Abstract
Background
Autosomal dominant optic atrophy type 1 (DOA) is the most common form of hereditary optic atrophy in human. We have previously identified the OPA1 gene and shown that it was mutated in patients with DOA. OPA1 is a novel member of the dynamin GTPase family that play a role in the distribution of the mitochondrial network. The Bst (belly spot and tail) mutant mice show atrophy of the optic nerves and previous mapping data raise the possibility that Bst and OPA1 are orthologs. In order to analyse the Bst mouse as a model for DOA, we therefore characterized mouse Opa1 and evaluated it as a candidate for the Bst mutant mouse.
Results
Comparison of mouse and human OPA1 sequences revealed 88% and 97% identity at the nucleotide and amino acid levels, respectively. Presence of alternatively spliced mRNAs as seen in human was conserved in the mouse. Screening of the whole mRNA coding sequence and of the 31 exons of Opa1 did not reveal any mutation in Bst. Using a radiation hybrid panel (T31), we mapped Opa1 to chromosome 16 between genetic markers D16Mit3 and D16Mit124, which is 10 cM centromeric to the Bst locus.
Conclusion
On the basis of these results we conclude that Opa1 and Bst are distinct genes and that the Bst mouse is not the mouse model for DOA.
Background
Autosomal dominant optic atrophy (DOA), Kjer type [1], is the most common form of hereditary optic neuropathies with an estimated prevalence of 1:50 000 in most populations [2] and prevalence as high as 1:10 000 in Denmark [3]. The disease appears with an insidious onset of variable visual loss, optic nerve pallor, caecocentral visual field scotoma, and color vision deficit. Histopathological [4,5] and electrophysiological [6,7] studies suggest that the underlying defect is a retinal ganglion cell degeneration. Most families of DOA have been shown to map to 3q28-29 (OPA1, MIM 165500) [8-14]. The gene named OPA1 was subsequently identified [15,16] with more than 70 mutations described today [17-21]. One single family was found to determine a second locus (OPA4, MIM 605293) at 18q12.2-q12.3 [22]. OPA1 codes a mitochondrial dynamin-related GTPase that may play a role in the maintenance of mitochondrial morphology and DNA. Tissue specific expression of OPA1 alternatively spliced exons may underlie some specific neuronal requirements in OPA1 [18].
In 1977, Southard et al. [23] identified a dominant mutation, belly spot and tail (Bst), which arose spontaneously in the C57BLKS mouse strain with in utero death of homozygotes. Heterozygous mice have a kinky tail, white feet and white spots at the ventral midline. In addition, approximately 50% of the Bst/+ mice, show a reduction or a complete absence of the pupillary light reflex in one or both eyes [24]. This neurological phenotype is associated with unilateral or bilateral atrophy of the optic nerves. The severity of the atrophy of the optic nerves is highly variable ranging from a slight reduction in the number of ganglion cell axons in only one optic nerve to a complete elimination of both optic nerves. This is reminiscent of the human situation in which DOA patients show variable expression ranging from asymptomatic carriers to patients with legal blindness. The aspect of the retinal surface and the appearance of the inner and outer nuclear layers in Bst/+ mice are qualitatively normal [24]. However, the ganglion cell numbers appear to be reduced because of a failure of the ganglion cell axons to reach the optic nerve head in early development [25].
The Bst gene is located on mouse chromosome 16 in a region that is partially conserved on human chromosome 3q28-qter [26-28]. Based on its reported mapping to the syntenic region of OPA1 and on some phenotypic similarities of the Bst mouse and human DOA, the Bst gene could be the murine ortholog of OPA1, and therefore the Bst mouse could be proposed as a model for DOA. To address this question we determined the Opa1 gene sequence including its promoter region, as well as its chromosomal localization and that of the markers of the Bst interval. Absence of mutations in the mouse gene and distinct localization of Opa1 and Bst definitely excludes Opa1 as the gene responsible for the Bst phenotype.
Results and Discussion
cDNA sequence and gene structure of mouse Opa1
In a previous study we isolated OPA1 [15], a human dynamin-related protein mutated in DOA. To isolate and characterise the mouse ortholog of the human OPA1 gene, we amplified an adult mouse brain cDNA using primers designed from the human cDNA sequence. 5' RACE-PCR experiments extended the sequence 238 bp upstream from the ATG triplet. The comparison of the RACE-PCR product with the genomic sequence upstream from the start codon revealed that the newly identified 5'cDNA sequence is contiguous with the previously defined exon 1 in the Opa1 genomic DNA, thereby only extending exon 1 without forming a new exon. The open reading frame was found to be 2883 bp in size encoding a mouse Opa1 protein of 960 amino acids. As in human OPA1, mouse Opa1 contains a putative mitochondrial targeting signal, GTPase and dynamin central region domains and two predicted coiled-coil structures (upstream the GTPase domain and in the C-terminal domain) with 97 % overall identity with the human sequence. Human and mouse sequences are somewhat divergent in the first 200 N-terminal amino acids (83 % identity) that mostly contains the mitochondrial leading sequence.
To examine the genomic structure of mouse Opa1 we performed a Blast search against the mouse genomic database with the full-length mouse Opa1 cDNA sequence. A mouse "working draft" BAC clone was identified that contained the entire mouse Opa1 genomic sequence. By rearranging the sequences in this BAC, the intron/exon boundaries and most intron sequences of mouse Opa1 were determined. We established that Opa1 consists of 31 exons and 30 introns extending over 68 kb (Fig. 1). We found that exon/intron boundaries of human and mouse OPA1 were preserved and followed the GT/AG rule. The sizes of all exons are remarkably conserved between the murine and human gene but 16 mouse Opa1 introns are smaller than human ones. As for the human gene, mouse Opa1 exhibits 8 transcripts that result from the alternative splicing of exons 4, 4b and 5b with a predominance of the transcript without exons 4b and 5b in neuronal tissues (retina and brain) (Fig. 2).
Figure 1.
Genomic structure of mouse Opa1. The bars and numbers represent coding exons and the lines represent introns. Exons encoding the different domain of the protein are in brackets. The 5' UTR and the 3' UTR (white bar) are indicated. The conserved region in intron 8 is indicated with an asterisk.
Figure 2.
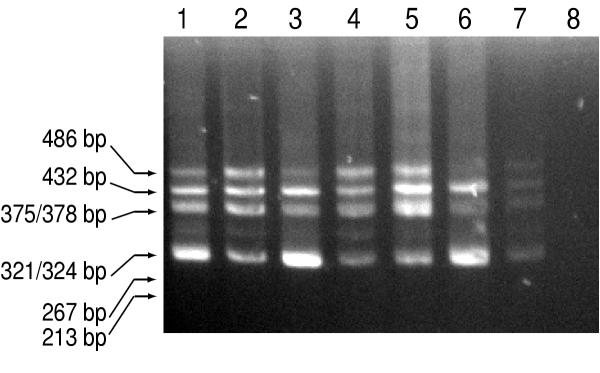
RT-PCR analysis demonstrating multiple Opa1 transcripts in mouse tissues. Use of ME3S and MK5bAS primers resulted in the amplification of 6 distinct fragments, the size of which is indicated on the left. RT-PCR was performed on mouse lung (lane 1), kidney (lane 2), brain (lane 3), liver (lane 4), testis (lane 5), retina (lane 6) spleen (lane 7) and H2O (lane 8).
In search for conserved putative regulatory sequences, we performed a comparative analysis of the Opa1 introns. We found small conserved regions (ranging from 50 to 155 bp in length) in almost all introns but the largest homologous fragment was found in intron 8. This 155 bp-long fragment is 79% identical in both species and covers 1/3 of the intron length. Polymorphisms IVS8 + 4 C/T and IVS8 + 32 T/C putatively involved in normal tension glaucoma [29] are located upstream of this fragment. We sequenced this fragment in DOA patients (n = 3) who had no mutations in the exons and splice junctions of OPA1 and found no mutations.
Promoter regions of mouse and human Opa1
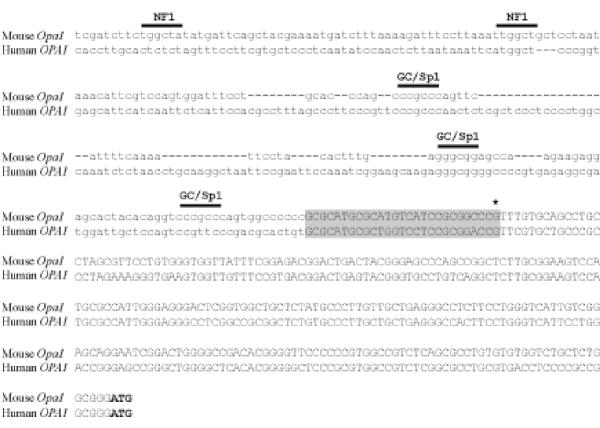
Comparison of the 5'genomic sequences of mouse and human genes revealed a 26 bp-long stretch that exhibits 80 % homology (as found for the rest of exon 1) and lies next to the 5' limit of our RACE-PCR sequence (Fig. 3). It is possible that this stretch corresponds to the 5' end of the mRNA that could have been missed by the RACE-PCR experiment. Mouse and human upstream regions did not contain any binding sites for RNA polymerase II core promoter elements such as CAAT and TATA boxes. This was not unexpected since TATA boxes are found in only 32 % of the promoters [30]. However, we did found several conserved GC boxes, 3 in mouse and 2 in human sequences, that are binding sites for the SP1 transcription activator (Fig. 3). The presence of these GC boxes is typical of housekeeping gene promoters, corroborating previous findings showing ubiquitous expression of OPA1 [15,16]. A sequence matching Nuclear Factor 1 (NF1) binding site was also identified and conserved between human and mouse. As for GC boxes, genes regulated by NF1 proteins are widely expressed.
Figure 3.
Comparison of the genomic sequence upstream of the human and mouse OPA1 gene. The transcribed sequence is shown in capital letters. Asterisk indicates the 5' limit found by RACE-PCR and highlighted in grey is the conserved sequence that may represent the actual 5' end of the mRNA. The initiation codon for translation (ATG) is in bold. Putative binding sites conserved between human and mouse as identified by computer searches are shown with horizontal bars on top of the sequence line.
Mutation screening of Opa1 in Bst mouse
Because OPA1 mutations presumably cause the degeneration of retinal ganglion cells in DOA, we evaluated it as a candidate gene in the mutant mouse Bst which is phenotypically comparable to the DOA in man [24,25]. Indeed, both OPA1 and Bst mutations are inherited as dominant phenotypes with variable expressivity, and both target the retinal ganglion cells. We therefore sequenced the entire cDNA, the genomic DNA (exons and flanking sequences) and the putative promoter region of Opa1 in Bst/+ heterozygous mice and in the +/+ background strain. No mutation was detected in the coding region or intron/exon boundaries. No deletion was detected by amplification of the full length cDNA. This virtually excluded point mutations in amino acid coding sequences and in splice site regions but large deletions or mutations present in unexplored regions of the gene were still possible.
Mapping of mouse Opa1 to chromosome 16 in a region distinct from the Bst locus
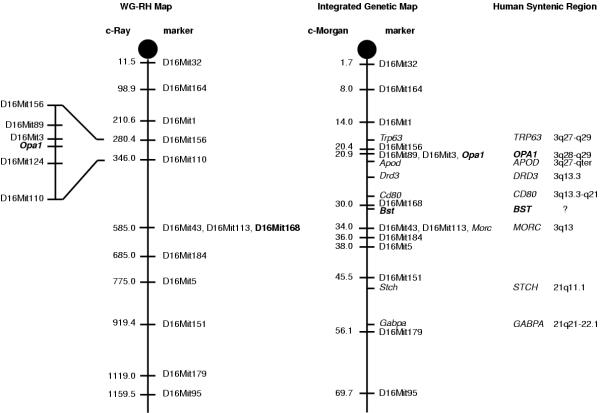
Using radiation hybrid mapping, we found that Opa1 is located on chromosome 16 between genetic markers D16Mit3 and D16Mit124, 10.09 cR distal to the marker D16Mit3 (lod>3) which correspond to 20.9 cM from the centromere (Fig. 4). Previous studies have mapped the Bst locus to mouse chromosome 16, 1.9+/- 1.1 cM from D16Mit168 [25,27]. In order to compare the location of D16Mit168 on genetic map with the physical map developed with the T31 radiation hybrid panel, we mapped the marker D16Mit168 with the T31 RH panel and found it at 2.12 cR from D16Mit113, which is approximately at 33 cM from the centromere. This position fits with the location of this marker on the consensus map for mouse Chromosome 16 at 30 cM from the centromere [28]. According to the placement of D16Mit168, Bst lies approximately 31.5 cM from the centromere, in a region syntenic with human 3q13. In the integrated genetic map, Opa1 locus is approximately 10 cM centromeric to the Bst locus, indicating that Opa1 and Bst are not allelic. This excludes Opa1 as the candidate gene for the Bst mutant mouse and shows that the Bst mouse is not the corresponding animal model for DOA.
Figure 4.
Mouse Opa1 mapped to Chromosome 16 outside the Bst interval. On the WG-RH mapping panel T31 (left diagram), mouse Opa1 locus maps to Chromosome 16 between genetic markers D16Mit3 and D16Mit124. On the integrated genetic map (middle diagram) prepared from the mouse gene database (Mouse Chromosome 16 Linkage Map: http://informatics.jax.org/searches/linkmap_form.shtml), Opa1 mapped 10 cM centromeric to Bst locus. Analysis of human syntenic regions of Opa1 and of the Bst locus (right diagram) indicates that the Bst locus (the synteny of which being unknown, question mark) is flanked by human 3q13 syntenic regions.
The exclusion of Opa1 from the Bst locus is in accordance with some phenotypic discrepancies between the DOA patients and Bst mice. Indeed, there are a variety of organs consistently affected by the Bst mutation. The Bst mouse is smaller than its normal counterpart, shows depigmentation of hair and exhibits numerous skeletal abnormalities attributed to developmental delay: polydactyly of fingers and toes and a short kinked tail [26]. In addition, some Bst mice have colobomas of the optic nerve and retina with subretinal neovascularisation [31], which is never seen in DOA patients. These defects suggest that the normal Bst gene is essential to development and may regulate cellular differentiation during organogenesis. Nevertheless the mitochondrial origin of the Leber Hereditary optic atrophy, DOA type OPA1 and Type III 3-Methylglutaconic Aciduria (Costeff optic atrophy syndrome) suggests that the Bst gene could encode a mitochondrial protein [32]. Moreover, a mutation in a mitochondrial transmembrane protein has been described in a mouse with flexed tail, white belly spots and skeletal abnormalities resembling the Bst phenotype [33]. We found a human gene, TOMM70A (OMIM#606081) that encode a translocase of the outer mitochondrial membrane and that map to 3q12.3 in the syntenic region of the Bst locus. We sequenced the full genomic DNA of the Tom70 mouse homolog of Bst mice and found no changes, thereby excluding also this gene as the Bst gene.
Conclusions
In this work, we report the genomic structure and the position of the mouse Opa1 gene by radiation hybrid mapping. The exclusion of the Opa1 candidate gene for the Bst phenotype suggests that the Bst mouse was not the mouse model of DOA. Considering these data, the development of a mouse model will permit further understanding of Opa1 function and the study of the pathophysiological mechanisms of DOA, as well as the future evaluation of treatment strategies for the disease.
Methods
RNA and DNA purification
The mouse strains C57BLKS (control) and C57BLKS Bst/+ (heterozygous belly spot and tail) were purchased from The Jackson Laboratory (Bar Harbor, ME). Adult Bst/+ mice exhibit a kinked tail and a white belly spot. Total RNA of various tissues was extracted with the Rneasy kit (Qiagen, Germany) following the manufacturer's instructions. First strand cDNA synthesis was performed with Super Script II reverse transcriptase (Invitrogen, The Netherlands) using 1 μg of RNA and 50 pmol of random primers (Promega, USA). DNA was isolated from tail snips by standard methods.
DNA amplification
Genomic DNA and overlapping RT-PCR products were amplified under standardised conditions as follows: denaturation at 94°C for 3 min followed by 35 cycles of 30 sec at 94°C, 30 sec at 62°C, 1 min at 72°C and a final elongation at 72°C for 10 min. Each PCR fragment was purified using the Qiaquick purification kit (Qiagen, Germany) and sequenced on an ABI 310 capillary sequencer (Applied Biosystems, USA). The alternatively spliced transcripts of mouse Opa1 were amplified in several tissues (lung, kidney, brain, liver, testis, retina and spleen) using primers located in exon 3 (ME3S: 5'-GTGACTATAAGTGGATTGTGCCT-3') and in exon 7 (MK5BAS: 5'-CGCTCCAAGATCCTCTGATAC-3'). The obtained RT-PCR products were cloned into the pGEM-T vector (Promega, USA) and sequenced using primers M13F and M13R. The 5'-untranslated region of Opa1 was obtained by RACE-PCR amplification of cDNA sequences generated from brain mRNA (GeneRacer Kit, Invitrogen, The Netherlands) according to the manufacturer's instructions. The gene-specific primer B9AS (5'-AAGAGTCTTGCAGCTAACCTTGC-3') located in exon 2 was used for the initial reverse transcription. The second PCR was performed with the provided GeneRacer 5' nested primer and a nested gene-specific primer located in the 5' region of exon 2 (B8AS: 5'-TTGAAGCTTGAGGGCAGGATGAT-3'). PCR products were then subcloned in the pPCR4Blunt-TOPO vector (Invitrogen, The Netherlands) and sequenced using the nested gene-specific primer.
Radiation hybrid mapping
The 100 radiation hybrid (RH) DNAs of the T31 mouse/hamster RH panel [34] (Research Genetics, USA) were used as templates for PCR amplification of Opa1 and D16Mit168, this marker being close to the Bst locus [26,27]. For Opa1, we designed forward (MK5S: 5'-TTGCCAGTTTAGCTCCCGACTT-3') and reverse (MK6AS: 5'-CAGATCCATGATCTGTTGCTCG-3') primers to amplify a 1438-bp PCR fragment from mouse genomic DNA. No fragment was obtained when hamster genomic DNA was subjected to amplification. For the D16Mit168 marker, we used forward (D16Mit168F: 5'-TGTGGTAGATGGATAAAAGAATGTG-3') and reverse (D16Mit168F: 5'-CATGGAGAAGTTCCTGTAAGCA-3') primers to amplify a 152-bp amplicon. No signal was detected from hamster DNA amplification. The results were submitted to the linkage database at the Whitehead Institute/MIT Center for Genome Research http://www.genome.wi.mit.edu.
Sequence analysis
The human and mouse putative promoter regions were compared by BLAST2 sequences [35]. Potential transcription regulatory sites were detected using the MatInspector program http://transfac.gbf.de/cgi-bin/matSearch/matsearch.pl as implemented by the Baylor College of Medecine Search Launcher. This software accesses the TRANSFAC MATRIX database which contains the consensus binding sites for a variety of transcription factors.
Acknowledgments
Acknowledgments
We thank Benjamin Delprat for help in computational analysis. Research funds came from SOS Rétinite (Montpellier, France), Retina France (Colomiers, France), Fondation pour la Recherche Médicale, CNRS and INSERM (France). C.D. has a research fellowship from Fondation de France, France.
Contributor Information
Cécile Delettre, Email: delettre@pasteur.fr.
Guy Lenaers, Email: glen@cict.fr.
Pascale Belenguer, Email: belengue@cict.fr.
Christian P Hamel, Email: hamel@montp.inserm.fr.
References
- Kjer P. Infantile optic atrophy with dominant mode of inheritance: a clinical and genetic study of 19 Danish families. Acta Ophthalmol Scand. 1959;37:1–146. [PubMed] [Google Scholar]
- Lyle WM. Genetic risks. University of Waterloo Pres Waterloo Ontario. 1990.
- Kjer B, Eiberg H, Kjer P, Rosenberg T. Dominant optic atrophy mapped to chromosome 3q region II Clinical and epidemiological aspects. Acta Ophthalmol Scand. 1996;74:3–7. doi: 10.1111/j.1600-0420.1996.tb00672.x. [DOI] [PubMed] [Google Scholar]
- Johnston PB, Gaster RN, Smith VC, Tripathi RC. A clinicopathological study of autosomal dominant optic atrophy. Am J Ophthalmol. 1979;88:668–675. doi: 10.1016/0002-9394(79)90565-8. [DOI] [PubMed] [Google Scholar]
- Kjer P, Jensen OA, Klinken L. Histopathology of eye optic nerve and brain in a case of dominant optic atrophy. Acta Ophthalmol. 1983;61:300–312. doi: 10.1111/j.1755-3768.1983.tb01424.x. [DOI] [PubMed] [Google Scholar]
- Elenius V. Rod thresholds in dominantly inherited juvenile optic atrophy. Ophtalmologica. 1991;202:208–212. doi: 10.1159/000310202. [DOI] [PubMed] [Google Scholar]
- Holder GE, Votruba M, Carter AC, Bhattacharya SS, Fitzke FW, Moore AT. Electrophysiological findings in dominant optic atrophy (DOA) linking to the OPA1 locus on chromosome 3q 28-qter. Doc Ophthalmol. 1999;95:1998–217. doi: 10.1023/a:1001844021014. [DOI] [PubMed] [Google Scholar]
- Eiberg H, Kjer B, Kjer P, Rosenberg T. Dominant optic atrophy (OPA1) mapped to chromosome 3q region 1. Linkage analysis. Hum Mol Genet. 1994;3:977–980. doi: 10.1093/hmg/3.6.977. [DOI] [PubMed] [Google Scholar]
- Lunkes A, Hartung U, Magarino C, Rodriguez M, Palmero A, Rodriguez L, Heredero L, Weissenbach J, Weber J, Auburger G. Refinement of the OPA1 gene locus on chromosome 3q28-q29 to a region of 2–8 cM in one Cuban pedigree with autosomal dominant optic atrophy type Kjer. Am J Hum Genet. 1995;57:968–970. [PMC free article] [PubMed] [Google Scholar]
- Bonneau D, Souied E, Gerber S, Rozet JM, D'Haens E, Journel H, Plessis G, Weissenbach J, Munnich A, Kaplan J. No evidence of genetic heterogeneity in dominant optic atrophy. J Clin Genet. 1995;32:951–953. doi: 10.1136/jmg.32.12.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RL, Burdon MA, Spalton DJ, Bryant SP, Behnam JT, Seller MJ. Dominant optic atrophy Kjer type Linkage analysis and clinical features in a large British pedigree. Arch Ophthalmol. 1997;115:100–103. doi: 10.1001/archopht.1997.01100150102017. [DOI] [PubMed] [Google Scholar]
- Votruba M, Moore AT, Bhattacharya SS. Genetic refinement of dominant optic atrophy (OPA1) locus to within a 2 cM interval of chromosome 3q. J Med Genet. 1997;34:117–121. doi: 10.1136/jmg.34.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Jr, Fingert JH, Taylor CM, Lake M, Sheffield VC, Stone EM. Clinical and genetic analysis of a family affected with dominant optic atrophy (OPA1) Arch Ophthalmol. 1997;115:95–99. doi: 10.1001/archopht.1997.01100150097016. [DOI] [PubMed] [Google Scholar]
- Votruba M, Moore AT, Bhattacharya SS. Demonstration of a founder effect and fine mapping of dominant optic atrophy locus on 3q28-qter by linkage disequilibrium method: a study of 38 British Isles pedigrees. Hum Genet. 1998;102:79–86. doi: 10.1007/s004390050657. [DOI] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin J-M, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel C. Nuclear gene OPA1 encoding a mitochondrial dynamin-related protein is mutated in dominant optic atrophy. Nature Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1 encoding a dynamin-related GTPase is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nature Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Pesch UE, Leo-Kottler B, Mayer S, Jurklies B, Kellner U, Apfelstedr-Sylla E, Zrenner E, Alexander C, Wissinger B. OPA1 mutations in patients with autosomal dominant optic atrophy and evidence for semi-dominant inheritance. Hum Mol Genet. 2001;10:1359–1368. doi: 10.1093/hmg/10.13.1359. [DOI] [PubMed] [Google Scholar]
- Delettre C, Griffoin JM, Kaplan J, Dollfus H, Lorenz B, Faivre L, Lenaers G, Belenguer P, Hamel C. Mutation spectrum and splicing variants in the OPA1. Hum Genet. 2001;109:584–591. doi: 10.1007/s00439-001-0633-y. [DOI] [PubMed] [Google Scholar]
- Toomes C, Marchbank NJ, Mackey DA, Craig JE, Newbury-Ecob R, Bennet CP, Vize CJ, Desai SP, Black GCM, Patel N, Teimory M, Markham AF, Inglehearn CF, Churchill AJ. Spectrum frequency and penetrance of OPA1 mutations in dominant optic atrophy. Hum Mol Genet. 2001;10:1369–1378. doi: 10.1093/hmg/10.13.1369. [DOI] [PubMed] [Google Scholar]
- Thiselton DL, Alexander C, Morris A, Broooks S, Rosenberg T, Eiberg H, Kjer B, Kjer P, Bhattacharya SS, Votruba M. A frameshift mutation in exon 28 of the OPA1 gene explains the high prevalence of dominant optic atrophy in the Danish population: evidence for a founder effect. Hum Genet. 2001;109:498–502. doi: 10.1007/s004390100600. [DOI] [PubMed] [Google Scholar]
- Thiselton DL, Alexander C, Taanman J-W, Brooks S, Rosenberg T, Eiberg H, Andreasson S, Van Regemorter N, Munier FL, Moore AT, Bhattacharya SS, Votruba M. A comprehensive survey of mutations in the OPA1 gene in patients with autosomal dominant optic atrophy. Invest Ophthalmol Vis Sci. 2002;43:1715–1724. [PubMed] [Google Scholar]
- Kerrison JB, Arnould VJ, Ferraz Sallum JM, Vagefi MR, Barmada MM, Li Y, Zhu D, Maumenee IH. Genetic heterogeneity of dominant optic atrophy Kjer type: identification of a second locus on chromosome 18q12.2-12.3. Arch Ophthalmol. 1999;117:805–810. doi: 10.1001/archopht.117.6.805. [DOI] [PubMed] [Google Scholar]
- Southard JL, Eicher EM. Bst. Mouse News Lett. 1977;56:40. [Google Scholar]
- Rice DS, Williams RW, Davisson MT, Harris BS, Goldowitz D. A new mutant phenotype of retinal ganglion cell dysgenesis discovered in the mouse. Soc Neurosc Abs. 1993;19:51. [Google Scholar]
- Rice DS, Tang Q, Williams RW, Harris BS, Davisson MT, Goldowitz D. Decreased retinal ganglion cell number and misdirected axon growth associated with fissure defects in Bst/+ mutant mice. Invest Ophthalmol Vis Sci. 1997;38:2112–2124. [PubMed] [Google Scholar]
- Epstein R, Davisson M, Lehmann K, Akeson EC, Cohn M. Position of Igl-1 md and Bst loci on chromosome 16 of the mouse. Immunogenetics. 1986;23:78–83. doi: 10.1007/BF00377965. [DOI] [PubMed] [Google Scholar]
- Rice DS, Williams RW, Ward-Bailey P, Johnson KR, Harris BS, Davisson MT, Goldowitz D. Mapping the Bst mutation on mouse chromosome 16: a model for human optic atrophy. Mamm Genome. 1995;6:546–548. doi: 10.1007/BF00356174. [DOI] [PubMed] [Google Scholar]
- Reeves RH, Citron MP. Mamm Genome. 1994;5 Spec No:S229–37. [PubMed] [Google Scholar]
- Aung T, Ocaka L, Ebenezer ND, Morris AG, Krawczak KM, Thiselton DL, Alexander A, Votruba M, Brice G, Child AH, Hitchings RA, Lehmann OJ, Bhattacharya SS. A major marker for normal tension glaucoma: association with polymorphisms in the OPA1 gene. Hum Genet. 2002;110:52–6. doi: 10.1007/s00439-001-0645-7. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, Suyama A, Sakaki Y, Morishita S, Okubo K, Sugano S. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.GR-1640R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RS, John SW, Zabeleta A, Davisson MT, Hawes NL, Chang B. The bst locus on mouse chromosome 16 is associated with age-related subretinal neovascularization. Proc Natl Acad Sci U S A. 2000;97:2191–2195. doi: 10.1073/pnas.040531597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anikster Y, Kleta R, Shaag A, Gahl WA, Elpeleg O. Type III 3-methylglutaconic aciduria (optic atrophy plus syndrome or Costeff optic atrophy syndrome): identification of the OPA3 gene and its founder mutation in Iraqi Jews. Am J Hum Genet. 2001;69:1218–1224. doi: 10.1086/324651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MD, Campagna DR, Haslett JN, Trenor CC, 3rd, Andrews NC. A mutation in a mitochondrial transmembrane protein is responsible for the pleiotropic hematological and skeletal phenotype of flexed-tail (f/f) mice. Genes Dev. 2001;15:652–657. doi: 10.1101/gad.873001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy LC, Terrett J, Davis ME, Knights CJ, Smith AL, Critcher R, Schmitt K, Hudson J, Spurr NK, Goodfellow PN. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences – a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1016/S0378-1097(99)00149-4. [DOI] [PubMed] [Google Scholar]