1994; Van Pett et al., 2001; Sanchez et al., 1999), dense CRFR2 binding was observed in the lateral septum in meadow voles. However, unlike rat and mouse, meadow voles also showed dense binding in the hippocampal fields. Meadow voles showed significantly higher CRFR2 binding in the dorsal caudal lateral septum (cLSD) than prairie voles (P < 0.05 following a significant species by brain region interaction (F = 9.6, P < 0.01) (Fig. 3E,F). In addition, meadow voles showed significantly higher CRFR2 binding than prairie voles in all the hippocampal fields, with the most dramatic species differences in the CA1, and to a lesser extent the CA2 and CA3 fields (P < 0.001, P < 0.01, P < 0.01, respectively) (Fig. 3H,I).
Striatum and habenula
Unlike other rodent and primate species, monogamous prairie voles very strongly express CRFR2 binding in the striatum. A significant species by brain regions interaction (F = 45.8, P < 0.001) indicated that prairie voles had significantly higher CRFR2 binding than meadow voles in the spNAcc, and CPUv (P < 0.01 in all cases) (Fig. 3B,C).
It is interesting again to note that there are several regions where prairie voles exceed meadow voles, and vice versa. Therefore, it is unlikely that global species differences in CRFR2 affinity exist. In fact, no sex or species differences exist in the nonspecific binding values obtained when [125I-Tyr0]-sauvagine is incubated with the cold ligand (as seen previously in Fig. 1).
Distribution of CRFR1 in pine and montane voles
Mean values (DPM/mg) for CRFR1 from several brain regions in both male and female promiscuous montane and monogamous pine voles are shown in Table 3. Selected CRFR1 montane and pine vole species comparisons are shown in Figure 4.
TABLE 3.
Comparison of Pine and Montane Voles: CRFR1 Binding Using 125-I-Sauvagine and CRFR2 Subtraction
| Region | Abbreviation | Meadow Female Mean ± SEM | Meadow Male Mean ± SEM | Prairie Female Mean ± SEM | Prairie Male Mean ± SEM |
|---|---|---|---|---|---|
| Cortical | |||||
| Prefrontal | PFCtx** | 1944 ± 345 | 1951 ± 124 | 777 ± 364 | 974 ± 184 |
| Insular | IFCtx** | 2839 ± 572 | 2904 ± 252 | 1246 ± 362 | 1165 ± 77 |
| Orbitofrontal | OFCtx* | 2873 ± 324 | 2813 ± 218 | 1261 ± 519 | 1757 ± 372 |
| Cingulate rostral | CgCtx1** | 1943 ± 119 | 1838 ± 225 | 980 ± 217 | 1145 ± 162 |
| Cingulate middle | CgCtx2** | 1553 ± 122 | 1700 ± 132 | 837 ± 307 | 959 ± 53 |
| Cingulate caudal | CgCtx3 | 1395 ± 59 | 1733 ± 227 | 711 ± 297 | 1137 ± 303 |
| Retrosplanial | RSCtx** | 1681 ± 339 | 1213 ± 159 | ND | 499 ± 151 |
| Septal-hippocampal | |||||
| Lateral septum | LS | ||||
| dorsal rostral | rLSD | 3445 ± 895 | 4009 ± 861 | 3149 ± 2178 | 1949 ± 764 |
| intermediata rostral | rLSI | 3845 ± 1191 | 5609 ± 1001 | 5427 ± 4015 | 4297 ± 1540 |
| dorsal caudal | cLSD | 4821 ± 960 | 4470 ± 456 | 8864 ± 5168 | 7790 ± 2827 |
| intermediata caudal | cLSI | 6817 ± 1208 | 6657 ± 338 | 4054 ± 2508 | 8395 ± 3563 |
| Hippocampus | HC | ||||
| CA1 | CA1 | 651 ± 65 | 527 ± 44 | ND | 573 ± 50 |
| CA2 | CA2 | 549 ± 120 | 533 ± 69 | ND | 465 ± 45 |
| CA3 | CA3 | 657 ± 104 | 766 ± 102 | ND | 538 ± 85 |
| dentate gyrus | DG | 547 ± 144 | 775 ± 27 | ND | ND |
| Extended amygdala | |||||
| Bed nucleus of the stria terminals | BnST | ||||
| medial | mBnST | 835 ± 185 | 907 ± 331 | 836 ± 326 | 1247 ± 601 |
| lateral | lBnST | 1582 ± 378 | ND | ND | 453 ± 199 |
| caudal | cBnST | 3863 ± 2189 | 2206 ± 556 | 3082 ± 1761 | 4059 ± 507 |
| Amygdala | Amyg | ||||
| central | CeA | 691 ± 135 | 740 ± 160 | 672 ± 179 | 980 ± 497 |
| basolateral rostral | rBLA | 589 ± 456 | 1340 ± 679 | 492 ± 86 | 813 ± 475 |
| medial rostral | rMeA | 1848 ± 738 | 1281 ± 318 | 952 ± 679 | ND |
| cortical rostral | rCoA | 1944 ± 279 | 2071 ± 380 | 970 ± 258 | 947 ± 178 |
| basolateral caudal | cBLA | 1221 ± 171 | 1430 ± 175 | ND | 498 ± 169 |
| medial caudal | cMeA | 2115 ± 544 | 1375 ± 43 | 858 ± 537 | 681 ± 208 |
| cortical caudal | cCoA | 1834 ± 169 | 2425 ± 296 | 799 ± 195 | 1148 ± 260 |
| Striatum and habenula | |||||
| Nucleus accumbens | NAcc | ||||
| shell | shNAcc** | 4335 ± 608 | 4598 ± 719 | 977 ± 106 | 1043 ± 83 |
| septal pole | spNAcc | 1512 ± 348 | 1956 ± 198 | 2344 ± 944 | 3307 ± 729 |
| Caudate-putamen | CPu | ||||
| dorsal | CPuD* | 2032 ± 468 | 2000 ± 435 | 967 ± 233 | 955 ± 181 |
| ventral | CPuV | 1353 ± 247 | 1612 ± 195 | 1137 ± 293 | 1247 ± 259 |
| Habenula | |||||
| medial | mHab | 781 ± 84 | 658 ± 75 | ND | 540 ± 163 |
| lateral | lHab | 730 ± 177 | 584 ± 124 | 883 ± 388 | 458 ± 170 |
| Thalamus | |||||
| laterodorsal | ldThal** | 3380 ± 552 | 3384 ± 802 | ND | 563 ± 127 |
| Olfactory and hindbrain | |||||
| Olfactory bulb | |||||
| external plexiform | OB* | 11849 ± 1897 | 9461 ± 3408 | 5080 ± 1477 | 5787 ± 2041 |
| Superior colliculus | SColl** | 8202 ± 1908 | 7970 ± 945 | 1522 ± 279 | 1982 ± 319 |
| Periaqueductal gray | PAG | 752 ± 47 | 888 ± 50 | 599 ± 174 | ND |
| Dorsal raphe | DR | 3891 ± 1250 | 2991 ± 1109 | 1728 ± 1311 | 10495 ± 2245 |
| Locus coeruleus | LC* | 1365 ± 205 | 1226 ± 112 | 540 ± 389 | 484 ± 130 |
| Pontine dorsal tegmental n. | PDTg* | 796 ± 140 | 971 ± 170 | ND | 452 ± 188 |
| Principal motor nucleus of 5 | Pr5 | 1834 ± 276 | 1951 ± 630 | 2826 ± 879 | 1969 ± 742 |
| Cerebellum | |||||
| granular layer | CBg | 2222 ± 424 | 3399 ± 279 | 2396 ± 323 | 1966 ± 565 |
Values represent mean DPM/mg tissue from n = 4 per group.
ND, not detected (<2 S.D. nonspecific binding);
P < 0.05 species effect;
P < 0.01 species effect;
P < 0.05 sex effect.
Fig. 4.
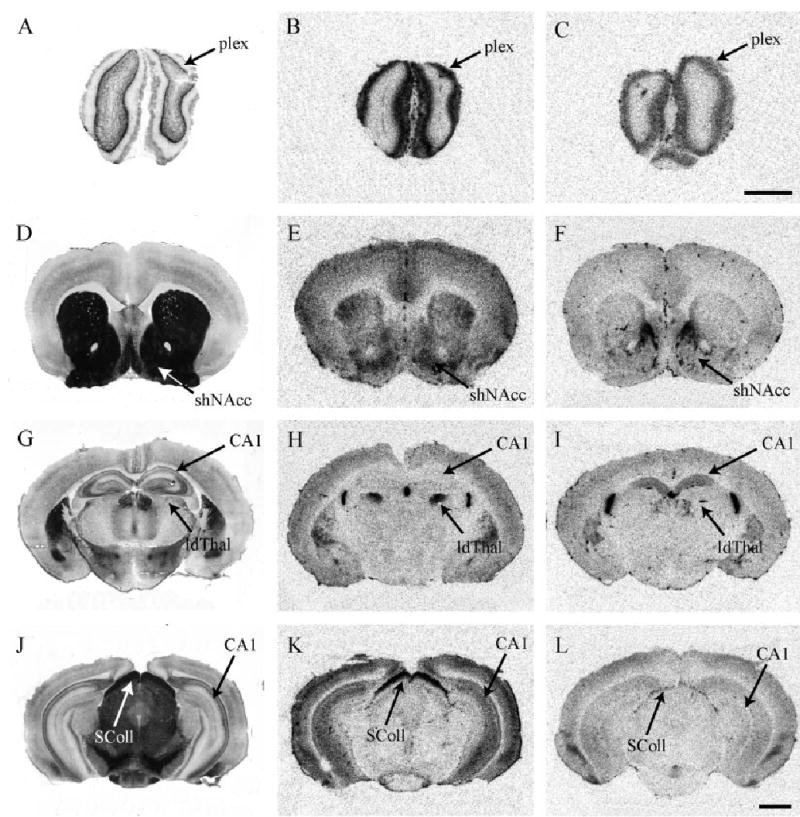
Total CRF receptor binding, highlighting differences in CRFR1 between promiscuous montane and monogamous pine vole species. Montane vole (B) and pine vole (C) brain sections through the olfactory bulb. Montane vole (E) and pine vole (F) brain sections through the nucleus accumbens. Note the strikingly dense CRFR1 binding in the shell of the NAcc in the montane vole. Montane vole (H) and pine vole (I) brain sections showing the species difference in CRFR1 binding in the laterodorsal thalamus. Montane vole (K) and pine vole (L) hindbrain section highlighting the superior colliculus. Acetylcholinesterase-stained adjacent sections are shown to the left of each panel. Scale bar = 1 mm in C (applies to A,B,C), L (applies to D–L).
There were no differences between pine and montane voles in CRFR1 binding in the septal-hippocampal system or extended amygdala. CRFR1 binding in these regions was comparable to prairie and meadow voles.
Cortical regions
Like meadow and prairie voles, montane and pine voles also showed moderate to dense CRFR1 binding throughout the cortex. However, in contrast to the meadow/prairie comparison, an overall significant species by brain region interaction (F = 3.4, P < 0.005) followed by simple effects tests indicated that montane voles demonstrated greater binding in the prefrontal cortex (PFCtx), insular cortex (IFCtx), orbitofrontal cortex (OFCtx), cingulate cortices (CgCtx1 and CgCtx2), and the retrosplenial cortex (RSCtx) (P < 0.05).
Striatum and habenula
The striatal and habenular regions demonstrated a significant species by brain region interaction (F = 18.3, P < 0.001). Simple effects tests revealed that montane voles demonstrated greater CRFR1 binding in the shNAcc and CPu relative to monogamous pine voles (Fig. 4E,F). There were striking species differences in the laterodorsal thalamus (ldThal), a novel brain region which did not show CRFR1 binding in meadow or prairie voles. Montane voles had very dense CRFR1 binding in this region, while pine voles were virtually devoid of binding (P < 0.001) (Fig. 4H,I).
Olfactory and hindbrain
Montane voles had significantly higher CRFR1 binding in the olfactory bulb and the superior colliculus relative to pine voles (F = 3.0, P < 0.05, overall brain by species interaction) (Fig. 4B,C and 4K,L, respectively). In addition, montane voles also had higher CRFR1 binding in the locus coeruleus and pontine dorsal tegmental nucleus (P < 0.05). There were no species differences observed in the other hindbrain regions, including the PAG, DR, Pr5, and CBg.
Distribution of CRFR2 in montane and pine voles
Mean values (DPM/mg) for CRFR2 from several brain regions in both male and female montane and pine voles are shown in Table 4. Selected CRFR2 species comparisons are shown in Figure 5. Data will be discussed primarily in relation to the species differences found between prairie and meadow voles.
Fig. 5.
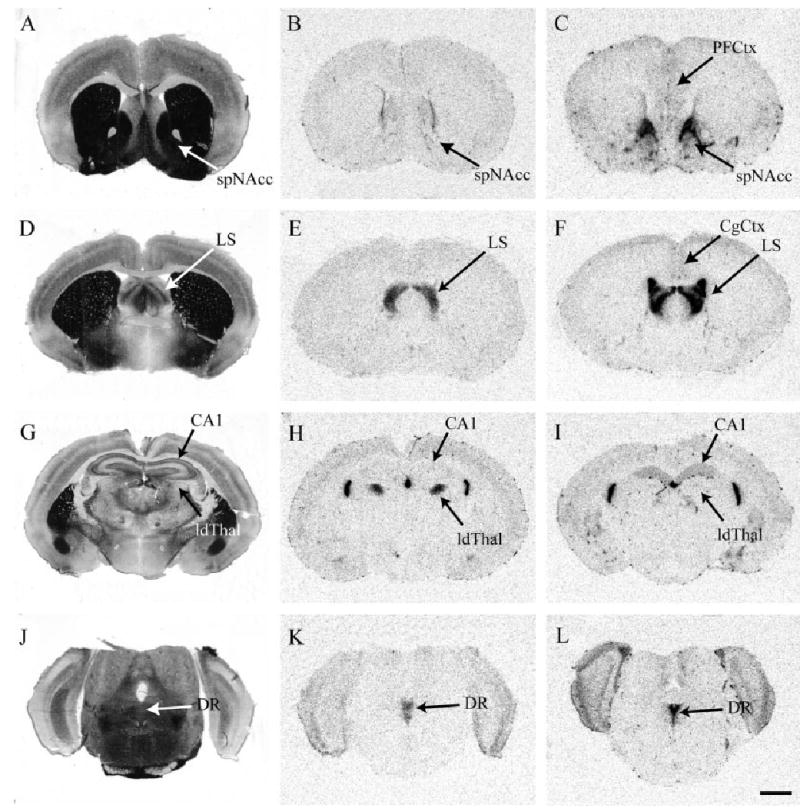
CRFR2 binding in promiscuous montane and monogamous pine vole species. Montane vole (B) and pine vole (C) sections through the striatum. Note that the species difference in CRFR2 in the septal pole of the NAcc is in the same direction as the meadow versus prairie comparison. Montane vole (E) and pine vole (F) brain sections through the lateral septum. Note that the species difference in CRFR2 in the lateral septum is in the opposite direction as the meadow versus prairie comparison. Montane vole (H) and pine vole (I) brain sections through the hippocampus. Note the species differences in the CA1 fields and the laterodorsal thalamus. Montane vole (K) and pine vole (L) hindbrain sections through the dorsal raphe nucleus. Acetylcholinesterase-stained adjacent sections are shown to the left of each panel. Scale bar = 1 mm in L (applies to A–L).
Cortical regions
Consistent with cortical regions in meadow and prairie voles, minimal to moderate CRFR2 binding was observed in montane and pine voles. However, unlike meadow and prairie voles, who showed no significant species differences in cortex, pine voles showed significantly higher CRFR2 than montane voles in the IFCtx, CgCtx1, CgCtx2, and CgCtx3 (F = 6.7, P < 0.05) (Fig. 5B,C, 5E,F).
Septal-hippocampal regions
Like meadow and prairie voles, there were also significant differences in CRFR2 in the lateral septum and hippocampus between montane and pine voles. However, whereas meadow voles had higher CRFR2 in these regions than prairie voles, the opposite was observed for pine and montane voles. Pine voles had nearly a 4-fold increase in lateral septum CRFR2 binding than montane voles, as well as significantly more CRFR2 in the CA1 and CA3 fields of the hippocampus (F = 22.7, P < 0.001) (Fig. 5E,F and 5H,I, respectively).
Extended amygdala
Similar to that observed in the meadow and prairie vole comparison, montane and pine voles expressed weak and diffuse CRFR2 binding through-out the bed nucleus of the stria terminalis and amygdala nuclei. No significant species differences were observed in the mBnST, lBnST, cBnST, or any of the subnuclei of the amygdala (P > 0.05).
Striatum and habenula
Montane and pine voles showed a significant species by brain region interaction in these subregions (F = 25.3, P < 0.001). Pine voles had significantly higher CRFR2 binding in the septal pole of the nucleus accumbens (spNAcc) and caudate-putamen than montane voles (Fig. 5B,C). Like meadow and prairie voles, there were no significant species differences in CRFR2 binding in the shNAcc or mHab (P > 0.05), although the lHab was marginally significantly different (P < 0.05). CRFR2 levels in the laterodorsal thalamus (ldThal) were significantly higher in montane voles compared to pine voles (P < 0.001) (Fig. 5H,I).
Olfactory and hindbrain
Unlike the meadow and prairie comparison, there were some olfactory and hind-brain regions that significantly differed between montane and pine voles (F = 15.3, P < 0.001). CRFR2 levels in the olfactory bulb were marginally significantly elevated in pine voles compared to montane voles (P < 0.05). CRFR2 levels in the dorsal raphe and cerebellum were also significantly elevated in pine voles compared to montane voles (P < 0.01) (Fig. 5K,L). There were no differences observed in the other hindbrain regions analyzed.
Like the meadow and prairie vole comparison, there are several regions for both CRFR1 and CRFR2 where montane voles exceed pine voles and vice versa. Therefore, it is unlikely that global species differences in CRF receptor affinity for the ligand exist between pine and montane voles.
Sex differences in CRFR1 and CRFR2 distribution
No sex differences have been previously reported in CRFR1 or CRFR2 in any other species mapping studies (Aguilera et al., 1987; De Souza et al., 1985; Potter et al., 1994; Primus et al., 1997; Sanchez et al., 1999; Van Pett et al., 2000). However, in vole species, large sex differences in CRFR2 binding were detected in the medial caudal subdivision of the BnST (cBnST), also known as the encapsulated or posteromedial subdivision, which expresses CRFR2 in both rats and mice (Van Pett et al., 2000). In the prairie and meadow vole comparison, males of both species displayed significantly higher CRFR2 binding than females of both species, approximating a 2–3-fold increase (P < 0.01, post-hoc Bonferroni adjusted t-test) (Fig. 6). In the pine and montane vole comparison, males of both species also showed significantly higher CRFR2 binding than females of both species, although to a somewhat lesser extent than in the prairie and meadow comparison (P < 0.05) (Fig. 7).
Fig. 6.

Sex differences in CRFR2 binding in the BnST in promiscuous meadow and monogamous prairie voles. A: Rat brain schematic of the medial caudal, or encapsulated, BnST and fornix (Paxinos and Watson, 1998). B: Meadow male. C: Prairie male. D: Acetylcholinesterase staining of an adjacent brain section through the BnST. E: Meadow female. F: Prairie female. Note the CRFR2 binding in the choroid plexus (ChP) is roughly equivalent across sex and species. Scale bar = 1 mm in F (applies to A–F).
Fig. 7.

Sex differences in CRFR2 binding in the BnST in promiscuous montane and monogamous pine voles. A: Rat brain schematic of the medial caudal, or encapsulated, BnST and fornix (Paxinos and Watson, 1998). (B) Montane male. (C) Pine male. (D) Acetylcholinesterase staining of an adjacent brain section through the BnST. (E) Montane female. (F) Pine female. Note the CRFR2 binding in the choroid plexus (ChP) is roughly equivalent across sex and species. Scale bar = 1 mm in F (applies to A–F).
Whole brain CRFR1 and CRFR2 binding did not significantly differ between the sexes, suggesting that sex differences in receptor expression were in fact specific to particular brain regions.
DISCUSSION
Given the remarkable species differences in social structure between the monogamous prairie and pine vole and promiscuous meadow and montane vole, species differences likely exist in the brain that might explain the proximate mechanisms of behavior. Past literature has focused predominantly on the neuropeptide systems oxytocin and vasopressin to explain the species differences in social behavior; however, recent studies have shown that corticosterone and CRF modulate partner preference in prairie voles as well (DeVries et al., 1995, 1996, 2002). Our results show that monogamous versus promiscuous vole species pairs in fact have dramatically different CRFR1 and CRFR2 distributions in the brain. These data support the hypothesis that CRF plays a role in the monogamous-typical behaviors of the prairie vole, insofar as central CRF release would potentially activate different neural circuits, depending on where CRFR1 and CRFR2 are expressed in the brain. For example, it is possible that CRFR1 and CRFR2 patterns may fall within specific neual circuits that elicit affiliative, monogamous social behavior in prairie and pine voles, whereas CRFR1 and CRFR2 may be absent in these neural circuits in promiscuous species. Identifying these differences may generate further hypotheses as to which brain regions are specifically involved in CRF-induced pair bond formation.
Vole species differences in CRFR1 brain distribution
Differences in CRFR1 distribution between meadow and prairie voles were observed throughout several brain regions. Most notably, large differences appeared in limbic areas such as the shell of the nucleus accumbens (shNAcc) and the medial habenula, and more subtle differences appeared in the olfactory bulb (OB) and the superior colliculus (SColl). Differences in CRFR1 distribution between montane and pine voles were also observed throughout the brain, including cortical regions, the striatum (including shNAcc), and similarly subtle differences in the OB and SColl. These global patterns of CRFR1 in the shNAcc, OB, and SColl, which are increased in promiscuous meadow and montane voles compared to monogamous prairie and pine voles, suggest that these patterns could underlie a neural circuit contributing to monogamous social behavior.
Vole species differences in CRFR2 brain distribution
We observed dramatic species differences between meadow and prairie voles in CRFR2 distribution in several brain regions, including the striatum (caudate and NAcc), the lateral septum, and CA1 field of the hippocampus. However, not all these species differences were replicated in our second comparison between montane and pine voles. In fact, some of the species differences went in the opposite direction from predicted. For example, in both the lateral septum and hippocampus, meadow voles had more CRFR2 than prairie voles, yet pine voles had more CRFR2 than montane voles in these regions. But in the striatum, and especially in the septal pole of the NAcc, both monogamous vole species had significantly higher CRFR2 than the two promiscuous vole species. The significance of CRFR2 in the NAcc in pair bond formation in monogamous species remains to be determined. It is interesting to note that the NAcc has already been implicated in regulating partner preference formation in prairie voles: Activation of oxytocin receptors and dopamine D2 receptors in the NAcc are both necessary and sufficient for pair bond formation in prairie voles (Aragona et al., 2003; Gingrich et al., 2000; Liu and Wang, 2003; Young et al., 2001). It is possible that CRFR2 in the NAcc may also interact with existing oxytocin and dopaminergic systems to facilitate pair bond formation.
Vole sex differences in the encapsulated BnST
Sex differences in CRFR2 binding in the medial caudal bed nucleus of the stria terminalis (cBnST), or encapsulated subdivision, were observed in all four vole species. Males of all species had significantly higher CRFR2 binding in the cBnST than females of all species. This is a particularly interesting finding in light of the fact that no other experiments in rats, mice, or nonhuman primates have reported sex differences in CRFR1 or CRFR2 in any brain region (Aguilera et al., 1987; De Souza et al., 1985; Potter et al., 1994; Primus et al., 1997; Sanchez et al., 1999; Van Pett et al., 2000). The cBnST is a prominent brain region that exhibits sex differences in gonadal steroid systems such as estrogen and androgen receptors and aromatase expression in rat (Herbison, 1995; Herbison and Fenelon, 1995; Wagner and Morrell, 1997). A recent study suggested that CRFR2 may also play a role in behavioral sex differences: Female CRFR2 knockout mice showed opposite behavioral responses to swim stress compared to male CRFR2 knockout mice (Bale and Vale, 2003). Interestingly, past studies in prairie voles have shown that the effects of exogenous corticosterone on pair bond formation are also sexually dichotomous, analogous to the findings in CRFR2 knockout mice (DeVries et al., 1996). Since we observed sex differences in BnST CRFR2 binding in voles, it is possible that these brain differences might underlie the sexual dichotomy in pair bond formation in prairie voles. For example, canonical HPA axis feedback could potentially modulate central CRF release onto CRFR2 in the cBnST, which could then lead to inhibition or facilitation of partner preference, depending on CRFR2 density in the cBnST. The BnST has been implicated in depressive-like behaviors, and there is also a prominent gender difference in the epidemiology of depression in humans (Erb et al., 2001; Stout et al., 2000; Young, 1998). Perhaps vole species could represent a novel animal model in which sex differences in social behavior could be further studied.
CRF and social behavior
Despite the abundance of studies on CRF and stress/anxiety, relatively few studies in the literature have examined the role of the CRF system in social behavior. One study examining the behavioral effects of chronic CRF infusion on rhesus monkeys housed either singly or in groups found increased depressive-like behaviors only in the socially housed monkeys (Strome et al., 2002). Another recent study found lower concentrations of CRF in the cerebrospinal fluid of bonnet macaques, which are gregarious and affiliative, than pigtail macaques, which are more socially distant (Rosenblum et al., 2002).
How could CRF be acting to modulate social behavior? In a way, social behavior overlaps with stress and anxiety, especially in behaviors involving social support or coping with social isolation. Previous research in prairie voles has shown that plasma corticosterone levels elevate during social separation from the partner, and reunion with the familiar partner is followed by a return to baseline (Carter et al., 1997). In naïve prairie voles, both male and female prairie voles experience a reduction in corticosterone levels within an hour of being paired with an animal of the opposite sex, although males tend to show a greater reduction than females (DeVries et al., 1997). A recent preliminary study using prairie voles showed that recently separated pair-bonded prairie voles exhibited more “depressive-like” symptoms in the forced swim test than their sibling-separated counterparts (Bosch et al., 2004). These data suggest that CRF systems do have a role in modulating social behavior, and this is especially apparent in an animal model whose affiliative social behavior is its defining feature.
Evolutionary implications
Because it is currently unknown whether vole species have similar CRFR1 and CRFR2 to those which have been identified in rat, mouse, monkey, and human, it is possible that there could be other species differences that we have not reported. For example, it is unknown what the affinity of the endogenous ligands for these putative CRFR1 and CRFR2 is in voles. Thus, our results should be interpreted accordingly. However, our results do suggest that voles do have CRFR1- and CRFR2-like receptors, since [125I-Tyr0]-sauvagine bound to the tissue in all four species, and cold sauvagine competed the [125I-Tyr0]-sauvagine completely off. Furthermore, there are some core areas which are highly conserved across all species examined, such as the choroid plexus, cortex, raphe, cerebellum, and olfactory bulb.
Given the assumption that vole species have similar CRFR1 and CRFR2 as characterized in other species, a broader question is: What is the significance of these species similarities and differences in elucidating CRF receptor function? When one compares CRF distribution patterns in the four vole species to those in rat and nonhuman primate, CRF patterns appear to segregate into three different classes: 1) those that are conserved across mammalian species, including rat, monkey, and the four vole species; 2) those that are phylogenetically plastic across the six species, including within closely related vole species; and 3) those that segregate based on social organization. This conceptual framework, summarized in Tables 5 and 6, might be used as a general heuristic in understanding the functional and evolutionary significance of different CRF systems.
TABLE 5.
Relative Comparison of CRFR1 Binding across Six Species: Semiquantitative Analysis
| Region | Meadow | Prairie | Montane | Pine | Rat1 | Monkey2 |
|---|---|---|---|---|---|---|
| Cortex | ++++ | ++++ | ++ | ++ | ++++ | ++++ |
| Cerebellum | +++ | +++ | ++ | ++ | +++ | ++++ |
| Olfactory bulb | +++++ | ++++ | +++++ | +++ | +++ | nr |
| Superior colliculus | +++++ | ++++ | ++++ | + | ++ | nr |
| Lateral septum | ++ | ++ | ++++ | ++++ | 0 | 0 |
| Hippocampus | 0 | 0 | 0 | 0 | ++ | ++++ |
| Amygdala | +/++ | + | + | 0/+ | +++ | ++++ |
| Periaqueductal gray | + | + | 0 | 0 | ++++ | nr |
| Habenula | ++ | ++++ | 0 | 0 | 0 | nr |
| Laterodorsal thalamus | 0 | 0 | ++ | 0 | ++ | nr |
| Locus coeruleus | + | + | + | 0 | 0 | +++ |
| Dorsal raphe | 0 | 0 | ++ | ++++ | ++ | 0 |
| Nucleus accumbens, shell | +++ | 0/+ | +++ | 0 | ++ | nr |
Rat data are compiled from meta-analysis of CRFR1 receptor binding reports from de Souza et al. (1985), de Souza et al. (1987), Primus et al. (1997), and Steckler and Holsboer (1999).
Monkey data are summarized from Sanchez et al. (1999) and Grigoriadis et al. (1995).
For vole data, averaged across sex: 0, <1000 dpm/mg; +, 1000–2000 dpm/mg; ++, 2000–4000 dpm/mg; +++, 4000–6000 dpm/mg; ++++, 6000–10000 dpm/mg; +++++, >10000 dpm/mg; nr, not reported.
TABLE 6.
Relative Comparison of CRFR2 Binding across Six Species: Semiquantitative Analysis
| Region | Meadow | Prairie | Montane | Pine | Rat1 | Monkey2 |
|---|---|---|---|---|---|---|
| Choroid plexus | +++++ | +++++ | +++++ | +++++ | +++++ | +++++ |
| Dorsal raphe | ++++ | ++++ | ++++ | +++++ | +++ | ++ |
| Olfactory bulb | ++ | ++ | + | ++ | +++ | nr |
| Cortex | ++ | ++ | + | + | + | + |
| Amygdala | ++ | ++ | + | + | +++ | +++ |
| Periaqueductal gray | ++ | ++ | 0/+ | + | + | nr |
| Locus coeruleus | ++ | ++ | 0/+ | + | 0 | 0 |
| Cerebellum | ++ | ++ | 0/+ | + | 0 | 0 |
| Lateral septum | ++++ | +++ | ++++ | +++++ | ++++ | + |
| Hippocampus | ++++ | ++ | + | + | + | +++ |
| Laterodorsal thalamus | 0 | 0 | ++++ | 0/+ | 0 | nr |
| Superior colliculus | ++ | ++ | + | + | 0 | nr |
| Nucleus accumbens, septal pole | ++ | ++++ | + | +++++ | 0 | nr |
Rat data are compiled from meta-analysis of CRFR2 receptor binding reports from de Souza et al. (1987), Primus et al. (1997), Rominger et al. (1998), and Steckler and Holsboer (1999).
Monkey data are summarized from Sanchez et al. (1999) and Grigoriadis et al. (1995).
For vole data, averaged across sex: 0, <1000 dpm/mg; +, 1000–2000 dpm/mg; ++, 2000–4000 dpm/mg; +++, 4000–6000 dpm/mg; ++++, 6000–10000 dpm/mg; +++++, > 10000 dpm/mg; nr, not reported.
CRFR1 brain distribution across six species
Both similarities and differences in CRFR1 distribution were observed throughout the brain across species (Table 5). Data are summarized from meta-analysis of past reports of CRFR1 receptor binding in rat and rhesus macaque brains (Aguilera et al., 1987; Chalmers et al., 1995; De Souza, 1987; De Souza et al., 1985; Grigoriadis et al., 1995; Primus et al., 1997; Rominger et al., 1998; Sanchez et al., 1999; Steckler and Holsboer, 1999). The cortex, olfactory bulb, cerebellum, and superior colliculus appeared the most highly conserved in terms of similar relative CRFR1 binding, and had strong to moderate binding across all six species. Therefore, cortical, olfactory, and cerebellar CRFR1 may represent “classic,” or typical functions of CRFR1 since the dense binding is so highly conserved across rodent and primate species.
CRFR1 binding patterns in other regions appear to distinguish voles from rats and nonhuman primates, and hence may reflect the phylogenetic relatedness of the four vole species. For example, the lateral septum had moderate to dense CRFR1 binding in all four vole species, but is devoid of CRFR1 binding in rat and monkey. The hippocampus, amygdala, and periaqueductal gray demonstrate strong to moderate CRFR1 binding in rat and monkey, but little to no binding in all four vole species. It is possible that CRFR1 receptor function in these regions could play an important role in producing behaviors specific to vole species.
Some brain regions differ between species in a less predictable pattern: The habenula, laterodorsal thalamus, locus coeruleus, and dorsal raphe all show remarkable plasticity across species, but with no obvious pattern. Perhaps the functional significance of CRF receptors in these brain regions could reflect adaptation to a particular ecological niche, such as enhanced spatial memory or specific stress response to predation.
Additional comparisons can be made between species with different social organizations. Since prairie and pine voles are the only species with monogamous social organization out of the six species examined, one would predict that brain regions involved in monogamous behavior would yield CRFR1 patterns common to just prairie and pine voles. For example, the olfactory bulb, superior colliculus, and shell of the NAcc fit this criterion, as they all have higher CRFR1 binding in promiscuous vole species. It is possible that the olfactory bulb, superior colliculus, and NAcc comprise a CRF-relevant neural circuit that could relate to nonmonogamous behaviors. In support of this hypothesis, nonmonogamous rats do show minimal to moderate CRFR1 expression in these regions as well, although it is difficult to draw a solid conclusion from this single observation.
CRFR2 brain distribution across six species
Semiquantitative similarities and differences for CRFR2 brain distribution across all four vole species, rat, and monkey are summarized in Table 6. Data were compiled from meta-analysis of prior reports of CRFR2 receptor binding in rat and rhesus macaque (Aguilera et al., 1987; Chalmers et al., 1995; De Souza, 1987; De Souza et al., 1985; Grigoriadis et al., 1995; Primus et al., 1997; Rominger et al., 1998; Sanchez et al., 1999; Steckler and Holsboer, 1999). CRFR2 binding in several brain regions was highly conserved across the six species, including the choroid plexus, dorsal raphe, olfactory bulb, and cortex. CRFR2 functions in these regions could potentially represent prototypical CRFR2 physiology since they are so highly conserved across these unrelated species.
Some brain regions are conserved only within the four vole species, such as the amygdala, periaqueductal gray, locus coeruleus, and cerebellum, which show minimal to moderate binding in voles, but are different in rat and monkey. Other brain regions are extraordinarily plastic in their CRFR2 expression across species, such as the lateral septum, hippocampus, laterodorsal thalamus, and superior colliculus. CRFR2 in the lateral septum and hippocampus are particularly interesting in this regard because of their roles in modulating basic learning and memory systems (Bakshi et al., 2002; Radulovic et al., 1999). One would predict that if CRFR2 in these regions were extremely critical for basic learning and memory, their patterns might appear more conserved across mammals. It is possible that such wide variation in CRFR2 binding in these regions reflects species differences in adaptation to a particular ecological niche, which could require different specializations for spatial memory or fear learning of predators, for example.
Only one brain region cleanly segregates with social organization. CRFR2 binding in the septal pole of the nucleus accumbens is much higher in monogamous vole species compared to promiscuous vole species. In support of this observation, CRFR2 binding is similarly absent in the nonmonogamous rat (but was not reported in nonmonogamous rhesus macaque) (Potter et al., 1994; Primus et al., 1997; Sanchez et al., 1999). The significance of CRFR2 in the NAcc in pair bond formation remains to be determined, but it would be interesting to compare CRFR2 binding in the septal pole of the NAcc in additional monogamous-promiscuous species pairs beyond just vole species.
What are the evolutionary implications for this plasticity in neuropeptide systems in such closely related vole species, as well as across other species? Vasopressin, oxytocin, and CRF receptors all show tremendous variation in brain distribution across species (Barberis and Tribollet, 1996; Goodson and Bass, 2001). However, classical neurotransmitter systems such as dopamine receptors and serotonin transporter distributions are highly conserved within voles and other rodents, as well as cholinergic systems as measured by acetylcholinesterase staining (unpubl. data, Lim and Young). It appears that neuropeptide systems, which are typically more modulatory in nature and control social behaviors, are evolutionarily more plastic than these classical neurotransmitter systems. This tendency for rapid change in neuropeptide receptor distribution can in turn produce large variation in social behavior, which can then be shaped by natural selection depending on environment. Thus, neuropeptide receptor plasticity could be a critical substrate for the rapid evolution of social behavior within a species.
Footnotes
Grant sponsor: National Institutes of Health; Grant number: MH65050 (to M.M.L.); Grant number: MH64692 (to L.J.Y.); Grant sponsor: National Science Foundation; Grant number: STC IBN-9876754; Grant sponsor: Yerkes Center; Grant number: RR00165.
References
- Aguilera G, Millan MA, Hauger RL, Catt KJ. Corticotropin-releasing factor receptors: distribution and regulation in brain, pituitary, and peripheral tissues. Ann N Y Acad Sci. 1987;512:48–66. doi: 10.1111/j.1749-6632.1987.tb24950.x. [DOI] [PubMed] [Google Scholar]
- Aragona BJ, Liu Y, Curtis JT, Stephan FK, Wang Z. A critical role for nucleus accumbens dopamine in partner-preference formation in male prairie voles. J Neurosci. 2003;23:3483–3490. doi: 10.1523/JNEUROSCI.23-08-03483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi VP, Smith-Roe S, Newman SM, Grigoriadis DE, Kalin NH. Reduction of stress-induced behavior by antagonism of corticotropin-releasing hormone 2 (CRH2) receptors in lateral septum or CRH1 receptors in amygdala. J Neurosci. 2002;22:2926–2935. doi: 10.1523/JNEUROSCI.22-07-02926.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL, Vale WW. Increased depression-like behaviors in corticotropin-releasing factor receptor-2-deficient mice: sexually dichotomous responses. J Neurosci. 2003;23:5295–5301. doi: 10.1523/JNEUROSCI.23-12-05295.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis C, Tribollet E. Vasopressin and oxytocin receptors in the central nervous system. Crit Rev Neurobiol. 1996;10:119–154. doi: 10.1615/critrevneurobiol.v10.i1.60. [DOI] [PubMed] [Google Scholar]
- Bosch OJ, Nair HP, Neumann ID, Young LJ. 2004. Pair-bonded prairie voles display depression-like behavior after separation. Soc Neurosci Abstr Viewer/Itinerary Planner: 762.712.
- Carter CS, DeVries AC, Taymans SE, Roberts RL, Williams JR, Getz LL. Peptides, steroids, and pair bonding. Ann N Y Acad Sci. 1997;807:260–272. doi: 10.1111/j.1749-6632.1997.tb51925.x. [DOI] [PubMed] [Google Scholar]
- Chalmers DT, Lovenberg TW, De Souza EB. Localization of novel corticotropin-releasing factor receptor (CRF2) mRNA expression to specific subcortical nuclei in rat brain: comparison with CRF1 receptor mRNA expression. J Neurosci. 1995;15:6340–6350. doi: 10.1523/JNEUROSCI.15-10-06340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza EB. Corticotropin-releasing factor receptors in the rat central nervous system: characterization and regional distribution. J Neurosci. 1987;7:88–100. doi: 10.1523/JNEUROSCI.07-01-00088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza EB, Insel TR, Perrin MH, Rivier J, Vale WW, Kuhar MJ. Corticotropin-releasing factor receptors are widely distributed within the rat central nervous system: an autoradiographic study. J Neurosci. 1985;5:3189–3203. doi: 10.1523/JNEUROSCI.05-12-03189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries AC, DeVries MB, Taymans S, Carter CS. Modulation of pair bonding in female prairie voles (Microtus ochrogaster) by corticosterone. Proc Natl Acad Sci U S A. 1995;92:7744–7748. doi: 10.1073/pnas.92.17.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries AC, DeVries MB, Taymans SE, Carter CS. The effects of stress on social preferences are sexually dimorphic in prairie voles. Proc Natl Acad Sci U S A. 1996;93:11980–11984. doi: 10.1073/pnas.93.21.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries AC, Taymans SE, Carter CS. Social modulation of corticosteroid responses in male prairie voles. Ann N Y Acad Sci. 1997;807:494–497. doi: 10.1111/j.1749-6632.1997.tb51949.x. [DOI] [PubMed] [Google Scholar]
- DeVries AC, Guptaa T, Cardillo S, Cho M, Carter CS. Corticotropin-releasing factor induces social preferences in male prairie voles. Psychoneuroendocrinology. 2002;27:705–714. doi: 10.1016/s0306-4530(01)00073-7. [DOI] [PubMed] [Google Scholar]
- Erb S, Salmaso N, Rodaros D, Stewart J. A role for the CRF-containing pathway from central nucleus of the amygdala to bed nucleus of the stria terminalis in the stress-induced reinstatement of cocaine seeking in rats. Psychopharmacology (Berl) 2001;158:360–365. doi: 10.1007/s002130000642. [DOI] [PubMed] [Google Scholar]
- Getz LL, Carter CS, Gavish L. The mating system of the prairie vole Microtus ochragaster: field and laboratory evidence for pair-bonding. Behav Ecol Sociobiol. 1981;8:189–194. [Google Scholar]
- Gingrich B, Liu Y, Cascio C, Wang Z, Insel TR. Dopamine D2 receptors in the nucleus accumbens are important for social attachment in female prairie voles (Microtus ochrogaster) Behav Neurosci. 2000;114:173–183. doi: 10.1037//0735-7044.114.1.173. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Bass AH. Social behavior functions and related anatomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res Brain Res Rev. 2001;35:246–265. doi: 10.1016/s0165-0173(01)00043-1. [DOI] [PubMed] [Google Scholar]
- Grigoriadis DE, Dent GW, Turner JG, Uno H, Shelton SE, De Souza EB, Kalin NH. Corticotropin-releasing factor (CRF) receptors in infant rhesus monkey brain and pituitary gland: biochemical characterization and autoradiographic localization. Dev Neurosci. 1995;17:357–367. doi: 10.1159/000111306. [DOI] [PubMed] [Google Scholar]
- Grigoriadis DE, Liu XJ, Vaughn J, Palmer SF, True CD, Vale WW, Ling N, De Souza EB. 125I-Tyro-sauvagine: a novel high affinity radio-ligand for the pharmacological and biochemical study of human corticotropin-releasing factor 2 alpha receptors. Mol Pharmacol. 1996;50:679–686. [PubMed] [Google Scholar]
- Gruder-Adams S, Getz LL. Comparison of the mating system and paternal behavior in Microtus ochragaster and M. pennsylvanicus. J Mammal. 1985;66:165–167. [Google Scholar]
- Herbison AE. Sexually dimorphic expression of androgen receptor immunoreactivity by somatostatin neurones in rat hypothalamic periventricular nucleus and bed nucleus of the stria terminalis. J Neuroendocrinol. 1995;7:543–553. doi: 10.1111/j.1365-2826.1995.tb00791.x. [DOI] [PubMed] [Google Scholar]
- Herbison AE, Fenelon VS. Estrogen regulation of GABAA receptor subunit mRNA expression in preoptic area and bed nucleus of the stria terminalis of female rat brain. J Neurosci. 1995;15:2328–2337. doi: 10.1523/JNEUROSCI.15-03-02328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Shapiro LE. Oxytocin receptor distribution reflects social organization in monogamous and polygamous voles. Proc Natl Acad Sci U S A. 1992;89:5981–5985. doi: 10.1073/pnas.89.13.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Young LJ. The neurobiology of attachment. Nat Rev Neurosci. 2001;2:129–136. doi: 10.1038/35053579. [DOI] [PubMed] [Google Scholar]
- Insel TR, Wang ZX, Ferris CF. Patterns of brain vasopressin receptor distribution associated with social organization in microtine rodents. J Neurosci. 1994;14:5381–5392. doi: 10.1523/JNEUROSCI.14-09-05381.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MM, Hammock EA, Young LJ. A method for acetylcholinesterase staining of brain sections previously processed for receptor autoradiography. Biotech Histochem. 2004a;79:11–16. doi: 10.1080/10520290410001671344. [DOI] [PubMed] [Google Scholar]
- Lim MM, Wang Z, Olazabal DE, Ren X, Terwilliger EF, Young LJ. Enhanced partner preference in a promiscuous species by manipulating the expression of a single gene. Nature. 2004b;429:754–757. doi: 10.1038/nature02539. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang ZX. Nucleus accumbens oxytocin and dopamine interact to regulate pair bond formation in female prairie voles. Neuroscience. 2003;121:537–544. doi: 10.1016/s0306-4522(03)00555-4. [DOI] [PubMed] [Google Scholar]
- McEwen BS, Weiss JM, Schwartz LS. Selective retention of corticosterone by limbic structures in rat brain. Nature. 1968;220:911–912. doi: 10.1038/220911a0. [DOI] [PubMed] [Google Scholar]
- Nair HP, Collisson T, Gonzalez-Lima F. Postnatal development of cytochrome oxidase activity in fiber tracts of the rat brain. Brain Res Dev Brain Res. 1999;118:197–203. doi: 10.1016/s0165-3806(99)00149-2. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. 2001. The mouse brain in stereotaxic coordinates. New York: Academic Press.
- Paxinos G, Watson C. 1998. The rat brain in stereotaxic coordinates. New York: Academic Press.
- Potter E, Sutton S, Donaldson C, Chen R, Perrin M, Lewis K, Sawchenko PE, Vale W. Distribution of corticotropin-releasing factor receptor mRNA expression in the rat brain and pituitary. Proc Natl Acad Sci U S A. 1994;91:8777–8781. doi: 10.1073/pnas.91.19.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primus RJ, Yevich E, Baltazar C, Gallager DW. Autoradiographic localization of CRF1 and CRF2 binding sites in adult rat brain. Neuropsychopharmacology. 1997;17:308–316. doi: 10.1016/S0893-133X(97)00071-7. [DOI] [PubMed] [Google Scholar]
- Radulovic J, Ruhmann A, Liepold T, Spiess J. Modulation of learning and anxiety by corticotropin-releasing factor (CRF) and stress: differential roles of CRF receptors 1 and 2. J Neurosci. 1999;19:5016–5025. doi: 10.1523/JNEUROSCI.19-12-05016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rominger DH, Rominger CM, Fitzgerald LW, Grzanna R, Largent BL, Zaczek R. Characterization of [125I]sauvagine binding to CRH2 receptors: membrane homogenate and autoradiographic studies. J Pharmacol Exp Ther. 1998;286:459–468. [PubMed] [Google Scholar]
- Rosenblum LA, Smith EL, Altemus M, Scharf BA, Owens MJ, Nemeroff CB, Gorman JM, Coplan JD. Differing concentrations of corticotropin-releasing factor and oxytocin in the cerebrospinal fluid of bonnet and pigtail macaques. Psychoneuroendocrinology. 2002;27:651–660. doi: 10.1016/s0306-4530(01)00056-7. [DOI] [PubMed] [Google Scholar]
- Salo AL, Shapiro LE, Dewsbury DA. Affiliative behavior in different species of voles (Microtus) Psychol Rep. 1993;72:316–318. doi: 10.2466/pr0.1993.72.1.316. [DOI] [PubMed] [Google Scholar]
- Sanchez MM, Young LJ, Plotsky PM, Insel TR. Autoradiographic and in situ hybridization localization of corticotropin-releasing factor 1 and 2 receptors in nonhuman primate brain. J Comp Neurol. 1999;408:365–377. [PubMed] [Google Scholar]
- Sapolsky RM, Armanini MP, Packan DR, Sutton SW, Plotsky PM. Glucocorticoid feedback inhibition of adrenocorticotropic hormone secretagogue release. Relationship to corticosteroid receptor occupancy in various limbic sites. Neuroendocrinology. 1990;51:328–336. doi: 10.1159/000125357. [DOI] [PubMed] [Google Scholar]
- Schulz DW, Mansbach RS, Sprouse J, Braselton JP, Collins J, Corman M, Dunaiskis A, Faraci S, Schmidt AW, Seeger T, Seymour P, Tingley FD, 3rd, Winston EN, Chen YL, Heym J. CP-154,526: a potent and selective nonpeptide antagonist of corticotropin releasing factor receptors. Proc Natl Acad Sci U S A. 1996;93:10477–10482. doi: 10.1073/pnas.93.19.10477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro LE, Dewsbury DA. Differences in affiliative behavior, pair bonding, and vaginal cytology in two species of vole (Microtus ochrogaster and M. montanus) J Comp Psychol. 1990;104:268–274. doi: 10.1037/0735-7036.104.3.268. [DOI] [PubMed] [Google Scholar]
- Skelton KH, Nemeroff CB, Knight DL, Owens MJ. Chronic administration of the triazolobenzodiazepine alprazolam produces opposite effects on corticotropin-releasing factor and urocortin neuronal systems. J Neurosci. 2000;20:1240–1248. doi: 10.1523/JNEUROSCI.20-03-01240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckler T, Holsboer F. Corticotropin-releasing hormone receptor subtypes and emotion. Biol Psychiatry. 1999;46:1480–1508. doi: 10.1016/s0006-3223(99)00170-5. [DOI] [PubMed] [Google Scholar]
- Stevens J. 1996. Applied multivariate statistics for the social sciences. Hillsdale, NJ: Lawrence Erlbaum.
- Stout SC, Mortas P, Owens MJ, Nemeroff CB, Moreau J. Increased corticotropin-releasing factor concentrations in the bed nucleus of the stria terminalis of anhedonic rats. Eur J Pharmacol. 2000;401:39–46. doi: 10.1016/s0014-2999(00)00412-x. [DOI] [PubMed] [Google Scholar]
- Strome EM, Wheler GH, Higley JD, Loriaux DL, Suomi SJ, Doudet DJ. Intracerebroventricular corticotropin-releasing factor increases limbic glucose metabolism and has social context-dependent behavioral effects in nonhuman primates. Proc Natl Acad Sci U S A. 2002;99:15749–15754. doi: 10.1073/pnas.232480899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pett K, Viau V, Bittencourt JC, Chan RK, Li HY, Arias C, Prins GS, Perrin M, Vale W, Sawchenko PE. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Wagner CK, Morrell JI. Neuroanatomical distribution of aromatase MRNA in the rat brain: indications of regional regulation. J Steroid Biochem Mol Biol. 1997;61:307–314. [PubMed] [Google Scholar]
- Wigger A, Sanchez MM, Mathys KC, Ebner K, Frank E, Liu D, Kresse A, Neumann ID, Holsboer F, Plotsky PM, Landgraf R. Alterations in central neuropeptide expression, release, and receptor binding in rats bred for high anxiety: critical role of vasopressin. Neuropsychopharmacology. 2004;29:1–14. doi: 10.1038/sj.npp.1300290. [DOI] [PubMed] [Google Scholar]
- Young EA. Sex differences and the HPA axis: implications for psychiatric disease. J Gend Specif Med. 1998;1:21–27. [PubMed] [Google Scholar]
- Young LJ, Wang Z. The neurobiology of pair bonding. Nat Neurosci. 2004;7:1048–1054. doi: 10.1038/nn1327. [DOI] [PubMed] [Google Scholar]
- Young LJ, Nilsen R, Waymire KG, MacGregor GR, Insel TR. Increased affiliative response to vasopressin in mice expressing the V1a receptor from a monogamous vole. Nature. 1999;400:766–768. doi: 10.1038/23475. [DOI] [PubMed] [Google Scholar]
- Young LJ, Lim MM, Gingrich B, Insel TR. Cellular mechanisms of social attachment. Horm Behav. 2001;40:133–138. doi: 10.1006/hbeh.2001.1691. [DOI] [PubMed] [Google Scholar]


