Abstract
RecQ DNA helicases function in DNA replication, recombination and repair. Although the precise cellular roles played by this family of enzymes remain elusive, the importance of RecQ proteins is clear; mutations in any of three human RecQ genes lead to genomic instability and cancer. In this report, proteolysis is used to define a two-domain structure for Escherichia coli RecQ, revealing a large (∼59 kDa) N-terminal and a small (∼9 kDa) C-terminal domain. A short N-terminal segment (7 or 21 residues) is also shown to be sensitive to proteases. The effects of removing these regions of RecQ are tested in vitro. Removing 21 N-terminal residues from RecQ severely diminishes its DNA-dependent ATPase and helicase activities, but does not affect its ability to bind DNA in electrophoretic mobility shift assays. In contrast, removing the ∼9 kDa C-terminal domain from RecQ results in a fragment with normal levels of ATPase and helicase activity, but that has lost the ability to stably associate with DNA. These results establish the biochemical roles of an N-terminal sequence motif in RecQ catalytic function and for the C-terminal RecQ domain in stable DNA binding.
INTRODUCTION
RecQ DNA helicases play important and broad roles in cellular genome maintenance reactions (reviewed in 1,2). In Escherichia coli, RecQ acts in the RecF recombination pathway (3) which functions in plasmid recombination (4,5) and in helping bacteria recover from replication fork stalling at sites of ultraviolet light-induced DNA damage (6–11). Escherichia coli RecQ is also vital for suppression of illegitimate recombination or recombination between sequences with limited homology (12–14). In eukaryotes, RecQ proteins function in several cellular processes, including aging (15–17), silencing (18) and DNA replication, recombination and repair (1,2). The importance of RecQ function in cells is best exemplified by three human syndromes, Bloom’s, Werner’s and Rothmund–Thompson syndromes, which arise from mutation of human recQ genes, BLM, WRN or RECQ4, respectively (15,16,19). These mutations result in genomic instability (e.g. increased chromosomal rearrangements, chromosome breakage and/or hyper-recombination) and a predisposition to several cancers in all three syndromes. Since RecQ proteins play critical roles in cell function, a mechanistic understanding of their biochemical activities is of fundamental biological importance. However, the structural basis of RecQ family function has not been well defined.
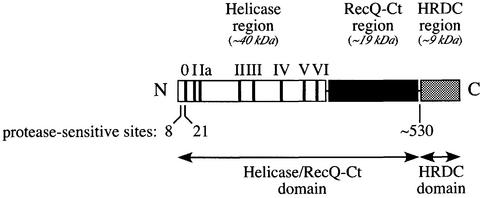
RecQ family members share three regions of conserved protein sequence referred to as the Helicase, RecQ-conserved (RecQ-Ct) and Helicase-and-RNase-D-C-terminal (HRDC) domains (20) (Fig. 1). The helicase region contains sequence motifs necessary for using the energy of ATP binding and hydrolysis to unwind DNA (21), and several RecQ proteins [Human WRN (22), BLM (23) and RecQL (24), Saccharomyces cerevisiae Sgs1 (25,26), Schizocaccharomyces pombe Rqh1 (27), Drosophila melanogaster RecQ5 (28,29), Xenopus laveis FFA-1 (30) and E.coli RecQ (31)] have been shown to act as ATP-dependent DNA helicases in vitro. Moreover, helicase activity is known to be important for the cellular functions of eukaryotic RecQ proteins. Five individual missense mutations that lead to Bloom’s syndrome result in sequence changes in the helicase region of the human BLM protein (19,32,33). Two of these mutations have been recapitulated in the murine BLM protein resulting in variants that lack ATPase and helicase activities in vitro (34). In addition, a point mutation that inactivates helicase activity in the S.cerevisiae RecQ homolog, Sgs1, leads to hypersensitivity to DNA damaging agents in the mutant cells that mimics the SGS1 null phenotype (35,36). Thus, helicase activity appears to be important for RecQ cellular functions.
Figure 1.
Schematic diagram of E.coli RecQ. Conserved regions, Helicase (white), RecQ-Ct (black) and HRDC (gray) (20) are shown as boxes. The locations of the conserved helicase motifs (I–VI) and a proteolytically labile motif identified in this study (motif 0) are indicated with bars in the Helicase box. The approximate molecular weight of each region is listed. Protease-sensitive sites and structural domains identified in this study are indicated below the schematic.
The RecQ-Ct region lays C-terminal to the helicase segment in RecQ proteins (20). Very little is known about the structure and function of the RecQ-Ct region in RecQ proteins, but mutational analyses have demonstrated its importance for activity in several RecQ homologs (19,34,37–40). Two point mutations that lead to Bloom’s syndrome alter conserved cysteines in the RecQ-Ct region of human BLM protein (19,37). Similarly to the BLM helicase mutations, analogous RecQ-Ct mutations made in murine BLM variants result in the loss of ATPase and helicase activity (34), suggesting the RecQ-Ct region could play an integral role in RecQ enzymatic activity.
The third region of homology in RecQ enzymes forms its C-terminal conserved element, called the HRDC region (20). This region is homologous with a domain of the RNase D family of nucleases but does not include the nuclease domain in RNase D. Since the HRDC region is present in two enzyme families involved in nucleic acid metabolism, it has been proposed to be involved in nucleic acid binding, the only known function in common between RecQ and RNase D (20). Hypersensitivity of S.cerevisiae SGS1-deletion strains to DNA damaging agents can be complemented equally well by either full-length Sgs1 or a C-terminally truncated variant lacking the HRDC region (36,41). Also, a recombinant fragment of Sgs1 lacking the HRDC region is active as an ATPase and DNA helicase in vitro (25,26). In contrast, an Sgs1 variant lacking the HRDC domain does not complement the aberrant growth phenotypes in strains lacking both SGS1 and either topoisomerase I or III, implying that this region is required for some cellular functions (25,36). Thus, the role of the HRDC domain in RecQ proteins remains unclear.
Although genetic and biochemical analyses have identified important regions within the RecQ family of proteins, connections linking RecQ protein structure to its biochemical functions have not been well defined. For example, whether regions of conserved sequence with the RecQ family correspond to true structural domains and how these domains cooperate in catalysis are important questions that have not been widely explored. In this report, we use limited proteolysis as an unbiased structural probe to define the domain structure of purified E.coli RecQ. Our results reveal a two-domain structure for E.coli RecQ, with a composite Helicase/RecQ-Ct N-terminal domain and a C-terminal HRDC domain. In addition, 21 residues at the N-terminus of the Helicase/RecQ-Ct domain were found to be sensitive to protease cleavage. Using the protease-sensitive sites as guides, we have constructed vectors to over-express three E.coli RecQ variants: (i) a C-terminal truncation that lacks the HRDC domain (RecQΔC); (ii) an N-terminal truncation that removes 21 residues (RecQΔN) and (iii) an N- and C-terminal truncation that lacks both the N-terminal 21 residues and the HRDC domain (RecQΔNΔC). The activities of the purified variants were compared with full-length E.coli RecQ in DNA binding, DNA-dependent ATPase and helicase activity assays. The results of these analyses showed the HRDC domain to be required for stable DNA binding, but dispensable for catalytic function (DNA-dependent ATPase and helicase activities). In addition, our analyses illustrate the contribution of a novel N-terminal sequence motif to RecQ catalytic function.
MATERIALS AND METHODS
Subcloning, over-expression and purification of E.coli RecQ, RecQΔN, RecQΔC and RecQDNΔC
Escherichia coli recQ was amplified from strain DH5α using PCR with primers specific for the 5′ and 3′ ends of the protein-coding region of the gene (31,42). The primers incorporated NdeI (5′) and BamHI (3′) restriction sites to allow subcloning into the pET15b bacterial over-expression plasmid, creating pJK100. The recombinant recQ gene was sequenced to confirm the proper DNA sequence for the gene. One minor discrepancy was found relative to published E.coli K12 strain recQ sequences (42,43). In our clone, the predicted protein sequence starting from residue 501 is Val-Leu-Arg-Gly-Glu, whereas the E.coli K12 recQ sequence in the genome database is Val-Leu-Ala-Glu with all subsequent sequences the same to the C-terminus. The predicted protein sequence of our clone is well conserved among most other bacterial RecQ proteins, including the closely related E.coli strain O157 (44). pET-15b derivative plasmids encoding for RecQΔN (residues 22 to the natural C-terminus, pJK100ΔN), RecQΔC (residues from the natural N-terminus to 530, pJK100ΔC) and RecQΔNΔC (residues 22–530, pJK100ΔNΔC) were constructed using a similar PCR-based approach and sequenced to confirm that no mutations were introduced during subcloning. Each of the plasmids allows inducible over-expression of a protein that incorporates an N-terminal hexahistidine Ni2+-affinity purification tag and a thrombin cleavage site (20 residues total).
Escherichia coli BL21(DE3) cells transformed with pLysS (Novagen) and either pJK100, pJK100ΔN, pJK100ΔC or pJK100ΔNΔC were grown at 37°C in Luria–Bertani medium (45) supplemented with 100 µg/ml ampicillin and 25 µg/ml chloramphenicol. Cells at early logarithmic phase (OD600 of 0.3–0.4) were induced to over-express RecQ (or a RecQ variant) by the addition of 1 mM isopropyl β-d-thiogalactopyranoside and were harvested by centrifugation after an additional 2.5 h of growth. Cells were resuspended in 20 mM Tris, pH 8.0, 20 mM imidazole, pH 8.0, 300 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol and lysed by sonication on ice. All subsequent purification steps were performed at 4°C. Soluble lysate was loaded on a Ni2+–NTA column and washed with lysis buffer until protein was undetectable in the eluent. His-tagged protein was then eluted by the addition of 20 mM Tris, pH 8.0, 100 mM imidazole, pH 8.0, 300 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol. Except for full-length protein used in proteolysis experiments, the eluent was dialyzed against lysis buffer, digested with thrombin to remove the His-tag (a Gly-Ser-His sequence remains on the N-terminus) and passed over a Ni2+–NTA column to remove E.coli proteins that fortuitously bind to Ni2+–NTA resin. The protein sample was then diluted to low-salt conditions and loaded onto one of two ion-exchange columns depending upon the protein being purified. For full-length RecQ and RecQΔN, the protein was diluted to 20 mM Tris, pH 8.0, 125 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol, loaded onto a MonoQ column (Pharmacia), and eluted with a linear NaCl gradient from 100 to 250 mM. RecQΔC and RecQΔNΔC were diluted to 20 mM Tris, pH 8.0, 20 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol, loaded onto a MonoS column (Pharmacia), and eluted with a linear NaCl gradient from 20 to 100 mM. Purified fractions of each protein were pooled, concentrated to ∼2 ml and dialyzed against 20 mM Tris, pH 8.0, 300 mM NaCl, 1 mM β-mercaptoethanol, 1 mM EDTA, 10% glycerol. Proteins were then subjected to size-exclusion chromatography over a Sephacryl S-200 column (Pharmacia) equilibrated in the same buffer. Highly purified fractions were identified by polyacrylamide gel electrophoresis (PAGE), pooled and concentrated to >1 mg/ml prior to storage at 4°C. Protein concentrations were determined by measuring their A280 in 6.0 M guanidine–HCl (46) (1 OD280 nm, 6 M Gdn–HCl = 21.8 µM for RecQ or RecQΔN, or 22.4 µM for RecQΔC or RecQΔNΔC).
Limited proteolysis of E.coli RecQ and identification of protease-sensitive sites
Escherichia coli RecQ (14 µM) was incubated with 0.14 µM protease (α-chymotrypsin or subtilisin) in 20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM β-mercaptoethanol, 1 mM EDTA, 0.1 mM nucleotide (in a subset of reactions as indicated in Fig. 2) at room temperature. Aliquots (10 µl) from the reaction were quenched by mixing with 10 mM PMSF and freezing on dry ice. Samples were then separated by SDS–PAGE on a 10% gel and stained with Coomassie Brilliant Blue. To determine the N-terminal sequence of the digested fragments, samples separated by PAGE were transferred to a PVDF membrane, stained briefly with Coomassie Brilliant Blue, then destained and washed extensively with water. Bands on PVDF membrane from chymotrypsin- or subtilisin-treated RecQ were sequenced by M. Byrne’s laboratory (Tufts University). To measure the mass of the chymotrypsin-resistant core fragment, chymotrypsin-treated apo E.coli RecQ was subjected to Electrospray Mass Spectroscopic analysis (Harvard Chemistry and Chemical Biology core facility).
Figure 2.
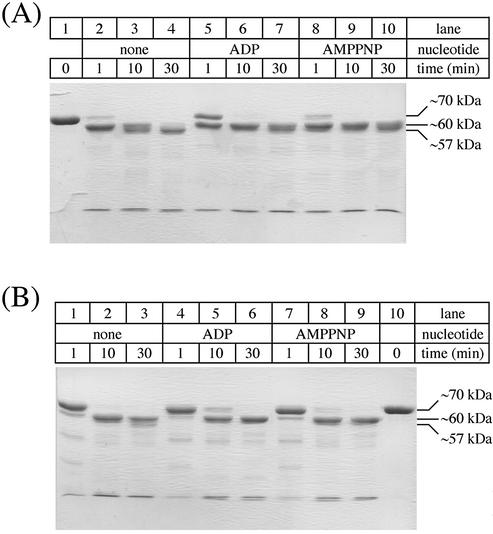
Limited proteolysis of E.coli RecQ. Purified E.coli RecQ (14 µM) was treated with 0.14 µM chymotrypsin (A) or subtilisin (B) as described in the Materials and Methods, quenched at the indicated times, separated by SDS–PAGE, and stained with Coomassie Brilliant Blue. In some reactions, ADP or AMPPNP was included at 0.1 mM. Aliquots of E.coli RecQ prior to the addition of protease are shown (A, lane 1, and B, lane 10). Molecular weight estimates from SDS–PAGE analyses are indicated.
Electrophoretic mobility shift assay (EMSA)
DNA substrates were created by phosphorylating the 5′ end of an 18-nt long oligonucleotide (oligo 1: AAGCACAATTA CCCACGC) in a T4 polynucleotide kinase reaction with [γ-32P]ATP and then annealing with a 30-nt oligonucleotide (oligo 2: GCGTGGGTAATTGTGCTTCAATGGACTGAC) by boiling and slowly cooling an equimolar mixture. The resulting DNA substrate has an 18-bp region and a 12-base 3′ single-stranded (ss) extension. Substrate was then purified via native PAGE on a 20% gel followed by electroelution. Purified RecQ variants were incubated with the substrate (∼1 nM, molecules) in 20 mM Tris, pH 8.0, 50 mM NaCl, 1 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 g/l bovine serum albumin, 4% glycerol for 30 min at 25°C with protein concentrations between 7.8 and 250 nM and then analyzed by PAGE on a 6% non-denaturing gel, dried onto Whatman paper and imaged using a phosphorimager.
DNA-dependent ATPase assays
Purified RecQ variants were mixed with 0–1000 nM dT28 in 20 mM HEPES, pH 8.0, 50 mM NaCl, 1 mM β-mercaptoethanol, 5 mM MgCl2, 0.1 g/l bovine serum albumin, 1 mM ATP, at 25°C in the presence of an ATP regeneration system that converts ADP to ATP in a reaction that is coupled to the conversion of NADH to NAD+ (47). This coupled reaction can be detected spectrophotometically by observing the decrease of A340 due to NADH oxidation. Steady-state ΔA340/Δt were measured and converted to Δ[ATP]/Δt to determine the ATPase rate and then normalized to the concentration of RecQ included in each reaction. For highly active RecQ variants (RecQ and RecQΔC), protein concentrations were 2–20 nM, and for poorly active variants (RecQΔN and RecQΔNΔC), protein concentrations were 50–500 nM.
Helicase assays
Purified RecQ variants were incubated with the substrate described for EMSA analysis (∼1 nM, molecules) in 20 mM Tris, pH 8.0, 50 mM NaCl, 1 mM β-mercaptoethanol, 1 mM MgCl2, 1 mM ATP, 0.1 g/l bovine serum albumin, 4% glycerol for 30 min at 25°C with protein concentrations between 0.001 and 100 nM. Reactions were terminated by the addition of 11% glycerol, 0.28% SDS (to denature RecQ) and 5 ng unlabeled oligo 1 (to prevent reannealing of the unwound radiolabeled DNA). The helicase products were analyzed by PAGE on a 12% non-denaturing gel, dried onto Whatman paper and imaged using a phosphorimager.
RESULTS
Limited proteolysis of recombinant E.coli RecQ
To probe the domain structure of E.coli RecQ, limited proteolysis was performed on purified His-tagged E.coli RecQ protein. His-tagged E.coli RecQ migrated as a ∼70 kDa protein by SDS–PAGE, consistent with its predicted monomeric molecular weight of 70 254 Da (Fig. 2A, lane 1, and B, lane 10). Upon the addition of limiting amounts of either chymotrypsin or subtilisin, however, E.coli RecQ was efficiently degraded to higher-mobility fragments (Fig. 2A, lanes 2–4, and B, lanes 1–3). Within 1 min of chymotrypsin treatment, nearly all of the full-length protein was degraded to a fragment with an estimated mass of ∼60 kDa. This fragment was then further proteolyzed to a minimal core structure (∼57 kDa) that was resistant to further protease digestion. A similar proteolysis pattern was observed when subtilisin was used as the protease, except that the reaction appeared to proceed more slowly than with chymotrypsin (compare Fig. 2A, lanes 2–4, with B, lanes 1–3).
Since E.coli RecQ can bind and hydrolyze ATP (31), we tested whether adding either ADP or AMPPNP (an ATP analog) might alter RecQ proteolytic patterns. ADP appeared to slow proteolytic digestion of RecQ by chymotrypsin, with a greater amount of full-length RecQ surviving in the 1 min reaction (Fig. 2A, lane 5) and with proteolysis stopping at the ∼60 kDa fragment size (Fig. 2A, lanes 6 and 7). Inclusion of AMPPNP in chymotrypsin reactions also appeared to limit proteolysis to the ∼60 kDa fragment size (Fig. 2A, lanes 9 and 10). In contrast, the addition of nucleotide to the subtilisin reactions did not affect the pattern or rate of proteolysis dramatically (Fig. 2B). The addition, and presumably binding, of nucleotide to RecQ therefore appeared to protect the protein from digestion by chymotrypsin.
To identify N-terminal protease cleavage sites, chymotrypsin- and subtilisin-derived E.coli RecQ core fragments were analyzed by protein sequencing as described in Materials and Methods. The N-terminal sequence of chymotrypsin-digested E.coli RecQ was determined to be (Gly or Asn)-Tyr-Gln-Gln-Phe while the subtilisin-digested RecQ N-terminus was (Gly or Asn)-Leu-Glu-Ser-Gly. These sequences corresponded uniquely to sequences starting at residues 22 (Gly-Tyr-Gln-Gln-Phe) or 8 (Asn-Leu-Glu-Ser-Gly) of E.coli RecQ for chymotrypsin or subtilisin cleavage, respectively. Cleavage at residues 22 or 8 accounted for only 4.6 or 3.0 kDa of the estimated mass change of ∼13 kDa observed in Figure 2, and implied that a second protease-sensitive site was located in the C-terminal region of the protein. To map the C-terminal protease cleavage site, the mass of the chymotrypsin-digested RecQ core fragment was measured by mass spectroscopic methods as described in Materials and Methods. The results showed two major species of 56 469 and 57 936 Da with many minor peaks also apparent between these peaks, leaving the precise identity of the C-terminal cleavage site (or sites) unclear. However, we estimated the second protease-sensitive region to be between residues 525 and 535, since the predicted masses of fragments that span from residue 22 to 525 or 535 were consistent with the changes in mobility observed by PAGE (Fig. 2) and the mass spectroscopic measurements. This region was predicted to form the N-terminal border of the E.coli RecQ HRDC domain based on an alignment of the protein with the sequence from the S.cerevisiae Sgs1 HRDC domain structure (48), and indicated that both proteases could cleave between the HRDC domain and the remainder of the protein, removing the ∼9 kDa HRDC domain.
Purification of RecQΔN, RecQΔC and RecQΔNΔC
Based on the N-terminal sequence of chymotrypsin-treated RecQ and on our estimate of the C-terminal cleavage site, we designed three recombinant variants of the E.coli recQ gene to test the biochemical effects of removing proteolytically labile sequences. These included variants that lacked the N-terminal 21 residues (RecQΔN), the C-terminal 79 residues, or HRDC domain (RecQΔC), or both (RecQΔNΔC). Each protein was purified to apparent homogeneity (Fig. 3). The electrophoretic mobility of RecQΔNΔC closely matched that of chymotrypsin-digested E.coli RecQ, indicating that our estimate of the C-terminal protease-sensitive site was accurate (data not shown).
Figure 3.
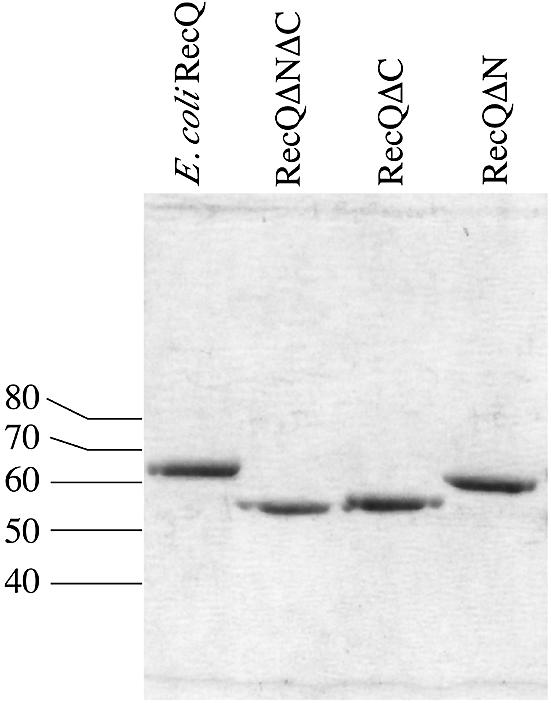
SDS–PAGE analysis of purified E.coli RecQ variants. Escherichia coli RecQ, RecQΔN, RecQΔC and RecQΔNΔC were purified as described in the Materials and Methods. Equimolar amounts of each were separated on a 10% SDS–PAGE gel and stained with Coomassie Brilliant Blue. Molecular weights from protein standards are indicated.
EMSA analysis of DNA binding by E.coli RecQ, RecQΔN, RecQΔC and RecQΔNΔC
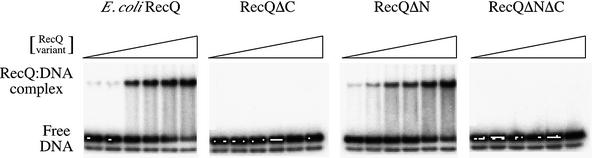
Escherichia coli RecQ is a 3′ to 5′ DNA helicase (31) and preferentially binds to partially duplex DNA with a 3′ ss extension (14). We therefore used a synthetic duplex DNA substrate with a 3′ ss extension to compare DNA binding ability among our RecQ variants. The same substrate had been used previously to measure DNA binding and helicase activity in the S.cerevisiae RecQ homolog, Sgs1 (49). Each RecQ variant was incubated with substrate in the absence of ATP, and DNA binding was analyzed by EMSA (Fig. 4). Full-length E.coli RecQ and RecQΔN were able to shift the electrophoretic mobility of the DNA at the same protein concentrations (half-maximal shift at ∼100 nM enzyme), indicating that removal of the N-terminal 21 residues did not perturb stable DNA binding. In contrast, neither RecQΔC nor RecQΔNΔC was able to shift the DNA substrate in the EMSA experiment even at the highest protein concentrations used (250 nM). This result shows that the HRDC domain is important for stable DNA binding.
Figure 4.
EMSA analysis of DNA binding by E.coli RecQ, RecQΔN, RecQΔC and RecQΔNΔC. Proteins at 8, 16, 31, 62, 125 or 250 nM were incubated with a ∼1 nM radiolabeled DNA substrate as described in the Materials and Methods and protein:DNA complexes were separated from free DNA by PAGE on a 6% non-denaturing gel. Radiolabeled DNA was observed using a phosphorimager. A small amount of the radiolabeled DNA strand from the ds substrate is observed in its ss, unannealed form in each lane. Free DNA and RecQ:DNA complex bands are indicated. Binding experiments were performed multiple times and the data shown are representative of all observations.
DNA-dependent ATPase activities of E.coli RecQ, RecQΔN, RecQΔC and RecQΔNΔC
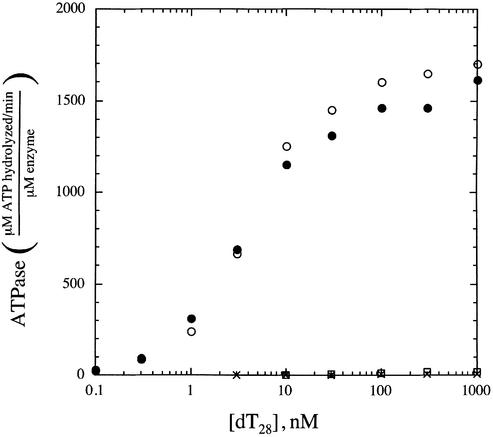
To further characterize the effects of N- and C-terminal deletions on RecQ activity, the ATPase activities of the RecQ variants were examined using a spectrophotometric ATPase assay (47). Escherichia coli RecQ has a weak ATPase activity that can be stimulated by the addition of a variety of DNA substrates, including ss and double-stranded (ds) DNA (31). We analyzed the DNA-dependent ATPase activity of the RecQ variants using a ss homopolymer (dT28) as a cofactor to allow measurement of ATPase function in the absence DNA unwinding. Consistent with previous results (31), full-length RecQ ATPase activity could not be detected in the absence of DNA but was stimulated upon the addition of DNA (Fig. 5). The mid-point of activation was ∼5 nM DNA. In spite of its apparent inability to bind DNA in EMSA experiments (Fig. 4), the RecQΔC variant also had essentially wild-type levels of DNA-dependent ATPase activity with a mid-point of activation at ∼5 nM DNA (Fig. 5). In contrast, the RecQΔN and RecQΔNΔC variants had very weak ATPase activities both in the absence and presence of DNA. Both of these proteins had maximal ATPase levels of ∼1% of the maximum ATPase rates of full-length RecQ or RecQΔC. These results indicate that elements in the N-terminal 21 residues of E.coli RecQ are important for stimulation of RecQ ATPase activity, which was unexpected since these residues are not part of the canonical helicase motifs of RecQ.
Figure 5.
Analysis of DNA-dependent ATPase activity in E.coli RecQ (filled circles), RecQΔN (‘X’ symbols), RecQΔC (open circles) and RecQΔNΔC (open boxes). Proteins were incubated with indicated concentrations of dT28 in the presence of 1 mM ATP in a coupled ATP regeneration/NADH oxidation system (47) as described in the Materials and Methods. For full-length RecQ and RecQΔC, protein concentrations were 1–8 nM whereas the weakly active RecQΔN and RecQΔNΔC variants required 50–500 nM enzyme concentrations to produce a measurable ATPase rate. Data points are normalized to the specific enzyme concentration used in each measurement.
Helicase activities of E.coli RecQ, RecQΔN, RecQΔC and RecQΔNΔC
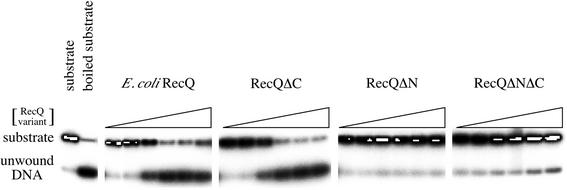
Finally, we compared helicase activities among the RecQ variants in conditions that mimicked the EMSA reactions, except with the addition of 1 mM ATP. In a concentration-dependent manner, full-length RecQ was able to unwind the 18-bp substrate with half-maximal unwinding at ∼0.1 nM enzyme (Fig. 6). Surprisingly, RecQΔC, which could not stably bind the same DNA substrate (Fig. 4), displayed an indistinguishable level of DNA unwinding activity to that of full-length RecQ. This shows that stable DNA binding and helicase activity can be separated in RecQ. In contrast to RecQ and RecQΔC, neither RecQΔN nor RecQΔNΔC was able to unwind the substrate to a measurable level even at the highest enzyme concentrations (100 nM), indicating that the N-terminal 21 residues are critical for helicase function.
Figure 6.
DNA helicase activity in E.coli RecQ, RecQΔN, RecQΔC and RecQΔNΔC. Proteins at 0.001, 0.01, 0.1, 1, 10 or 100 nM were incubated with a ∼1 nM radiolabeled DNA substrate in the presence of 1 mM ATP as described in the Materials and Methods and unwound DNA was separated from substrate by PAGE on a 12% non-denaturing gel. Radiolabeled DNA was observed using a phosphorimager. Substrate and boiled substrate controls are indicated. DNA unwinding experiments were performed multiple times and the data shown are representative of all observations.
DISCUSSION
RecQ helicases comprise a family of enzymes that play important roles in genome maintenance reactions in bacterial and eukaryotic cells. Genetic and biochemical studies of several different RecQ proteins have yielded insights into RecQ-catalyzed reactions but a structural model of RecQ proteins is presently unclear. Escherichia coli RecQ is an excellent system for probing structural features of the RecQ family since the sequence elements that are conserved among RecQ homologs are present in the E.coli protein and it has proven amenable to biochemical analysis (14,31,50–52). As a step towards resolving the structural basis of RecQ function, we have used limited proteolysis to map structural domains in E.coli RecQ and then used in vitro assays to determine the effects of removing proteolytically sensitive regions of RecQ on its biochemical activities.
Proteolytic mapping of E.coli RecQ
Our limited proteolysis experiments have revealed a two-domain structure for E.coli RecQ, with the Helicase and RecQ-Ct regions forming one structural domain and the HRDC region forming a second. Proteolysis by chymotrypsin occurs in two stages, with the first cleavage event removing ∼10 kDa and the second removing ∼3 kDa (Fig. 2A). These observed mass changes are consistent with the expected masses of the HRDC domain (∼9 kDa, Fig. 1) and N-terminal 21 residues (∼3 kDa), and imply that the linkage connecting the C-terminus of the Helicase/RecQ-Ct domain to the N-terminus of the HRDC domain is exposed in the structure. An alternative explanation of this result is that the HRDC region is unstructured in E.coli RecQ and can be degraded by either chymotrypsin or subtilisin. However, the HRDC domain from S.cerevisiae Sgs1 is folded when expressed independently (48), and we have observed that the E.coli HRDC region is folded in isolation (D.A.Bernstein and J.L.Keck, unpublished results), indicating the HRDC region most likely comprises a bona fide structural domain in RecQ.
Importance of the N-terminal 21 residues of E.coli RecQ
Removal of 21 N-terminal residues from E.coli RecQ impairs both its ATPase and helicase activities (Figs 5 and 6) but not its ability to stably bind DNA (Fig. 4). A sequence comparison of 61 bacterial RecQ proteins shows great variability among their N-termini up to residue 17 (E.coli numbering, data not shown). Starting at residue 17 however, bacterial and eukaryotic RecQ proteins have a highly conserved sequence of 14 amino acids that are 15–16 residues away from the first conserved helicase motif sequence (motif I, residues 49–60 in E.coli RecQ). This N-terminal conserved motif, which we propose to call ‘motif 0’, is LxxxFGxxxFRxxQ, where underlined residues are invariant, italicized residues are conserved and x is any residue (Fig. 7). Our results demonstrate that chymotrypsin is able to cleave after the first conserved Phe of motif 0 but that this region is not cleaved by subtilisin. Moreover, binding of ADP or AMPPNP appears to protect against chymotryptic digestion within motif 0 (Fig. 2A), implying that nucleotide binding changes the conformation of E.coli RecQ, which in turn affects protease accessibility to this motif.
Figure 7.
Alignment of a conserved sequence motif (motif 0) in 12 bacterial RecQ proteins. The consensus ‘motif 0’ amino acid sequence is written with invariant (underlined) and conserved (italicized) residues shown using single-letter amino acid nomenclature, and with non-conserved residues given as ‘x’. Numbers in parentheses are the number of amino acids either N-terminal to motif 0 or between motif 0 and motif I. Motif 0 is shared among all predicted bacterial RecQ proteins with only a subset shown here to demonstrate the most diverse sequences observed.
The identification and characterization of motif 0 in E.coli RecQ helps to define the biochemical basis of a known human BLM missense mutation that can lead to Bloom’s syndrome and may relate this portion of RecQ to other known helicases. Mutation of the Gln codon in motif 0 to Arg in the human BLM gene is sufficient to cause Bloom’s syndrome (19) and analogous Gln→Arg substitutions in murine BLM protein (34) and S.cerevisiae SGS1 (39) result in biochemical and cellular defects as well. Motif 0 also shares similarity with the ‘Q motif’ of DEAD-box RNA helicases, in which conserved Gln and Phe residues pack against helicase motif I and directly bind the adenine base of ATP (53). Our data are consistent with motif 0 in RecQ participating directly in ATP binding in a manner similar to the Q motif, with its invariant glutamine and an upstream conserved aromatic residue forming part of an adenine-binding pocket. Alternatively, motif 0 could play some role in the RecQ ATPase reaction mechanism independent of ATP binding, or an indirect role in stabilizing the ATPase domain. Determining whether this motif folds similarly to the Q motif and participates in ATP binding will require further study to define the functional and structural roles of individual amino acids in RecQ.
The HRDC domain is required for stable DNA binding
Our proteolytic mapping results demonstrate that the HRDC domain of E.coli RecQ can be physically separated from the Helicase/RecQ-Ct domain. While our analysis did not reveal the precise location of cutting, we were able to use mass estimates from mass spectroscopic and PAGE analyses to map the site to a short stretch of residues. In both DNA-dependent ATPase and helicase activity assays, RecQΔC, which lacks the HRDC domain, had a very similar specific activity to that of full-length E.coli RecQ (Figs 5 and 6), indicating that removal of the HRDC domain did not compromise catalytic function. However, when we compared the DNA binding abilities of RecQΔC to full-length RecQ using EMSA, we found that RecQΔC could not stably bind our model DNA substrate (Fig. 4). Thus, our results with RecQΔC appear to be contradictory: in two experiments, helicase and DNA- dependent ATPase, the HRDC domain is dispensable for DNA binding while in a third, EMSA, the domain appears to be required for DNA binding. The difference between these methods that is likely to account for this discrepancy is that in our helicase and DNA-dependent ATPase experiments, the protein can freely dissociate and re-associate with DNA, whereas in our EMSA experiments, dissociation could be irreversible due to the physical separation of free DNA from the protein. Since this is only observed with E.coli RecQ variants that lack the HRDC domain, our data are consistent with the HRDC domain playing a role in stable, but not transient, DNA binding. Moreover, these data imply that there are at least two distinct DNA binding elements in RecQ—an HRDC domain-containing region that is required for stable DNA binding and an HRDC-independent element that is sufficient for more transient interactions with DNA and for DNA-dependent ATPase and helicase function. The HRDC domain might enhance RecQ DNA unwinding activity where stable DNA binding is required (e.g. for processive unwinding of long DNA), but is not necessary for its catalytic function.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Michael M. Cox, Richard J. Bennett, Patricia J. Kiley and members of the Keck lab for helpful discussions and James C. Wang for intellectual contributions at the beginning of this project. This work was supported by startup funds from the University of Wisconsin, a grant from the University of Wisconsin Medical School under the Howard Hughes Medical Institute Research Resources Program for Medical Schools, and the Jane Coffin Childs Memorial Fund for Medical Research. D.A.B. was supported in part by an NIH training grant in Molecular Biophysics (T32 GM08293).
REFERENCES
- 1.Frei C. and Gasser,S.M. (2000) RecQ-like helicases: the DNA replication checkpoint connection. J. Cell Sci., 113 (Pt 15), 2641–2646. [DOI] [PubMed] [Google Scholar]
- 2.Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama H., Nakayama,K., Nakayama,R., Irino,N., Nakayama,Y. and Hanawalt,P.C. (1984) Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol. Gen. Genet., 195, 474–480. [DOI] [PubMed] [Google Scholar]
- 4.Kolodner R., Fishel,R.A. and Howard,M. (1985) Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J. Bacteriol., 163, 1060–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luisi-DeLuca C., Lovett,S.T. and Kolodner,R.D. (1989) Genetic and physical analysis of plasmid recombination in recB recC sbcB and recB recC sbcA Escherichia coli K-12 mutants. Genetics, 122, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcelle J., Carswell-Crumpton,C. and Hanawalt,P.C. (1997) recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 3714–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcelle J. and Hanawalt,P.C. (1999) RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet., 262, 543–551. [DOI] [PubMed] [Google Scholar]
- 8.Courcelle J., Crowley,D.J. and Hanawalt,P.C. (1999) Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol., 181, 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- 10.Galitski T. and Roth,J.R. (1997) Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics, 146, 751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox M.M. (1997) Recombinational crossroads: eukaryotic enzymes and the limits of bacterial precedents. Proc. Natl Acad. Sci. USA, 94, 11764–11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanada K., Ukita,T., Kohno,Y., Saito,K., Kato,J. and Ikeda,H. (1997) RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc. Natl Acad. Sci. USA, 94, 3860–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanada K., Iwasaki,M., Ihashi,S. and Ikeda,H. (2000) UvrA and UvrB suppress illegitimate recombination: synergistic action with RecQ helicase. Proc. Natl Acad. Sci. USA, 97, 5989–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmon F.G. and Kowalczykowski,S.C. (1998) RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev., 12, 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 16.Kitao S., Shimamoto,A., Goto,M., Miller,R.W., Smithson,W.A., Lindor,N.M. and Furuichi,Y. (1999) Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nature Genet., 22, 82–84. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair D.A., Mills,K. and Guarente,L. (1997) Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science, 277, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 18.Cogoni C. and Macino,G. (1999) Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science, 286, 2342–2344. [DOI] [PubMed] [Google Scholar]
- 19.Ellis N.A., Groden,J., Ye,T.Z., Straughen,J., Lennon,D.J., Ciocci,S., Proytcheva,M. and German,J. (1995) The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 20.Morozov V., Mushegian,A.R., Koonin,E.V. and Bork,P. (1997) A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem. Sci., 22, 417–418. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya A.E. and Koonin,E.V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Biol., 3, 419–429. [Google Scholar]
- 22.Gray M.D., Shen,J.C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nature Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- 23.Karow J.K., Chakraverty,R.K. and Hickson,I.D. (1997) The Bloom’s syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- 24.Tada S., Yanagisawa,J., Sonoyama,T., Miyajima,A., Seki,M., Ui,M. and Enomoto,T. (1996) Characterization of the properties of a human homologue of Escherichia coli RecQ from xeroderma pigmentosum group C and from HeLa cells. Cell Struct. Funct., 21, 123–132. [DOI] [PubMed] [Google Scholar]
- 25.Lu J., Mullen,J.R., Brill,S.J., Kleff,S., Romeo,A.M. and Sternglanz,R. (1996) Human homologues of yeast helicase. Nature, 383, 678–679. [DOI] [PubMed] [Google Scholar]
- 26.Bennett R.J., Sharp,J.A. and Wang,J.C. (1998) Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J. Biol. Chem., 273, 9644–9650. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad F., Kaplan,C.D. and Stewart,E. (2002) Helicase activity is only partially required for Schizosaccharomyces pombe Rqh1p function. Yeast, 19, 1381–1398. [DOI] [PubMed] [Google Scholar]
- 28.Ozsoy A.Z., Ragonese,H.M. and Matson,S.W. (2003) Analysis of helicase activity and substrate specificity of Drosophila RECQ5. Nucleic Acids Res., 31, 1554–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozsoy A.Z., Sekelsky,J.J. and Matson,S.W. (2001) Biochemical characterization of the small isoform of Drosophila melanogaster RECQ5 helicase. Nucleic Acids Res., 29, 2986–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan H., Chen,C.Y., Kobayashi,R. and Newport,J. (1998) Replication focus-forming activity 1 and the Werner syndrome gene product. Nature Genet., 19, 375–378. [DOI] [PubMed] [Google Scholar]
- 31.Umezu K., Nakayama,K. and Nakayama,H. (1990) Escherichia coli RecQ protein is a DNA helicase. Proc. Natl Acad. Sci. USA, 87, 5363–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barakat A., Ababou,M., Onclercq,R., Dutertre,S., Chadli,E., Hda,N., Benslimane,A. and Amor-Gueret,M. (2000) Identification of a novel BLM missense mutation (2706T>C) in a Moroccan patient with Bloom’s syndrome. Hum. Mutat., 15, 584–585. [DOI] [PubMed] [Google Scholar]
- 33.Rong S.B., Valiaho,J. and Vihinen,M. (2000) Structural basis of Bloom syndrome (BS) causing mutations in the BLM helicase domain. Mol. Med., 6, 155–164. [PMC free article] [PubMed] [Google Scholar]
- 34.Bahr A., De Graeve,F., Kedinger,C. and Chatton,B. (1998) Point mutations causing Bloom’s syndrome abolish ATPase and DNA helicase activities of the BLM protein. Oncogene, 17, 2565–2571. [DOI] [PubMed] [Google Scholar]
- 35.Saffi J., Pereira,V.R. and Henriques,J.A. (2000) Importance of the Sgs1 helicase activity in DNA repair of Saccharomyces cerevisiae. Curr. Genet., 37, 75–78. [DOI] [PubMed] [Google Scholar]
- 36.Mullen J.R., Kaliraman,V. and Brill,S.J. (2000) Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics, 154, 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foucault F., Vaury,C., Barakat,A., Thibout,D., Planchon,P., Jaulin,C., Praz,F. and Amor-Gueret,M. (1997) Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom’s syndrome. Hum. Mol. Genet., 6, 1427–1434. [DOI] [PubMed] [Google Scholar]
- 38.Neff N.F., Ellis,N.A., Ye,T.Z., Noonan,J., Huang,K., Sanz,M. and Proytcheva,M. (1999) The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells. Mol. Biol. Cell., 10, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onoda F., Seki,M., Miyajima,A. and Enomoto,T. (2000) Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom’s syndrome gene. Mutat. Res., 459, 203–209. [DOI] [PubMed] [Google Scholar]
- 40.Ui A., Satoh,Y., Onoda,F., Miyajima,A., Seki,M. and Enomoto,T. (2001) The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Gen., 265, 837–850. [DOI] [PubMed] [Google Scholar]
- 41.Mullen J.R., Kaliraman,V., Ibrahim,S.S. and Brill,S.J. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics, 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irino N., Nakayama,K. and Nakayama,H. (1986) The recQ gene of Escherichia coli K12: primary structure and evidence for SOS regulation. Mol. Gen. Genet., 205, 298–304. [DOI] [PubMed] [Google Scholar]
- 43.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 44.Perna N.T., Plunkett,G.,III, Burland,V., Mau,B., Glasner,J.D., Rose,D.J., Mayhew,G.F., Evans,P.S., Gregor,J., Kirkpatrick,H.A. et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature, 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J. and Russell,D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Edn. CSHL Press, Cold Spring Harbor.
- 46.Edelhoch H. (1967) Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry, 6, 1948–1954. [DOI] [PubMed] [Google Scholar]
- 47.Morrical S.W., Lee,J. and Cox,M.M. (1986) Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of recA protein and single-stranded DNA. Biochemistry, 25, 1482–1494. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z., Macias,M.J., Bottomley,M.J., Stier,G., Linge,J.P., Nilges,M., Bork,P. and Sattler,M. (1999) The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Struct. Fold Des., 7, 1557–1566. [DOI] [PubMed] [Google Scholar]
- 49.Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S.cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- 50.Umezu K. and Nakayama,H. (1993) RecQ DNA helicase of Escherichia coli. Characterization of the helix-unwinding activity with emphasis on the effect of single-stranded DNA-binding protein. J. Mol. Biol., 230, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 51.Harmon F.G., DiGate,R.J. and Kowalczykowski,S.C. (1999) RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell., 3, 611–620. [DOI] [PubMed] [Google Scholar]
- 52.Harmon F.G. and Kowalczykowski,S.C. (2001) Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase. J. Biol. Chem., 276, 232–243. [DOI] [PubMed] [Google Scholar]
- 53.Tanner N.K., Cordin,O., Banroquse,J., Doere,M. and Linder,P. (2003) The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell., 11, 127–138. [DOI] [PubMed] [Google Scholar]