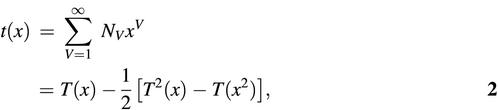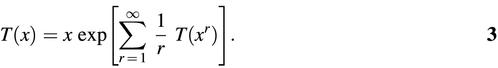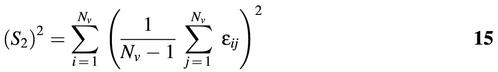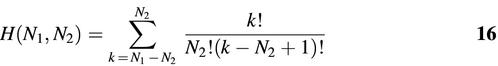Abstract
Understanding the structural repertoire of RNA is crucial for RNA genomics research. Yet current methods for finding novel RNAs are limited to small or known RNA families. To expand known RNA structural motifs, we develop a two-dimensional graphical representation approach for describing and estimating the size of RNA’s secondary structural repertoire, including naturally occurring and other possible RNA motifs. We employ tree graphs to describe RNA tree motifs and more general (dual) graphs to describe both RNA tree and pseudoknot motifs. Our estimates of RNA’s structural space are vastly smaller than the nucleotide sequence space, suggesting a new avenue for finding novel RNAs. Specifically our survey shows that known RNA trees and pseudoknots represent only a small subset of all possible motifs, implying that some of the ‘missing’ motifs may represent novel RNAs. To help pinpoint RNA-like motifs, we show that the motifs of existing functional RNAs are clustered in a narrow range of topological characteristics. We also illustrate the applications of our approach to the design of novel RNAs and automated comparison of RNA structures; we report several occurrences of RNA motifs within larger RNAs. Thus, our graph theory approach to RNA structures has implications for RNA genomics, structure analysis and design.
INTRODUCTION
RNA’s expanding repertoire
RNA molecules are integral components of the cellular machinery for protein synthesis and transport, transcriptional regulation, chromosome replication, RNA processing and modification, and other fundamental biological functions (1–3). Current research continues to provide growing evidence for RNA’s important roles in regulating protein-coding genes (4) and catalysis (5–8). The expanding list of RNA’s repertoire is stimulating research in RNA genomics or ribonomics (9), the large-scale characterization and analysis of RNA structures and functions (10,11). A central goal in ribonomics is to determine all the distinct structural motifs or three-dimensional (3D) folds. Unlike proteins, not many distinct functional RNA classes are currently known (2,5,12); for example, the Nucleic Acid Database (13) (NDB, http://ndbserver.rutgers.edu/NDB/) and The RNA Structure Database (www.rnabase.org) reveal, as of April 2003, ∼600 3D-RNA structures representing only about 20 major functional classes (Table 1).
Table 1. Survey of 3D structures of functional RNAs in NDB (http://ndbserver.rutgers.edu/NDB/index.html); small RNA fragments are not included.
| NDB code | RNA |
|---|---|
| TRNA12 | tRNA |
| PRV022 | RNA pseudoknot inhibitor complexed with HIV-1 reverse transcriptase |
| Ribozyme structures | |
| UR0003 | Large subunit (LSU) ribosomal RNA group I intron |
| UR0019 | Group II self-splicing intron |
| URX053 | P4-P6 RNA ribozyme domain |
| URX057 | RNA hammerhead ribozyme |
| PR0005 | HDV genomic ribozyme |
| PR0038 | Hairpin ribozyme |
| Single stranded RNA | |
| UR0004 | Ribosomal frameshifting viral pseudoknot |
| UR0018 | JIIIabc RNA Tertiary Domain |
| Structural RNAs | |
| PR0041 | Signal recognition particle RNA |
| Ribosomal RNAs | |
| PR0018 | 5S ribosomal RNA |
| RR0003 | 70S ribosome functional complexes |
| RR0006 | 16S ribosomal RNA |
| RR0011 | Domain V of 23S ribosomal RNA |
| RR0017 | 30S ribosomal subunit |
| RR0031, RR0032 | Ribosome at 5.5 Å resolution |
Current computational efforts in the identification of RNA-coding genes in genomes—i.e., DNA sequences that are transcribed into RNAs but do not encode proteins—are, however, limited to variants of known RNA classes (e.g., transfer and ribosomal RNAs) (14); a few potentially novel RNAs have also been identified in recent works (15,16). Finding novel RNA sequences in genomes is challenging because the start and stop codons that specify protein genes are not present in RNA genes (1,16); thus, RNA genes must be found based on sequence or structural homology to existing RNAs using programs like tRNAscan-SE, FAStRNA and Snoscan (14,17,63), for example. Another factor affecting the progress of ribonomics is the small allocation of resources to the RNA field compared with proteomics.
As Tinoco and Bustamante opine in 1999, ‘If 10% of protein fold researchers switched to RNA, the [RNA folding] problem could be solved in one or two years’ (19). Interestingly, the authors argue that the hierarchical nature of RNA folding, which implies that the two-dimensional (2D) RNA fold is stable independently of its 3D fold, may make the problem of predicting RNA tertiary structures more tractable than that of proteins. Another advantage in RNA structure is modularity, allowing assembly of different functional units, as in rational design of ribozymes (catalytic RNAs) (20).
Finding novel RNAs
Besides computational approaches to ribonomics, novel RNAs can be found using experimental in vitro selection of functional RNAs from large (1015) random sequence pools. Essentially, the RNA sequence space (4N nucleotide sequence combinations for N nucleotides) is explored by various strategies to select RNAs with novel functional properties, such as binding to target molecules (ligands, peptides, drugs). Such in vitro selection of functional nucleic acids has advanced impressively our known range of RNA’s functional capabilities (21–26). The selected ribozymes and target-binding RNAs (or aptamers) have potential applications as molecular switches, molecular sensors and therapeutic agents (20,27,28). Still, large complex ribozymes >200 nt are difficult to isolate from such random sequence pools due to the diminishing probability of finding functional RNAs with increasing sequence length (N) (23); this is because the size of the sequence space (4N) rises exponentially.
Both the search for naturally occurring RNAs in genomes and experimental selection of functional RNAs via in vitro methods are complementary approaches for exploring the space of RNA’s structural/functional possibilities. From this perspective, understanding RNA’s structural possibilities is likely to impact ribonomics and the search for novel RNA structures. Broadly speaking, the objective of ribonomics is to delineate the relationship among RNA sequence, secondary topology, tertiary structure and function. Due to current theoretical and experimental limitations (and, possibly, the greater emphasis on protein genomics and design), the extent of RNA’s structural repertoire is not known. We conjecture that the RNA structures available in current databases [e.g., NDB (13)] represent only a small subset of existing and possible structures.
Exploiting graph theory for RNA description
Despite these limitations, the challenges of ribonomics and the modularity of RNAs provide a fertile ground for new conceptual and mathematical approaches, such as the graph theory approach we introduce here. Indeed, there has been a growing recognition that mathematics and computer science provide promising tools for structural biology research. Biologist Dennis Bray asserted recently that ‘Theory needs to be embraced and to become part of the mainstream of biological research. The quality and accuracy of predictions will then inexorably rise’ (29).
Specifically, to address some of the limitations of current approaches to RNA structures, we develop a graph theory approach combined with modeling and computational biology tools for exploring RNA’s secondary-structural repertoire. A greater understanding of RNA’s 2D structural repertoire will provide important leads for the search for novel RNAs, since RNAs with dissimilar 2D motifs generally have different folded 3D structures and functions (9,10,15). To enumerate all possible 2D motifs, we represent RNA secondary structures schematically as planar graphs; we use tree graphs to represent RNA tree structures and dual graphs to represent any RNA secondary structures, including trees and pseudoknots. The simpler tree representation allows exploitation of key graph-theory results for RNA analysis, but a separate representation is required for pseudoknots since these more complex topologies cannot be represented as trees. (See Supplementary Material for graph theory terms.)
RNA tree and dual graphs provide discrete representations of 2D RNA motifs whose secondary elements (loops, bulges, stems, junctions) are defined by graph vertices (•) and edges (—). All possible 2D RNA motifs, both natural and hypothetical, can be enumerated using such discrete graphical representations. Although RNA tree graphs were developed earlier by Le et al. (30) and Benedetti and Morosetti (31) for identifying structural similarity between RNAs, our work develops graphical representations for both RNA trees and pseudoknots and describes RNA applications to 2D motif enumeration, design and structural comparison.
The graph theory framework introduced here allows an estimation of the size of RNA’s structural repertoire and immediately suggests a survey of RNAs in public databases and the literature to determine existing and missing motifs. Significantly, we find that about 35 distinct tree and pseudoknot motifs in our enumerated sets exist in solved RNAs but that many larger motifs are ‘missing’; this implies that natural RNAs represent only a small subset of all possible mathematically enumerated motifs. Some of the missing motifs may thus represent undiscovered natural RNAs or RNAs that may be generated synthetically in the laboratory, while others may correspond to energetically unfavorable motifs. These possibilities pose intriguing experimental and theoretical challenges for the systematic search of novel RNAs guided by enumerated topologies.
The RNA graphical representations further suggest three important applications: clustering of functional RNA classes, finding RNA motifs within substructures of larger RNAs and designing novel 2D RNA motifs.
Our clustering plot shows that existing RNAs are sparsely distributed within a narrow range of the possible topological characteristics (e.g., degree of secondary-structural branching). This observation can be interpreted in terms of physiochemical factors.
Our application of graph isomorphism to find smaller RNA motifs within larger RNAs reveals many occurrences of small RNA motifs (e.g., 5S ribosomal RNAs) within larger RNAs, such as 16S and 23S ribosomal RNAs, as expected, but also unexpected relations, such as the hepatitis delta virus (HDV) RNA motif within tmRNA (an RNA involved in protein biosynthesis). Such a computationally efficient method (and an alternative to manual inspection) of establishing structural relationships among existing RNAs is valuable for understanding the modularity of RNA for design applications.
Finally, we develop a procedure for designing novel RNA motifs using missing motifs, modular assembly of existing RNA subunits and 2D RNA folding algorithms (18,32), as the first step in the search for novel functional RNAs.
Article outline
The remainder of this article is organized as follows. In the next section, Concepts and Methods, we introduce the basic elements of graph theory representation and analysis of RNA structures. The Results section consists of the following parts: estimating the size of RNA space; survey of existing RNA topologies; clustering functional RNA classes; RNA substructure analysis; and search for novel RNAs: design and prediction. We conclude with a summary of this work. We elaborate in the Appendix upon: (A) the relationship between tree and dual graphs; (B) the limitations of graphical representations; (C) the algebraic properties of RNA topologies; and (D) an algorithm for finding structurally similar RNA graphs. The Supplementary Material contains a glossary that defines various terms in RNA structure and graph theory. The database we develop (RAG, for RNAs As Graphs) will be available on our group’s web site (please check monod. biomath.nyu.edu).
CONCEPTS AND METHODS
This section provides a brief background on RNA secondary structures (some of which is well known to biologists but possibly unfamiliar to mathematical researchers interested in this work), as well as available 2D folding algorithms, and key concepts in graph theory which we use to describe and analyze RNA structures. We then introduce the tree and dual graph rules for representing RNA secondary structures, and present graph-theory enumeration formulas.
RNA secondary structures and folding algorithms
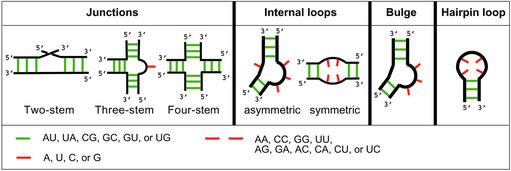
RNA biopolymers are made of four nucleotide bases denoted by letters A, C, G and U. The linear RNA chain molecule, running from a 5′ to a 3′ end, each distinguished chemically, can fold upon itself to form (2D) secondary and tertiary (3D) structures. An RNA secondary structure refers to a network of structural motifs such as helical stems, loops, bulges and junctions (Fig. 1). RNA stems are self-complementary base-paired regions (e.g., AU, UA, GC, CG), whereas loops and bulges are regions in the double-stranded RNA with mismatched (e.g., AG, UC) or unmatched (unpaired) bases; RNA junctions are constructs where two or more stems meet, and they usually contain unmatched bases (see details in Fig. 1). The overall molecular architecture of the secondary structure is stabilized by Watson–Crick (GC and AU) and other (e.g., GU wobble) base pairing motifs (33).
Figure 1.
RNA secondary structural elements: junctions, stems, loops and bulges. The double-strands (black) of RNA stems are stabilized by complementary base pairs (e.g., AU, GC, GU), shown schematically as green lines. The number of unmatched bases (e.g., A, C, U, or G, red lines) and mismatched base pairs (e.g., AG, GA, AC, CA, red lines) in junctions, bulges and loops can vary. Several types of RNA junctions (two-, three- and four-stem) and internal loops (symmetric and asymmetric) are illustrated.
The secondary structural motifs, aided by the presence of ions, can interact to form 3D folds of biologically active (functional) RNA molecules (33–36). These interactions produce a complex hierarchical relationship between secondary and tertiary structures (see Fig. 2D). According to the hierarchical view of RNA folding (19), the secondary structure is stabilized relatively fast and is followed by slow folding of the tertiary structure, which can take minutes or longer (37).
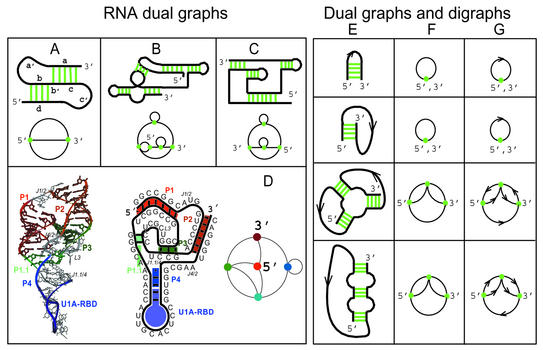
Figure 2.
(Left) Dual graph representations of existing RNAs. Schematic 2D pseudoknots (top) and their planar dual graph representations (bottom) for (A) simple pseudoknot configuration (with labeled segments a–d), (B) class III ligase ribozyme and (C) HDV ribozyme. Pseudoknots (B) and (C) are prototype ribozymes engineered by Schultes and Bartel (59). The lower panel shows the 3D tertiary structure, 2D secondary structure and schematic 2D dual graph representation of HDV ribozyme (PDB no. 1DRZ). The HDV ribozyme has two pseudoknots in regions P1/P2 and P1.1/P3. (Right) Comparing dual (column F) and digraph (G) representations of hypothetical RNA secondary structures (E). Digraphs are graphs whose edges have flow directions. The ambiguities in representing the topology of RNA secondary structures using dual graphs are resolved by employing digraphs; however, the topologies of single-stem structures (rows 1 and 2 of column E) cannot be differentiated by both dual and digraph representations.
RNA trees and pseudoknots are two major types of 2D RNA secondary structures, distinguished by the topology of their base pairing patterns. An RNA tree is a branching network of helical stems interrupted by bulges and junctions that end in loops, except at the 3′ and 5′ ends. An RNA pseudoknot has a stretch of nucleotides within a hairpin loop that pairs with nucleotides external to that loop (38). More precisely, a pseudoknot forms when a consecutive single-stranded domain with segments a, a′, b, b′, c, c′, and d (a′, b′, and c′ are connectors) fold to form two regions with Watson–Crick base pairing: a with c, and b with d (see Fig. 2A).
RNA secondary structures can be predicted from sequence using folding algorithms. Available programs based on dynamic programming algorithms (e.g., MFOLD and PKNOTS) predict base pairing patterns, the presence of base-pair mismatches, and regions with unpaired bases. Optimal solutions are obtained by minimizing the overall RNA free energy on the basis of experimentally derived free energy parameters for base pairs (39). For RNA tree structures, the widely used 2D folding algorithm by Zuker and coworkers (32,39) (MFOLD) is available at http://bioinfo.math.rpi.edu/∼zukerm/. Other related 2D RNA prediction algorithms have been developed by McCaskill (40), Wuchty et al. (41) and Rivas and Eddy (PKNOTS) (18). Notably, PKNOTS can predict small (<100 nt) pseudoknots, a capability lacking in other RNA folding programs; the computational cost of predicting large pseudoknots (>200 nt) is generally prohibitive (18). In our work, we use MFOLD and PKNOTS to predict small tree and pseudoknot structures; data for larger structures are taken from experimentally solved structures available in databases and the literature.
Basic aspects of graph theory
Since RNA secondary structures are essentially 2D networks, they may be represented using planar graphs to facilitate analysis of RNA structures; indeed, modeling network motifs using graphs has proven to be fruitful for many complex systems in biochemistry, neurobiology, ecology and engineering (42,43). Such ideas have also been previously applied to characterize and compare RNA structures at the base pair (44) and secondary-structural (30,31) levels.
Graph theory is a branch of mathematics that deals with configurations described by nodes and connections. The configurations may represent physical networks, such as electrical circuits or chemical compounds (45), where atoms and bonds are modeled as nodes and connections, respectively. Formally, such configurations are modeled by graphs consisting of vertices or nodes (•) and edges or lines (—), which connect the vertices. In our work, we use exclusively planar graphs whose vertices and edges are drawn in a 2D plane.
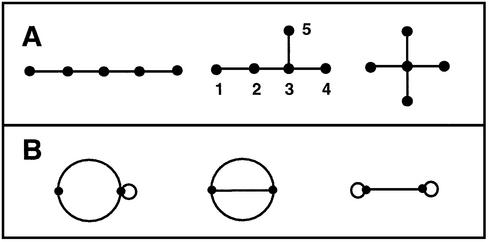
Figure 3 shows examples of tree (Fig. 3A) and non-tree (Fig. 3B) planar graphs. These are connected graphs since every pair of vertices in the graph is connected by one or more paths; a path is a ‘walk’ from a vertex to another with no repeated edges or vertices. A tree is a connected graph whose vertex connections do not form closed paths (e.g., no triangles). Hydrocarbon molecules, such butane and isobutane, can be represented using trees or tree graphs. In fact, the use of tree graphs to count possible hydrocarbon structures played a significant role in the development of graph theory through the graphical enumeration theorem of A. Cayley (46). We will exploit this and other tree enumeration theorems to count RNA’s topological motifs (see section below on enumeration of RNA graphs). For RNA pseudoknots, non-tree graphs are required to describe their complex patterns of connectivity involving closed paths or faces (Fig. 3B).
Figure 3.
Basic concepts of tree and dual graphs. (A) Three five-vertex planar tree graphs (the middle graph is labeled); (B) three two-vertex non-tree graphs. A planar graph is a set of nodes or vertices (•) and a set of line segments or edges (—) in a plane where each of the segments either joins two vertices or joins a vertex to itself. In a labeled graph, the graph vertices are labeled, as in the middle graph of set (A). Graphs with no vertex labels are called unlabeled graphs. Graphs in sets (A) and (B) are connected graphs since all vertices are linked. The tree and non-tree graphs are distinguished by the absence/presence of closed paths (faces), where a path is a ‘walk’ between vertices with no repeated edges. A tree is a connected graph with no closed paths. The non-tree graphs in (B) are also connected graphs, but closed paths between vertices are present in these graphs; the closed paths are the self-loops and multiple (two or more) edges connecting two vertices. The edges incident on, or emanating from, a vertex are called incident edges. For example, vertex 3 of the middle graph of set (A) has three incident edges; vertex 2 has two incident edges and vertices 1, 4 and 5 each have one incident edge. In set (B), a self-loop of a vertex counts two incident edges. For example, each vertex of the right graph of set (B) has three incident edges.
Graphical representation of RNA structures
Unlike graphs for chemical structures, where atoms are vertices and bonds are edges, our RNA graphs are RNA secondary topologies where a vertex or an edge can represent multiple nucleotide bases or base pairs, which are themselves composed of multiple atoms and bonds. To allow graphical representation of complex RNA secondary topologies, we state below rules for defining RNA graphs and provide justifications for these rules. The rules specify how to represent RNA loops, bulges, junctions and stems as vertices or edges in a graph. Essentially, the tree and dual graph rules simplify RNA secondary motifs to allow their representation as mathematical graphs; the ‘RNA graphs’ specify the skeletal connectivity of the secondary motifs.
We use tree graphs to represent RNA trees and dual graphs to represent any RNA secondary structures, including trees and pseudoknots, since pseudoknots cannot be represented as trees. Still, the tree representation is advantageous because of its intuitive appeal and the existence of applicable tree enumeration theorems, especially those by Cayley and by Harary and Prins (47,50). In the Appendix, we elaborate on the relationship between tree and dual representations (A) and the limitations of graphical representations (B).
Planar tree graph rules. To represent RNA trees as planar graphs, we use the following rules to assign edges and vertices.
T1. A nucleotide bulge, hairpin loop or internal loop is considered a vertex (•) when there is more than one unmatched nucleotide or non-complementary base pair. The special case of the GU wobble base pair is regarded as a complementary base pair.
T2. The 3′ and 5′ ends of a helical stem are considered a vertex (•).
T3. An RNA stem is considered an edge (—); we define an RNA stem to have two or more complementary base pairs.
T4. An RNA junction is a vertex (•).
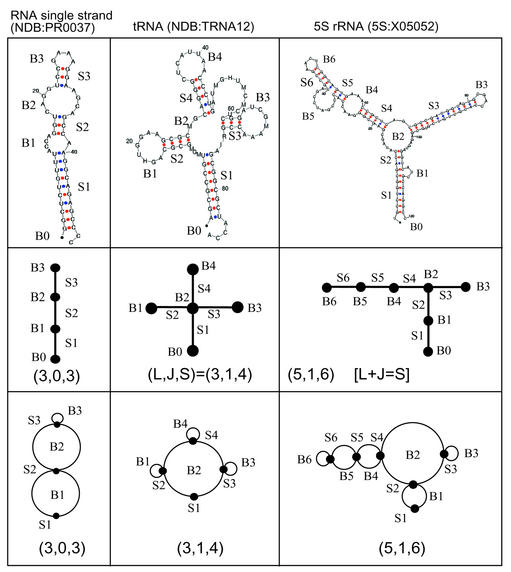
As shown in Figure 4, the above rules reduce the 2D structures of single-strand RNA, transfer RNA (tRNA), and 5S ribosomal RNA (rRNA) to tree graphs.
Figure 4.
Schematic graphical representations for three RNA secondary topologies computed using Zuker’s MFOLD algorithm (54) (top row). For these RNAs, our two types of graphical representations are shown: tree (middle row) and dual (bottom row) graphs. We use corresponding labels (B0, B1, etc. and S1, S2, etc.) on the secondary structures and graphs to denote the secondary structural elements that are represented as vertices (•) or edges (— or ∩); B0, denoting chain ends, is not represented in dual graphs. Note that the vertices and edges in tree and dual graphs represent different RNA secondary elements (see Table 2). In tree graphs, unmatched base pairs are represented as vertices and helical stems are edges. In contrast, in dual graphs, the unmatched base pairs are considered as edges and the vertices are helical stems. For all RNA graphs, the numbers L (loops plus bulges), J (junctions) and S (stems) obey a simple relation, L + J = S, derived from Euler’s formula (60) in algebraic topology (see discussion on algebraic properties of RNA topologies in Appendix C). The validity of the formula is illustrated for the three RNA topologies shown. The NDB codes are given for all structures (ndbserver.rutgers.edu/NDB/ index.html). Other examples of RNA tree graphs are shown in Figure 7.
Note that we consider the 3′ and 5′ ends of a double-stranded helical stem as a vertex (•). This assignment of the ends of a stem is required because graph theory stipulates that an edge must join two vertices or a vertex to itself. Not all stem ends are the same, however. In some cases, the 3′ end is part of a flexible single-stranded region (see Fig. 4). This and other variations of RNA stem ends are not captured by our vertex representation. In rule T2 above, when the 3′ and 5′ ends of an RNA structure do not belong to the same helical stem, the RNA cannot be represented as a tree. For such cases, we use the dual graph rules discussed below.
In our tree graph rules T1–T3, we have defined for physical reasons the minimal numbers for matched, mismatched or unmatched base pairs: >1 bp mismatch for a vertex and ≥2 bp matches for an edge. Our vertices (•)—which represent unmatched nucleotides or mismatched (non-complementary) base pairs (e.g., AG, AC and CU)—could participate in base pairing with unpaired bases in other parts of the RNA molecule through tertiary interactions, as in kissing hairpin and bulge-loop motifs (33). Such interactions stabilize 3D RNA structures, and they usually involve more than a single base pair (33). Rule T1—that RNA graph vertices represent more than one unmatched nucleotide or non-complementary base pair—reflects these significant features of RNA structure and interaction. Thus, RNA bulges, loops and junctions, which we represent as vertices, are determinants of RNA interaction, flexibility and tertiary structure.
Rule T3 requires that RNA stems—which are represented as edges (—)—have a minimum of two complementary base pairs; generally, RNA stems can have anywhere from two to dozens of base pairs. A minimum of two base pairs ensures that the RNA stem is stable against thermal fluctuations (∼0.6 kcal/mol at room temperature). Formation of a single base pair can interrupt a loop region, but such an isolated base pair may be unstable thermodynamically. We regard such a configuration as a loop region, which is represented as a vertex.
Certainly, our tree graph rules (T1–T4) may be modified as necessary to reflect other desired features of RNA structure; the simpler they are, the smaller the topological space implied by them. The more complex labeled RNA tree graphs of Le et al. distinguish various types of loops, bulges, junctions and stems (30).
Planar dual graph rules. To represent trees, pseudoknots and other RNA secondary topologies as planar graphs, we use the following general rules.
D1. A vertex (•) represents a double-stranded helical stem.
D2. An edge (— or ∩) represents a single strand that may occur in segments connecting the secondary elements (e.g., bulges, loops, junctions and stems).
D3. No representation is required for the 3′ and 5′ ends.
Implicit in the above rules is the requirement that a stem has two or more complementary base pairs and a bulge has more than one unmatched nucleotide or non-complementary base pair, as in tree-graph rules T1–T4. In contrast to tree-graph rules, a vertex (•) now represents a stem instead of a bulge/loop/junction, and an edge (— or ∩) represents a strand in bulge/loop/junction instead of a stem; we elaborate upon the relationship between tree and dual graphs in Appendix A; see Table 2.
Table 2. Representation of edges and vertices in tree and dual graphs of 2D RNA motifs.
| Secondary motif | Tree graph | Dual graph |
|---|---|---|
| Stem | Edge (—) | Vertex (•) |
| Loop/bulge | Vertex (•) | Edge (— or ∩) |
| Junction | A vertex with ≥3 incident edges | A face with ≥3 vertices |
| 3′, 5′ ends | Vertex (•) | Not represented |
We call the RNA graphs drawn using the above rules (D1–D3) dual graphs. Figure 2 shows the RNA secondary structures of four pseudoknots and their corresponding dual graphs: (A) a simple pseudoknot, (B) class III ligase ribozyme (engineered), (C) HDV ribozyme (engineered) and (D) HDV ribozyme (PDB no. 1DRZ).
RNA secondary structures represented as dual graphs have specific connectivity properties. A double-stranded RNA stem is connected to at most two strands on either ends for a total of four strands. This means that the maximum number of incident edges at any vertex (i.e., edges emanating from a vertex, see Fig. 3) of a dual graph is four, except for one or two vertices which may have two or three incident edges. The vertices with two/three incident edges indicate where one/both ends (3′ and 5′) is/are located. If two vertices have less than four incident edges, both must have three incident edges. On the other hand, if all vertices have four incident edges except one, that vertex must have two incident edges. These properties imply a total of 2V –1 edges for any V-vertex RNA dual graph.
The dual graph rules for RNAs with double-helical stems can be generalized to allow enumeration of RNAs with triple or quadruple helices. This generalization is important because RNA and DNA triple helices are found in nature (48,49). Another generalization of dual graph is to use the more precise directed graphs or digraphs RNA representation. The advantages of digraphs are discussed in Appendix B. Brifley, digraphs specify the flow directions of edges which can resolve ambiguities in the dual graph representation.
Enumeration of RNA graphs
To estimate the size of RNA’s structural space and to aid in finding new RNA folds, we now consider the enumeration of tree and dual graphs. Specifically, we seek to describe all possible RNA topologies (NV) for a fixed number of vertices (V); the number of vertices is a measure of RNA chain length.
Enumeration of RNA tree graphs. The number of possible RNA graphs (NV) for a given number of vertices (V) can be counted using tree enumeration theorems of Cayley for labeled trees (46) and Harary–Prins for unlabeled trees (trees with equivalent vertices) (46,47,50). These theorems are cornerstones in the subarea of graph theory that deals with graphical enumeration. Labeled trees refer to graphs with labeled vertices, as illustrated in Figure 3; graph vertices are not labeled in an unlabeled graph. Enumeration of unlabeled trees considers the number of non-isomorphic graphs, i.e., topologically distinct trees irrespective of the vertex identity. The labeled trees, on the other hand, allow distinction of specific bulges, loops, junctions and ends in RNA graphs. Both Cayley and Harary–Prins approaches are relevant to the counting of RNA’s structural repertoire.
The number of labeled trees for any V is given by the Cayley formula (46)
NV = VV – 2.1
For unlabeled trees, Harary and Prins obtained the counting polynomial t(x) (47,50) whose coefficient NV is the number of distinct graphs with V vertices:
where
The counting polynomial (equations 2 and 3) up to the first 12 terms is
t(x) = x + x2 + x3 + 2x4 + 3x5 + 6x6 + 11x7 + 23x8 + 47x9 + 106x10 + 235x11 + 551x12 + …4
In this polynomial, the coefficients of the first, second and third terms, for example, indicate that there is only one distinct graph each for V = 1, 2 and 3; the Harary–Prins enumeration polynomial is derived based on the Pölya–Burnside method (51,52). Clearly, the number of distinct graphs (NV) as a function of vertex number according to Cayley’s formula (1) for labeled trees grows faster than Harary–Prins’s formula (2) for unlabeled trees.
Based on the counting polynomial (equation 4), we can estimate the number of distinct secondary motifs for a given RNA size. Since a tree edge roughly corresponds to 20 nt, a tree with six vertices (five edges or 100 nt) has six possible motifs, whereas an 11-vertex (10 edges or 200 nt) tree has 235 possible motifs. As the RNA size increases from 100 to 200 nt, the number of possible motifs increases by a factor of 39, indicating the potential of large RNAs to form many more novel secondary motifs.
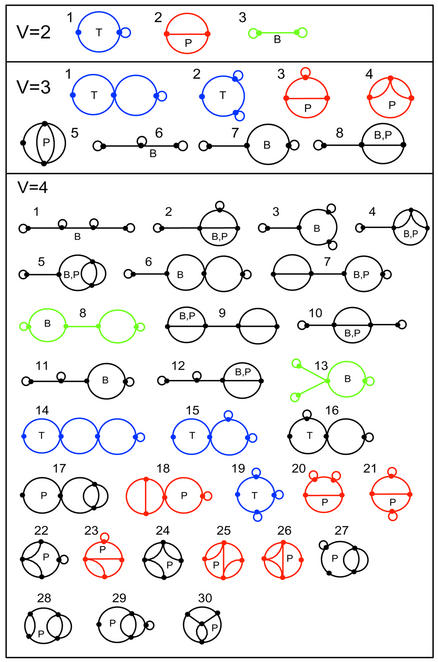
Enumeration of RNA dual graphs. The enumeration of dual graphs, unlike trees, simultaneously yields tree, pseudoknot and other possible topological motifs as defined by the dual graph rules (D1–D3). We have heuristically enumerated all such graphs for the cases of V = 2, 3 and 4, which correspond to 3, 8 and 30 possible dual graphs, respectively (Fig. 5). In addition to RNA trees (T) and pseudoknots (P), enumerated motifs in Figure 5 reveal graphs involving single-edge connectors; we call such motifs bridge graphs (B) or simply bridges. Bridges are biologically important since they suggest existence of independent RNA submotifs and thereby help in the modular design of RNAs. Examples of RNA bridges are box H/ACA snoRNA, hepatitis C virus (HCV) RNA and group I intron (Fig. 6).
Figure 5.
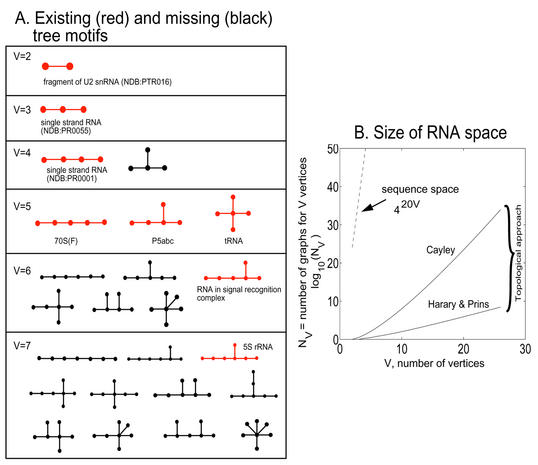
All possible topologies for RNAs represented by dual graphs having 2, 3 and 4 vertices. There are three types of graphs in each set: tree (T), pseudoknot (P) and bridge (B). Each graph is index by (V, i) where is V is the number of vertices. We color the existing RNA trees, pseudoknots and bridges in these graphs as blue, red and green, respectively. The existing RNA trees are: (2,1)—single strand RNA (NDB code PTR016), (3,1)—single strand RNA (NDB code PR0055), (4,14)—70S(F), (4,15)—P5abc, and (4,19)—tRNA; the existing RNA pseudoknots are: (2,2)—Simian Retrovirus type-1, (3,3)—P3/P7 pseudoknot of the Tetrahymena ribozyme, (3,4)—tmRNA pseudoknot 2 (Pk2) of Escherichia coli, (4,18)—tRNA-like structure bulge pseudoknot (PSEUDOBASE no. PKB143), (4,20)—viral frameshift (PSEUDOBASE no. PKB174), (4,21)—class III ligase ribozyme (engineered), (4,23)—pseudoknot E23–9/12 of 18S ribosomal RNA (PSEUDOBASE no. PKB205), (4,25)—pseudoknot PK1 of Legionella pneumophila tmRNA (PSEUDOBASE no. PKB67), (4,26)—pseudoknot of signal recognition particle RNA (PSEUDOBASE no. PKB163); and the existing RNA bridges are: (2,3)—bulged hairpin (analog of pAT1), (4,8)—box H/ACA snoRNA, and (4,13)—viral frameshift RNA (PSEUDOBASE no. PKB217).
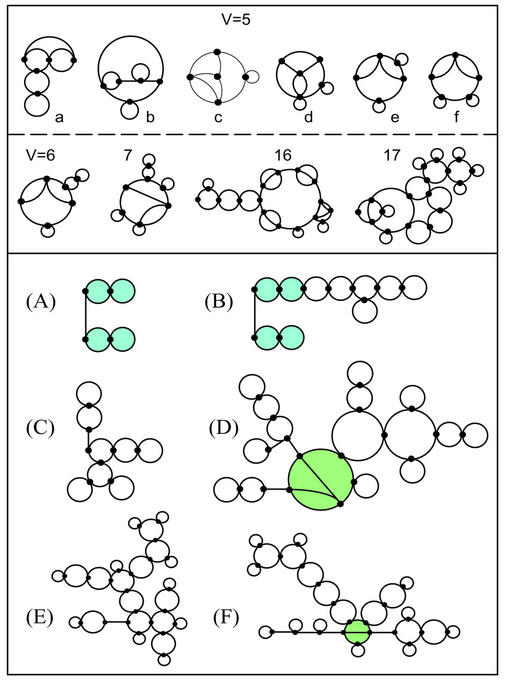
Figure 6.
Upper panel: RNA pseudoknot topologies for 5 ≤ V ≤ 17 found in the literature and the PSEUDOBASE database (www.bio.LeidenUniv.nl/∼Batenburg/PKBGet.html#s3). (The pseudoknots for V < 5 are listed in the enumerated sets in Fig. 5.) For V = 5, the pseudoknots found are: (a) attenuator pseudoknot of the hammerhead ribozyme (PSEUDOBASE no. PKB173); (b) class III ligase ribozyme (engineered) (59); (c) HDV ribozyme (PDB no. 1DRZ); (d) HDV ribozyme (‘Italy’ variant; PSEUDOBASE no. PKB75); (e) dicistronic cricket paralysis virus RNA (PSEUDOBASE no. PKB223); and (f) viral tRNA-like of alfalfa mosaic virus (PSEUDOBASE no. PKB191). The pseudoknots with V > 5 are: broad bean mottle virus tRNA-like (V = 6, PSEUDOBASE no. PKB135); brome mosaic virus tRNA-like (V = 7, PSEUDOBASE no. PKB134); tmRNA (V = 16) (61); and RNase P RNA (V = 17) (62). Lower panel: survey of RNA bridge motifs in the literature. The RNAs found are: (A) box H/ACA snoRNA; (B) hTR (bases 211–451); (C) Neurospora VS ribozyme; (D) HCV; (E) U19H; and (F) group I intron. The isomorphic subgraphs of (A) and (B) are shaded blue; the pseudoknot subgraphs of (D) and (F) are shaded green.
RNA trees, pseudoknots and bridges can be categorized as topological types differentiated by the order of connectivity between the vertices or, as known in graph theory, the edge-cut property; an edge-cut is a set of edges whose removal results in a disconnected graph. RNA trees are characterized by minimal edge-cuts with two edges; RNA pseudoknots have at least a minimal edge-cut with three edges (e.g., Fig. 2A); and RNA bridge graphs become disconnected graphs upon removal of an edge (or unpaired RNA strand), a property known in graph theory as one-edge-connected graph. A bridge graph can also be a pseudoknot because it can contain a pseudoknot subgraph. However, some existing RNA bridges have no pseudoknot subgraphs, for example, bulged hairpin, box H/ACA snoRNA and viral frameshift RNA (green graphs in Fig. 5). Thus, dual graphs describe three topological RNA types: tree (edge-cut with two edges), pseudoknot (edge-cut with three edges), and bridge (one-edge-connected); see Figures 5 and 6.
The distribution of tree, pseudoknot and bridge types in V = 2, 3 and 4 motif sets is as follows. For the V = 2 motif set, three graphs correspond to one tree, one pseudoknot and one bridge; for V = 3 set, eight graphs correspond to two trees, three pseudoknots and three bridges; and for V = 4 set, 30 graphs include four trees, 20 pseudoknots and 13 bridges (thus seven graphs are both pseudoknots and bridges). These enumeration results imply that the number of bridges (NVbridge), trees (NVtree), and pseudoknots (NVpseudo) within a given topological set follow:
The complexity and number of the dual graphs increase quickly with vertex number, making it non-trivial to determine the number of topological possibilities for a given V. General enumeration theorems for RNA dual graphs are not available.
RESULTS AND DISCUSSION
Estimating the size of RNA space
The possibility to enumerate 2D RNA motifs provides a unique tool to estimate the size of RNA’s structural repertoire or the RNA space. Even though we do not expect most enumerated graphs to lead to natural RNAs, some motifs will certainly be naturally occurring or theoretically possible to generate in the laboratory. Those that are likely to be unphysical may be excluded by geometric, energetic and functional considerations. Thus, enumerating RNA motifs will provide an upper bound of the number of possible unique 3D RNA structures or functions. Below, we compare the RNA sequence space with estimates of the RNA topology space from Cayley and Harary–Prins tree enumeration formulas; we also discuss the implications of these results.
For an RNA of sequence length N, the sequence space size is 4N. Since a vertex in RNA graphical representation corresponds to ∼20 nt (based on our survey of existing RNAs), sequence space grows with vertex number as 420V. Hence, the sequence space (420V) is much larger than the tree topology space (NV): 420V >> NV, whether NV is counted using Cayley’s formula (VV – 2) (46) or using Harary–Prins’s formula (47), which can be approximately parametrized as 2.5V – 3 for V > 3; we obtain the dependence of these estimates on sequence length by setting V = N/20.
Figure 7B shows that the sequence space is significantly larger than Cayley’s topology space, which in turn is larger than Harary–Prins’s topology space. For example, for V = 10 or N ≈ 200, the sequence space contains 4200 = 2.6 × 10120 elements whereas topology space contains 106 trees (Harary–Prins) or 108 labeled trees (Cayley). Clearly, the RNA topology space is vastly smaller than the sequence space because many sequences can fold to the same topology; Reidys et al. (53) have estimated the number of sequences having a given secondary structure varies as 0.673 N3/2 2.164N. The small dimension of the RNA topology space implies an advantage in the search for novel RNAs. For example, rather than exploring the (random) sequence space, as in current in vitro selection technology for functional RNAs, we suggest searching for new RNA folds or functions corresponding to selected novel 2D motifs from enumerated repertoire.
Figure 7.
(A) Enumerated tree topologies or motifs having up to seven vertices (V). The motifs represented by existing RNAs in the literature and databases (NDB, http://ndbserver.rutgers.edu/NDB/index.html; 5S, http://rose.man.poznan.pl/5SData/) are shown in red; the missing motifs are in black. (B) Comparison of the number of tree graphs from Cayley and Harary–Prins enumeration formulas (equations 1 and 4). Also compared is the expected number of sequences in the random sequence space as a function of vertex number (dashed line); we estimated that ∼20 bases represent an edge in an RNA graph. The plots clearly show that selection of RNAs from random-sequence libraries is hampered by the rapidly rising sequence space with RNA size. In contrast, the number of RNA topologies is much smaller, especially with the Harary and Prins formula.
Survey of existing RNA topologies
The enumeration formulas above provide theoretical bounds on the number of possible RNA topologies for our graphical representations. To determine how many of these topologies are represented by natural RNAs, we survey existing RNA sequences and structures in public databases and the literature. Existing RNA structures can also be used to develop criteria for discriminating RNA-like from non-RNA motifs.
To describe RNA secondary motifs, we employ experimental secondary structure information where available, and 2D folding algorithms [e.g., MFOLD (54) and PKNOTS (18)] where necessary, to determine small RNA topologies from sequences. Such algorithms are expected to be reliable for small RNAs (<100 nt) (55); minor errors (e.g., in the size of stems, loops, bulges) in the predicted 2D structures should not affect our survey, which deals with global topological characteristics. Except for small RNAs (<100 nt), no effective prediction algorithms are available for folding pseudoknots from sequence. Thus we use published experimental structures for pseudoknots. Many of our experimental structures are RNAs in the NDB (see Table 1), which archives 3D RNA structures (2D motifs), sequences from 5S rRNA (http://rose.man.poznan.pl/5SData/) and PSEUDOBASE (http://www.bio.leidenuniv.nl/∼EBatenburg/PKB.html) for pseudoknots.
Existing RNA trees. We present our findings of the existing RNA trees together with missing trees for V < 8 in Figure 7. We found eight distinct RNA trees (red images) representing small RNAs (e.g., tRNA, 70S RNA, 5S rRNA, RNA in signal recognition and P5abc domain of group I intron); not shown is the large 23S rRNAs with V >> 8. Except for the smallest trees (V < 4), we immediately see that many distinct motifs are not found in RNA databases.
Specifically, the V = 2, 3 and 4 trees are represented by fragment or single strand RNAs. All three motifs for V = 5 are found: in tRNAs (NDB code TRNA12), P5abc domain and the 70S ribosome unit (NDB code RR0003). Only the RNA in signal-recognition complex NDB code PR0021 is represented in the set of six possible topologies for V = 6; only one of the total 11 motifs in the V = 7 set is represented, by 5S rRNA. Thus, while we find that several possible topologies are found in RNA databases, many others are not. As V increases, the number of possible trees increases rapidly and the number ‘missing’ motifs is expected to be larger. We are currently compiling such a topology database as a tool for cataloging, analyzing and identifying RNA sequences with similar topologies and/or functions (J.Zorn et al., unpublished).
Tree (and non-tree) motifs that correspond to real RNAs have a moderate degree of branching: the number of edges emanating from a vertex averages three or four, high-order junctions (more than five incident edges) found in large RNAs (e.g., 16S and 23S rRNA, see Fig. 9) (6,7). Branching promotes tertiary interactions between RNA secondary elements and reduces the entropic cost associated with folding into compact 3D structures. This advantage of branching also likely explains the absence of long ‘linear chain’ topologies. The rarity of high-order junctions may be explained by unfavorable energetic considerations due to geometric or steric factors. Their occurrence in large RNAs would thus require stabilization by special tertiary interactions in other parts of the molecule.
Figure 9.
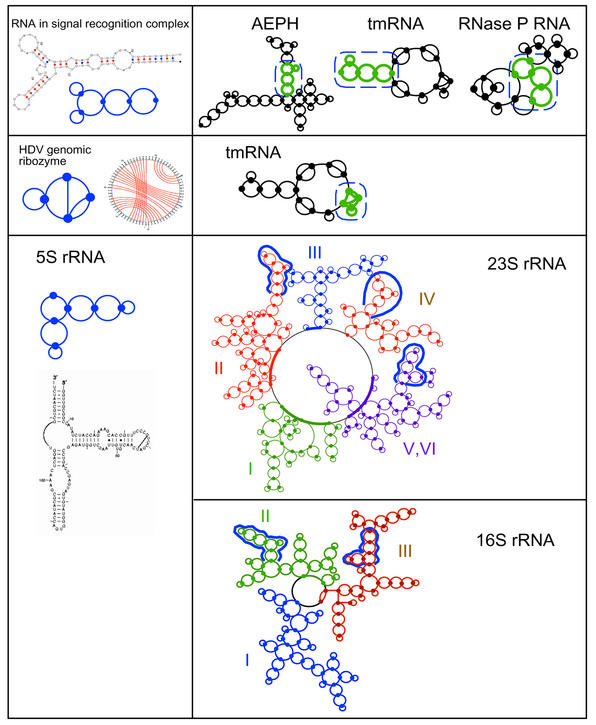
Identification of smaller RNA topologies (left panels) within larger RNA topologies (right panels, highlighted by thick green lines and broken/curly blue lines) using a graph similarity search algorithm (Appendix D). Left panels also show the secondary topologies of the probe RNAs. The probe RNAs are signal recognition complex RNA (NDB code PR0042), HDV ribozyme (NDB code PR0005), and 5S rRNA of S.cerevisiae; the larger RNA motifs (right panels) are aepH (GenBank accession no. S74077), tmRNA, RNase P RNA, and 16S and 23S rRNAs of S.cerevisiae. The 2D structures of signal recognition complex RNA and HDV ribozyme were predicted using the PKNOTS algorithm; the HDV fold differs slightly from crystal structure (64).
Existing RNA pseudoknots. We found a total of 22 distinct pseudoknot topologies in the literature and the PSEUDO BASE database. The topologies found are distributed as follows: nine for V = 2, 3, 4 (Fig. 5, red graphs); 12 for V = 5 – 18, 22 (Fig. 6, including D and F); and one for 16S rRNA pseudoknot (V = 87) (Fig. 9). The only pseudoknot topology for V = 2 is found in viral RNAs (e.g., actate dehydrogenase-elevating virus, strain C, Berne virus, potato leafroll virus, Porcine reproductive and respiratory syndrome virus). Two out of four possible pseudoknot topologies for V = 3 are found in pseudoknot of odontoglossum ringspot virus (PSEUDOBASE no. PKB28) and Neurospora VS ribozyme (PSEUDOBASE no. PKB178). For V = 4, we find six out of the 20 possible pseudoknot topologies, as shown in Figure 5.
Thus, for V < 5 the number of pseudoknots found in nature increases with the vertex number as predicted by our theoretical enumeration of topologies. Topology enumeration suggests there are many more pseudoknot motifs for V ≥ 5. However, our survey yields only six pseudoknots for V = 5 and one each for V = 6, 7, 16 and 17 (Fig. 6). This situation partly reflects our incomplete knowledge of pseudoknots and partly because many possible pseudoknots likely do not exist in nature.
Existing RNA bridges. Recall that bridge topologies are biologically interesting since they define modular units of RNAs that may be exploited for RNA design. Figure 6A–F displays six examples of naturally occurring RNA bridges with 4–22 vertices that we have identified. Among these RNAs, the HCV and group I intron are also pseudoknots (their pseudoknot substructures are shaded green in Fig. 6).
The four-vertex box H/ACA snoRNA motif in Figure 6A is the bridge graph (4,8) in Figure 5 (green); this snoRNA has a ACA trinucleotide and is involved in site selection for RNA modification by pseudouridine formation. Interestingly, the box H/ACA snoRNA motif is a subgraph of human telomerase (hTR) RNA (bases 211–451) in Figure 6B (shaded blue), and they have similar functional properties (56). The largest bridge graph is the group I intron (Fig. 6F); it has 22 vertices, four bridge edges and a pseudoknot subgraph (shaded green). Of the 30 enumerated dual graphs with four vertices (Fig. 5), there are 13 bridge graphs, seven of which have pseudoknot subgraphs. Thus, graphical enumeration alone suggests that naturally occurring bridge graphs with pseudoknots may not be rare.
Clustering functional RNA classes
Our topological characterization of RNAs provides an avenue for cataloging or classifying RNA structures. Understanding RNA topological characteristics can aid in identifying novel RNA-like topologies as candidates for RNA design. To describe the range of topological characteristics prevalent in natural RNAs—for example, degree of branching, ratio of loops to stems—we perform a ‘clustering analysis’ with dual graphs to span both RNA trees and pseudoknots. We define a simple topological characterization of RNA graphs using the number of vertices V and the number of ‘exterior’ loops T (including branches ending in loops and the chain ends) [Though our ‘exterior loops’ are called terminal loops in the graph theory literature, we avoid the latter since it may lead to confusion with RNA’s chemical terminal (5′ and 3′) ends.]; for example, the three RNA graphs in Figure 4 for an RNA single strand, tRNA and 5S rRNA have two, four and three exterior loops, respectively. For any dual RNA graph, T/V ≤ 1. Since we can derive the relation V = S = L + J from Euler’s formula, where S, L and J are, respectively, stem, loop/bulge and junction numbers (equation 10 in Appendix C), and the set of exterior loops (T) is a subset of all loops (L), i.e., T ≤ L, the following inequality holds:
This inequality defines the range of T/V [or L/(L + J)] in which RNA topologies can be found, but it does not provide information about the distribution of natural RNAs within the domain.
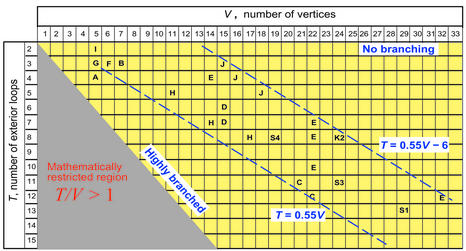
Figure 8 shows a range of T/V combinations with filled elements corresponding to existing RNAs. This map shows the distribution or clustering of functional RNA classes according to their 2D topological characteristics. For RNAs in 5 ≤ V ≤ 32, we find that the T and V values lie in a narrow range bounded by two lines of slope 0.55 (T has a range of 6 between the lines). This limited range of T, V values for real RNAs shows that RNAs tend to have a moderate degree of branching; the constant T = 2 value corresponds to topologies with no branching, and T/V ∼ 1 indicates highly branched topologies. RNAs with a high degree of branching are rare, except for the small tRNA with T/V = 0.8; substructures of large RNAs can also exhibit high branching structures. The 5S rRNA, for example, has a low branching ratio of T/V = 0.43.
Figure 8.
A 2D clustering map of functional RNA classes using their topological characteristics. Every RNA dual graph (tree, pseudoknot or bridge) has a fixed number of vertices (V) and exterior (or terminal) loops T; we also count a helix ending in 3′ and 5′ ends as an exterior loop. We use V versus T plot to map various functional RNA classes denoted by the letters: A—tRNA; B—5S rRNA; C—group I introns; D—LSU rRNA; E—noncoding RNAs: Ecc aepH, U19H, NTENOD40, BC200, U17HG; F—RNA in signal recognition complex; G—P5abc domain; H—RNase P RNA; I—70S (F) RNA; J—tmRNA; K2—domain II of 16S rRNA; and S1, S3, S4 are domains I, III and IV, respectively, of 23S rRNAs. No topologies are mathematically allowed in the T/V > 1 (shaded) region because the number of exterior loops exceeds the number of vertices. The dashed lines define the boundaries within which functional RNAs are found. The topologies for RNAs (A, B, …, S3, S4) are obtained from experimental structures in the literature, except for the small 70S (F) RNA (denoted by letter I) which is predicted using the MFOLD algorithm. Topologies with T = 2 are unbranched, whereas those with the T/V values close to 1 are highly branched.
Within the observed range of T and V values (Fig. 8), there are still many topologies with no RNA representation. These topologies may represent possible RNA structures, some of which are listed in our enumerated graphs (Figs 5 and 7). Thus, the missing RNA topologies suggest a new way to predict novel RNAs through RNA design and folding (see section below), with more promising candidate motifs lying within the typical range. Ideally, we would like to rank all possible topologies for a given V in subgroups (e.g., starshaped trees) and provide the elements in increasing likelihood to be natural RNAs. Our recent analysis of the typical ratio of total unpaired to paired bases (0.75) and the fraction of bases in stems, junctions, hairpin loops, and bulges/internal loops based on ribosome structure (J. Zorn et al., unpublished) may be useful in this regard. We are pursuing experimental/theoretical collaborations to exploit these RNA design proposals.
RNA substructure analysis
Structural similarity can occur between RNA substructures due to their common evolutionary origin. A known example is the occurrence of smaller snoRNA motifs within the larger hTR RNA structure, indicating a functional relation between these RNAs (56). In proteins, the development of algorithms for finding 3D substructure similarity has led to discoveries of novel functional relationships and structural classification of proteins (57). However, efficient techniques for finding structural similarity between RNAs are not well developed.
The topological framework described here offers a systematic way to search for similar substructures/submotifs in RNAs through the concept of graph isomorphism; isomorphic graphs are structurally equivalent graphs, i.e., those having the same pattern of connectivity between vertices. Similarity search techniques using graph isomorphism have already been used for establishing relationships among chemical compounds (45) and 2D RNA tree motifs (31). This approach may help also identify structural, functional and evolutionary relationships among RNAs that are not easily achieved by other methods (e.g., sequence alignment). Below, we summarize the concepts involved in the RNA graph comparison algorithm we have developed and illustrate its applications with several examples that reveal occurrences of RNAs within larger RNAs. The algorithm is sketched in Appendix D and detailed separately (S.Pasquali, H.H.Gan and T.Schlick, unpublished).
The mathematical task involves identifying a graph as a subgraph of a larger graph or, in biological terms, an existing RNA motif contained in a larger RNA. The computational complexity of identifying two structurally equivalent (i.e., isomorphic) graphs with V vertices is directly related to the number of ways the graph vertices can be labeled, which is of the order of V factorial (V!). Thus, the brute force method for finding isomorphic graphs is prohibitive except for small graphs (<10 vertices or RNAs with <200 nt). This is known as the graph isomorphism problem (46). We have developed an efficient method for testing graph isomorphisms based on graph topological numbers or invariants, as elaborated in Appendix D. Essentially, we associate each graph or subgraph with one or more topological numbers, which are computed based on the patterns of connectivity among graph vertices. By this method, isomorphic graphs have the same topological numbers while dissimilar graphs have different topological numbers. The similarity or dissimilarity between graphs can be thus established by comparing their topological numbers.
Our computational scheme can compare graphs having up to 40 vertices, or roughly corresponding to 840 nt RNAs. To date, the largest RNA graph known, i.e., 23S rRNA, has ∼160 vertices (3200 nt). Since the 23S rRNA is made of six domains (7,6), its average domain has ∼28 graph vertices (or ∼560 nt). Thus, a comprehensive search of topological similarities among all major RNA classes can be performed by using whole RNAs and RNA domains. Below, we present several examples of the occurrence of RNA motifs within larger RNAs found using such a computational search method.
Figure 9 shows occurrences of our three selected small probe RNA topologies (<100)—signal recognition RNA, 5S rRNA and HDV ribozyme—within larger target RNA topologies: aepH, tmRNA, RNase P RNA, 16S rRNA and 23S rRNA. The search was performed using the general RNA dual graph representation.
We find that the 2D RNA motif of our first probe, the five-vertex signal recognition complex (NDB code PR0042), occurs within the structures of aepH, tmRNA and RNase P RNA; these RNAs are functionally different from each other and from the probe RNA. In these matches, the probe RNA and the substructures of larger RNAs have the same graph connectivity, but the end (terminal) loops of stems may differ because these loops do not contribute to our topological number (see Appendix D). Further analyses by pair sequence alignments between the probe and matched substructures of larger RNAs do not reveal any significant sequence similarity; since the sequence identity is ∼30 to 40%, these 2D structural similarities may not be expected based on sequence comparison. These matches between small RNA motifs and RNA subdomains are likely due to general architectural similarity.
The search for the occurrence of our second probe, 5S rRNA, in the ribosomal 16S and 23S RNAs of Saccharomyces cerevisiae (U53879, http://www.rna.icmb.utexas.edu), employed a partitioning of the 16S and 23S rRNAs into three and five domains, respectively, roughly according to domains defined in the literature (6,7). The six-vertex 5S rRNA motif is found in two of the three domains of 16S rRNA as well as three of the five domains of 23S rRNA. These findings indicate that the 5S rRNA is a common motif in the substructures of the 16S and 23S ribosomal RNAs.
As with alignment of 3D protein structures (57), the significance of the matches found in RNA topologies increases with the size of RNA fragments or RNA graphs matched. The probes (signal recognition RNA and 5S rRNA) are small RNAs with simple topologies; each has a three-stem junction. We expect the significance of the matches found to also depend on the complexity of the RNA secondary topology. For example, a match involving a rare RNA topology is likely to be significant from a functional and evolutionary perspective. Ultimately, a statistical score quantifying the degree of significance is required. Such a score should incorporate information about RNA size and topological complexity.
Surprisingly, for our third probe, HDV ribozyme, we find a match motif within the pseudoknot substructure of tmRNA; our search among other RNA pseudoknots did not yield any positive occurrence. Although the HDV ribozyme is a small RNA (a 70 nt four-vertex graph), its topology is more complex than the simplest pseudoknot. The simplest pseudoknot is represented by two graph vertices and three edges (see Fig. 2), whereas the HDV ribozyme has four vertices and six edges, excluding the end loop (see Fig. 9). A sequence alignment of the matched RNAs demonstrates a low sequence identity of ∼35%. However, sequence alignment (not shown) indicates that three base paired regions of the HDV ribozyme and tmRNA have sequence segments that match perfectly, which strongly suggests a close structural, functional or evolutionary relationship between these RNAs. Our identification of HDV ribozyme motif in tmRNA warrants further experimental and theoretical analysis.
The positive matches found by our graph isomorphism technique could be a useful first step in the analysis of RNA structural and functional similarity since such similarity is often not revealed by sequence comparison. We will report this analysis of RNA substructures separately (S.Pasquali, H.H.Gan and T.Schlick, unpublished).
Search for novel RNAs: design and prediction
Graphical enumeration demonstrates that the RNA topology space is immensely smaller than the nucleic acid sequence space, which makes the search for novel RNAs plausible. Moreover, our survey and clustering analysis of existing RNAs help narrow this search by indicating the probable RNA topological characteristics (Fig. 8). However, how can we apply these tools and analyses to design RNAs in practice? The gap between candidate topologies and sequence proposals is clearly large, not to mention prediction of their 3D structures and functional properties.
We offer the following strategy for designing tree topologies not found in RNA databases as candidate templates for the design of RNA sequences. The hierarchical or modular nature of RNAs can be exploited for determining the sequences that lead to target motifs by assembling fragments or ‘building blocks’ from existing RNAs; the outcome of this assembly can be tested to a first approximation using secondary folding algorithms. Ultimately, the structures must be determined experimentally.
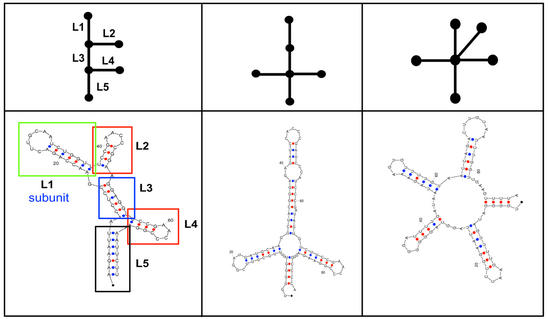
We illustrate our design approach in Figure 10 with three examples of designed sequences that fold to the novel 2D topologies we targeted. These targets originate from the missing motifs of V = 6 (see Fig. 7). The designed sequences by modular assembly are then folded using Zuker’s MFOLD algorithm (32). In fact, the designed sequences yield the target topologies with the lowest free energy. The sequence lengths of the three designed sequences only vary between 75 and 100 nt, but they have distinct topological characteristics: two three-stem junctions, four-stem junction and five-stem junction. Other design experiments in our laboratory suggest that determining the sequence from the target motif by modular library design is not dificult as judged by results of 2D folding algorithms. Of course, we cannot yet comment on the resulting tertiary structures at this stage, but this clearly represents a future goal. Work is underway to test these proposals by instrumentation. In particular, we propose that the combination of graph theory, sequence design protocol and in vitro selection is potentially a productive approach for finding novel RNAs.
Figure 10.
Three examples of novel 2D topologies with six vertices that have been constructed using a modular design approach from existing RNA fragments/submotifs. The target topologies (upper panels) and their designed 2D folds (lower panels) at the lowest energy states are shown. In the left panel, the 2D fold has five basic fragments labeled L1, …, L5 which were taken from tRNAs of various organisms. We used MFOLD to generate the 2D RNA folds.
SUMMARY AND CONCLUSIONS
We have developed two graphical representations of RNA secondary structures to allow exploration of RNA’s structural repertoire. We use tree graphs for representing RNA tree structures and the more general dual graphs for representing both RNA trees and pseudoknots. Such graphical representations provide a basis for enumerating, classifying, comparing and designing RNA motifs.
We estimate the number of distinct RNA tree motifs based on the Cayley and Harary–Prins enumeration theorems. These theorems imply that the RNA topology space is much smaller than the sequence space, which renders our topological approach potentially effective for finding novel RNAs. Our surveys of existing RNAs identified a number of motifs in nature (Figs 5–7) but showed that many hypothetical motifs do not exist. Since not all enumerated motifs are probable RNAs, energetic, functional and evolutionary aspects of RNA folds must be taken into consideration to provide better future estimates of RNA’s repertoire.
The graph theory approach for RNA trees and pseudoknots also aids in the search for structural similarity between RNAs using the concept of graph isomorphism. Significantly, we found many occurrences of the 5S rRNA motif in several large RNAs (including 16S and 23S rRNAs) and the HDV motif in tmRNA (Fig. 9). Such structural analyses may assist in the identification of functional similarity between RNAs.
The search for novel RNAs represents another intriguing area of application of graph theory. We suggest the promise of graph theory for the design of novel RNAs by showing that missing 2D motifs can be designed by modular assembly of existing RNAs’ subunits in combination with 2D folding algorithms. This design protocol is likely to be effective when the designed motifs have the topological characteristics of natural RNAs, i.e., in the range between highly branched and non-branched RNA structures (see Fig. 8) and contain a typical distribution of paired and unpaired bases among stems, loops, bulges and junctions as we recently estimated based on large ribosomal RNAs (J. Zorn et al., unpublished). However, the problem of bridging the gap between 2D and 3D structures remains a challenge for future investigations (58).
To integrate our efforts of cataloguing, comparing, and predicting RNA structures, we are currently building a database for archiving naturally occurring and hypothetical RNA motifs. We hope that the database will be exploited for topology searches, functional annotation of RNA sequences and RNA design. We invite interested researchers to contact us with suggestions and further information.
SUPPLEMENTARY MATERIAL
A glossary that defines various terms in RNA structure and graph theory is available as Supplementary Material at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Dinshaw J. Patel and Eric Westhof for constructive comments on aspects of this work. We are grateful to undergraduates Julie Zorn, Daniela Fera and Uri Laserson, and Michael Tang and Nahum Schiffeldrim for their assistance in our initiative; we also thank Dr Daniel Strahs for preparation of Figure 2. We are indebted to the generous support of the research, including database construction, by a Joint NSF/NIGMS Initiative to Support Research in the Area of Mathematical Biology (DMS-0201160). T.S. is an investigator of the Howard Hughes Medical Institute.
APPENDIX
A. Relationship between tree and dual graphs
RNA tree structures can be represented as either tree or dual graphs (see examples in Fig. 4); however, only dual graphs can represent pseudoknots. Here, we elaborate on the relation between these two graphical representations of RNA tree structures. For this purpose, we compare in Table 2 the tree and dual graph rules for vertices and edges.
As shown, a vertex in a tree graph represents an RNA loop/bulge/junction/ends, whereas it represents in a dual graph a double-helical stem; similarly, an edge in dual graphs (e.g., ∩) denotes a connecting strand between stems at loop/bulge/junction region, whereas an edge (—) in tree graphs is a double-helical stem. (Since these different meanings for the graph vertices and edges in the tree and dual graph representations may be a potential source of confusion, it is important to treat these two schemes for representing RNAs separately.) Another difference is that the 3′ and 5′ ends are not represented as graphical elements in dual graphs but are a vertex in tree graphs. This means that dual graphs can only be converted into tree graphs with loss information about the locations of the ends. In this respect, the dual representation of RNA is more precise than the tree representation. Another difference between these representations is that a V-vertex tree becomes a (V –1)-vertex dual graph; for example, a five-vertex tRNA tree becomes a four-vertex dual graph in the dual representation (see Fig. 4). Recall that although the dual representation suffices for RNA structures, the major advantage of the tree representation is that many mathematical results in graph theory can be exploited for the characterization, enumeration and construction of trees (46).
B. Limitations of RNA graphical representations
The tree and dual graph rules (T1–T4 and D1–D3) reduce RNA secondary structures to graphical objects in which only the connectivity of the secondary elements is specified. Such schematic representations necessarily retain only minimal information about the length of stems, the size of junctions and the number of unmatched nucleotides in loops and bulges; these are non-topological aspects of RNA. Still, the average dimensions of corresponding secondary elements can be estimated based on known RNA structures. For tree representations, there are ∼20 nt/edge, and the average size of an RNA graph can be estimated on that basis. For example, a small four-edge tRNA graph has ∼80 nt. This estimate becomes more accurate for larger RNAs.
Another issue is the accuracy of our graphical representations. Subtle differences in RNA secondary topologies can sometimes lead to the same dual graph. For example, Figure 2 shows the same dual graphs (column F) for two pairs of RNA examples (rows 1, 2 and 3, 4 of column E). RNA examples in rows 1 and 2 involve the hairpin-like structure representing two single-helical stem RNAs with different topological connectivities. Examples in rows 3 and 4 involve the three-vertex graph representing a three-stem junction and a secondary topology with three consecutive stems. The ambiguity in representing RNA topologies can be resolved by using directed graphs or digraphs. [Digraphs have been used for modeling food web of an ecosystem, Markov processes and many other applications involving analysis of networks (46)].
Digraphs remove topological ambiguity by specifying the direction of each edge. By representing the RNA examples in Figure 2 as digraphs (column G), the topologies of the second RNA pair (rows 3, 4 of column E) can be distinguished; however, the first pair (rows 1, 2 of column E) remains indistinguishable, implying that more detailed aspects of their structures need to be modeled to differentiate their topologies. Since the digraphs contain more information than simple dual graphs, the number of digraphs grows more rapidly with vertex number (47) compared with dual graphs, making it essential to use computer algorithms to analyze such graphs. Clearly, RNA secondary structures can be represented using various graphical representations depending on the level of specificity required. Such representations are worth exploring in future investigations.
C. Algebraic properties of RNA topologies
RNA bridges, trees and pseudoknots are special classes of 2D graphs or networks. For any 2D network, the number of vertices (V), number of edges (E) and number of faces (F) (or cycle rank) are related by Euler’s formula in graph theory (47). Indeed, by Euler’s formula, which is a basis of algebraic topological graph theory, we have the simple relation
V – E + F = 1.7
For polyhedrons, Euler’s formula becomes V – E + F = 2. We now translate this important relationship to RNA features known to structural biologists. Specifically, we use equation 7 to relate the number of loops/bulges (L), junctions (J) and stems (S) for any RNA secondary structure.
Since tree graphs have no faces (F = 0), we can simplify equation 7 to read
E = V – 1.8
The secondary elements of RNA trees (L, J and S) are related to vertices (V) and edges (E) by the formulas
S = E,
V = L + J + 1,9
since stems (S) are edges (E) and loops/bulges (L) and junctions (J) are vertices (V), and the chain ends are considered a vertex (add 1 to V). We thus combine equations 8 and 9 to relate the loop/bulge, junction and stem numbers of RNA trees as:
L + J = S.10
How does this relation hold for existing RNAs? Our survey of RNA secondary structures (i.e., tRNA, RNase and 5S, 16S, 23S rRNAs) shows that the ratio (L + J)/S is indeed unity for each RNA, implying that equation 10 accurately describes RNA tree structures. Figure 4 illustrates three secondary structures of single-strand RNA, tRNA and 5S rRNA obeying formula 10. For example, the tRNA molecule is described by L = 3 loops, J = 1 junction and S = 4 stems, as their sum.
We now show that equation 10 also holds for RNA dual graphs. Recall that for dual graphs (see dual graph rules section)
E = 2V –1.11
Substituting this relation into equation 7 yields
F = V.12
Equations 11 and 12 form the defining relations of RNA dual graphs. For RNA dual graphs, the following relations hold
V = S,13
F = L + J14
because stems (S) are vertices (V) and loops/junctions (L, J) are faces (F). Equations 12 and 13 imply that equation 10 is also true for RNA dual graphs. Equation 10 thus relates the secondary elements of any 2D RNA topology. It is useful for defining the number of loops, junctions or stems when one of these numbers is missing.
D. An algorithm for finding structurally similar graphs
Here we introduce topological invariants for graphs and use them to test structural similarity or isomorphism of RNA graphs, which is the basis of our computational scheme for finding RNA motifs within larger RNAs. Topological invariants are numbers that contain information about the connectivity of graphs; two identical graphs have the same topological invariants. To calculate topological invariants, we use the adjacency matrix to quantitatively represent graph connectivity (46). An adjacency matrix, whose columns and rows correspond to vertex labels of the graph, specifies the connectivity between graph vertices. For example, if the i and j vertices of a graph are connected by two edges, then the (i, j) element of the matrix is 2.
Our general procedure for calculating the topological number of a graph is to decompose the graph into all possible two, three, four-vertex, etc. configurations in a manner similar to atomic interactions in physics and chemistry. For each graph, we associate the topological numbers S2, S3, S4, … for two, three, four-vertex, etc. configurations; the topological numbers are calculated using the adjacency matrix, as elaborated below. Isomorphic (structurally equivalent) graphs have the same topological numbers S2, S3, S4, …. In practice, we only consider low-order topological numbers S2, S3 and S4 since these are more easily computed than higher-order ones.
We define S2, for example, as follows:
where εij is the ‘coupling’ parameter between the vertices i and j. We set εij = dij where dij is the number of edges separating vertices i, j. On the other hand, if the vertices i, j are connected by more than one edge, we set εij = 1/cij where cij is the number of connecting edges between the vertices. For i = j, we assign a zero coupling value, i.e., εii = 0, which implies that terminal loops do not contribute to the topological invariant. The above procedures can be generalized to compute S3, S4 and so on.
We test the similarity between RNA structures or substructures by comparing their topological numbers. In most cases comparing S2 values is a suficient test, but subtler dissimilarities between graphs require higher-order topological numbers to discriminate.
By using graph topological numbers to test the similarity or dissimilarity between RNA structures, we reduce the computational cost from about N! to
where N1 and N2 (< N1) are sizes of the (square) adjacency matrices of the two graphs compared. This computational cost is associated with the number of ways of constructing N2 × N2 submatrices within the larger N1 × N1 matrix. Assuming the worst case situation when N2 = N1/2, our computational scheme can compare graphs having up to 40 vertices, which correspond to ∼800 nt RNA.
REFERENCES
- 1.Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 2.Storz G. (2002) An expanding universe of noncoding RNAs. Science, 296, 1260–1263. [DOI] [PubMed] [Google Scholar]
- 3.Schlick T. (2002) Molecular Modeling and Simulation: An Interdisciplinary Guide. Springer-Verlag, New York, NY.
- 4.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 5.Doudna J.A. and Cech,T.R. (2002) The chemical repertoire of natural ribozymes. Nature, 418, 222–228. [DOI] [PubMed] [Google Scholar]
- 6.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 7.Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,N.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 8.Harms J., Schluenzen,F., Zarivach,R., Bashan,A., Gat,S., Agmon,I., Bartels,H., Franceschi,F. and Yonath,A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 292–302. [DOI] [PubMed] [Google Scholar]
- 9.Bourdeau V., Ferbeyre,G., Pageau,M., Paquin,B. and Cedergren,R. (1999) The distribution of RNA motifs in natural sequences. Nucleic Acids Res., 27, 4457–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doudna J. (2000) Structural genomics of RNA. Nature Struct. Biol. (Structural Genomics suppl.), 7, 954–956. [DOI] [PubMed] [Google Scholar]
- 11.Al-Hashimi H.M., Gorin,A., Majumdar,A., Gosser,Y. and Patel,D.J. (2002) Towards structural genomics of RNA: rapid NMR resonance assignment and simultaneous RNA tertiary structure determination using residual dipolar couplings. J. Mol. Biol., 318, 637–649. [DOI] [PubMed] [Google Scholar]
- 12.Doudna J.A. (1997) A molecular contortionist. Nature, 388, 830–831. [DOI] [PubMed] [Google Scholar]
- 13.Berman H.M., Olson,W.K., Beveridge,D.L., Westbrook,J., Gelbin,A., Demeny,T., Hsieh,S.H., Srinivasan,A.R. and Schneider,B. (1992) The Nucleic Acid Database: A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J., 63, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas E., Klein,R.J., Jones,T.A. and Eddy,S.R. (2001) Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol., 11, 1369–1373. [DOI] [PubMed] [Google Scholar]
- 16.Carter R.J., Dubchak,I. and Holbrook,S. (2001) A computational approach to identity genes for functional RNAs in genomic sequences. Nucleic Acids Res., 29, 3928–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Mabrouk N. and Lisacek,F. (1996) A very fast identification of RNA motifs in genomic DNA. Application to tRNA in the yeast genome. J. Mol. Biol., 264, 46–55. [DOI] [PubMed] [Google Scholar]
- 18.Rivas E. and Eddy,S.R. (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol., 285, 2053–2068. [DOI] [PubMed] [Google Scholar]
- 19.Tinoco I. Jr and Bustamante,C. (1999) How RNA folds. J. Mol. Biol., 293, 271–281. [DOI] [PubMed] [Google Scholar]
- 20.Soukup G.A. and Breaker,R.R. (2000) Allosteric nucleic acid catalysts. Curr. Opin. Struct. Biol., 10, 318–325. [DOI] [PubMed] [Google Scholar]
- 21.Ellington A.D. and Szostak,J.W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 22.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 23.Wilson D.S. and Szostak,J.W. (1999) In vitro selection of nucleic acids. Annu. Rev. Biochem., 68, 611–647. [DOI] [PubMed] [Google Scholar]
- 24.Soukup G.A. and Breaker,R.R. (1999) Nucleic acid molecular switches. Trends Biotechnol., 17, 469–476. [DOI] [PubMed] [Google Scholar]
- 25.Tang J.G. and Breaker,R.R. (2000) Structural diversity of self-cleaving ribozymes. Proc. Natl Acad. Sci. USA, 97, 5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breaker R.R. and Joyce,G.F. (1994) Inventing and improving ribozyme function: rational design versus iterative selection methods. Trends Biotechnol., 12, 268–275. [DOI] [PubMed] [Google Scholar]
- 27.Hermann T. and Patel,D.J. (2000) Adaptive recognition by nucleic acid aptamers. Science, 287, 820–825. [DOI] [PubMed] [Google Scholar]
- 28.Soukup G.A. and Breaker,R.R. (1999) Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA, 96, 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray D. (2001) Reasoning for results. Nature, 412, 863. [DOI] [PubMed] [Google Scholar]
- 30.Le S.Y., Nussinov,R. and Maizel,J.V. (1989) Tree graphs of RNA secondary structures and their comparisons. Comput. Biomed. Res., 22, 461–473. [DOI] [PubMed] [Google Scholar]
- 31.Benedetti G. and Morosetti,S. (1996) A graph-topological approach to recognition of pattern and similarity in RNA secondary structures. Biol. Chem., 59, 197–184. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology. NATO ASI Series, Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 11–43.
- 33.Burkhard M.E., Turner,D.H. and Tinoco,I.,Jr (1999) The interactions that shape RNA structure. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. New York, NY, pp. 233–264.
- 34.Hermann T. and Patel,D.J. (1999) Stitching together RNA tertiary architectures. J. Mol. Biol., 294, 829–849. [DOI] [PubMed] [Google Scholar]
- 35.Lilley D.M.J. (1998) Folding of branched RNA species. Biopolymers, 48, 101–112. [DOI] [PubMed] [Google Scholar]
- 36.Moore P.B. (1999) Structural motifs in RNA. Annu. Rev. Biochem., 68, 287–300. [DOI] [PubMed] [Google Scholar]
- 37.Thirumalai D., Lee,N., Woodson,S.A. and Klimov,D.K. (2001) Early events in RNA folding. Annu. Rev. Phys. Chem., 52, 751–762. [DOI] [PubMed] [Google Scholar]
- 38.Pace N.R., Thomas,B.C. and Woese,C.R. (1999) Probing RNA structure, function, and history by comparative analysis. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, pp. 113–141.
- 39.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 40.McCaskill J.S. (1990) The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers, 29, 1105–1119. [DOI] [PubMed] [Google Scholar]
- 41.Wuchty S., Fontana,W., Hofacker,I.L. and Schuster,P. (1999) Complete suboptimal of RNA and the stability of secondary structures. Biopolymers, 49, 145–165. [DOI] [PubMed] [Google Scholar]
- 42.Milo R., Shen-Orr,S., Itzkovitz,S., Kashtan,N., Chklovskii,D. and Alon,U. (2002) Network motifs: simple building blocks of complex networks. Science, 298, 549–564. [DOI] [PubMed] [Google Scholar]
- 43.Albert R. and Barabasi,A.-L. (2002) Statistical mechanics of complex networks. Rev. Mod. Phys., 74, 47–97. [Google Scholar]
- 44.Bermudez C.I., Daza,E.E. and Andrade,E. (1999) Characterization and comparison of Escherichia coli transfer RNAs by graph theory based on secondary structure. J. Theor. Biol., 197, 193–205. [DOI] [PubMed] [Google Scholar]
- 45.Mihalic Z. and Trinajstic,N. (1992) A graph-theoretic approach to structure-property relationships. J. Chem. Educ., 69, 701–712. [Google Scholar]
- 46.Gross J. and Yellen,J. (1999) Graph Theory and Its Applications. CRC Press, Boca Raton.
- 47.Harary F. (1969) Graph Theory. Addison-Wesley, Reading, MA.
- 48.Dinman J.D., Richter,S., Plant,E.P., Taylor,R.C., Hammell,A.B. and Rana,T.M. (2002) The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl Acad. Sci. USA, 99, 5331–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arya D.P., Coffee.R.L.,Jr, Willis,B. and Abramovitch,A.I. (2001) Aminoglycosidenucleic acid interactions: remarkable stabilization of DNA and RNA triple helices by neomycin. J. Am. Chem. Soc., 123, 5385–3595. [DOI] [PubMed] [Google Scholar]
- 50.Harary F. and Prins,G. (1959) The number of homeomorphically irreducible trees and other species. Acta Math., 101, 141–162. [Google Scholar]
- 51.Pölya G. (1973) Kombinatorische anzahlbestimmungen fur gruppen, graphen und cheische verbindungen. Acta Math., 68, 145–254. [Google Scholar]
- 52.Burnside W. (1911) Theory of Groups of Finite Order, 2nd Edn. Cambridge University Press, Cambridge, UK.
- 53.Reidys C., Stadler,P.F. and Schuster,P. (1997) Generic properties of combinatory maps: neutral networks of RNA secondary structures. Bull. Math. Biol., 59, 339–397. [DOI] [PubMed] [Google Scholar]
- 54.Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]
- 55.Zuker M. and Jacobson,A.B. (1995) Well-determined regions in RNA secondary structure prediction: analysis of small subunit ribosomal RNA. Nucleic Acids Res., 23, 2791–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell J.R., Cheng,J. and Collins,K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–603. [DOI] [PubMed] [Google Scholar]
- 58.Moore P.B. (1999) The RNA folding problem. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, pp. 381–401.
- 59.Schultes E.A. and Bartel,D.B. (2000) One sequence, two ribozymes: implications for the emergence of new ribozyme folds. Science, 289, 448–452. [DOI] [PubMed] [Google Scholar]
- 60.Devlin K.J. (1999) Mathematics – The New Golden Age. Columbia University Press, New York, NY.
- 61.Felden B., Massire,C., Westhof,E., Atkins,J.F. and Gesteland,R.F. (2001) Phylogenetic analysis of tmRNA genes within a bacterial subgroup reveals a specific structural signature. Nucleic Acids Res., 29, 1602–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown J.W. (1999) The Ribonuclease P Database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 64.Ferré-D’Amaré A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.