Abstract
The regB gene, from the bacteriophage T4, codes for an endoribonuclease that controls the expression of a number of phage early genes. The RegB protein cleaves its mRNA substrates with an almost absolute specificity in the middle of the tertranucleotide GGAG, making it a unique well-defined restriction endoribonuclease. This striking protein has no homology to any known RNase and its catalytic mechanism has never been investigated. Here, we show, using 31P nuclear magnetic resonance (NMR), that RegB produces a cyclic 2′,3′-phosphodiester product. In order to determine the residues crucial for its activity, we prepared all the histidine-to- alanine point mutants of RegB. The activity of these mutants was characterized both in vivo and in vitro. In addition, their binding capability was quantified by surface plasmon resonance and their structural integrity was probed by 1H/15N NMR correlation spectroscopy. The results obtained show that only the H48A and the H68A substitutions significantly reduce RegB activity without changing its ability to bind the substrate or affecting its overall structure. Altogether, our results define RegB as a new cyclizing RNase and present His48 and His68 as potent catalytic residues. The effect of the in vivo selected R52L mutation is also described and discussed.
INTRODUCTION
The RegB protein, encoded in the bacteriophage T4 genome, is responsible for the specific inactivation of several of the phage early mRNAs, favoring therefore the transition between the early and the middle phase. RegB is an RNase that cleaves very specifically in the middle of GGAG tetranucleotides (1), with a strong bias toward those found in the region responsible for the initiation of translation (the Shine–Dalgarno region). RegB has, in particular, little if any activity on the GGAG found in the coding regions or in the Shine–Dalgarno regions of late mRNAs. Several studies have established that this effect is due to a different activity of RegB towards different GGAG classes and not to an inactivation of the enzyme in the middle phase or a protection of GGAG in the coding sequence (by the ribosome, for example) (2). In fact, we demonstrated in a previous study that RegB preferentially cleaves the GGAG involved in a particular secondary structure, in which the last G of the tetranucleotide belongs to a short stem separating two non-structured loops (3). Another striking point is that RegB activity, as measured in vitro, is very low, but can be enhanced by a factor up to 100-fold in the presence of the ribosomal protein S1 (4), the S1 protein being involved in the recognition of the Shine–Dalgarno region by the Escherichia coli ribosome.
From the molecular point of view, RegB is a 153 amino acid protein whose structure is still unknown. Its sequence appears very conserved among the phages related to T4, but presents no homology to other proteins, in particular to the other known RNases. As a first step toward the determination of the molecular basis of RegB activity and specificity, we wanted to characterize the catalytic mechanism of the enzyme.
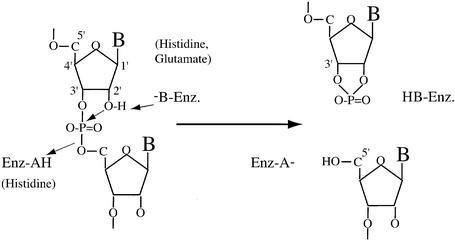
The RNases can be separated into two classes depending on whether they produce 5′- or 3′-phosphate extremities. The first class regroups many intracellular endo- and exoribonucleases such as those of the RNase H family (RNase H1, H2, HI, HII and HIII) (5,6), the prokaryotic RNases II, III, E and PNPase (7) or the mammalian RNases XRN1 and HKE1 (8). One of their characteristics is that they all possess a metal ion in their active site. The second class is mainly represented by the families of barnase (9), RNase A (10–12), RNase T1 (13) and RNase T2 (14,15). The catalysis is ensured by two side chains, one acting as a general base, the other as a general acid. The base activates the 2′-hydroxyl of the ribose which becomes able to attack the phosphodiester bond, generating a cyclic 2′,3′-phosphodiester. The acid gives its proton to the living 5′-hydroxyl group. It was proposed initially that the cyclic product would be an intermediate, immediately hydrolyzed in a second step necessary to recover the original state of ionization of the active site residues; however, it was shown that the cyclic 2′,3′-phosphodiester is the true reaction product, whose hydrolysis takes place only when all of the polynucleotide substrate has been used (16). In addition, many structural and mutational studies have shown that, in all known cases, the general acid catalyst is always a histidine, while the general base catalyst can be a histidine or a glutamate (17–21).
RegB produces a 3′-phosphate extremity (1). It thus seems reasonable to suppose that its mechanism is similar to that described previously. However, the RegB sequence is unrelated to those of barnase, RNase A, RNase T1 and RNase T2. In addition, its very low activity and very narrow specificity make it unique among RNases. Therefore, we could not exclude a completely different mechanism. We thus decided to check the production of a cyclic phosphate by RegB and the involvement of histidine residues in its mechanism.
MATERIALS AND METHODS
Site-directed mutagenesis
The ExSite™ PCR-based Site-directed Mutagenesis Kit (Stratagene) was used to construct the four histidine-to-alanine RegB mutants. The oligonucleotides, given hereafter 5′ to 3′, were purchased from Eurogentec and purified on a polyacrylamide gel: 421 (GCT CTA AAA TAT TCT CAA CAT CTT CTT GAT CGC G) and 422 (AAA CGA TGA GAC TCC TGC TGC TTT TG) for the H42A substitution; 481 (GCT CTT CTT GAT CGC GCA ATT CAA CGG) and 482 (TTG AGA ATA TTT TAG ATG AAA CGA TGA GAC TCC) for the H48A mutation; 681 (GCT AAA ATA AAA GAC CAT GTT TTA GAA GTT AAT G) and 682 (GAA TAA TTC AAA AAC GTA TGT CTC ATC AAT CTC) for the H68A mutation, and 731 (GCT GTT TTA GAA GTT AAT GAA TTC CTG AGT ATG C) and 732 (GTC TTT TAT TTT ATG GAA TAA TCA AAA ACG TAT G) for the H73A mutation. The DNA template was the plasmid pARNU2 (2) containing the wild-type copy of the regB gene. This plasmid yielded the plasmids pH42A, pH48A, pH68A and pH73A.
Purification of different RegB forms using plasmids pARNU2, pH42A, pH48A, pH68A and pH73A
Transformed E.coli strain XL1-Blue (Stratagene) was grown at 37°C in Luria–Bertani broth medium [1.0% nutriment broth (Difco), 0.5% yeast extract (Difco), 0.8% NaCl, 0.052% Tris] containing 100 µg ml–1 ampicillin (Sigma) and supplemented with 0.2% (w/v) maltose. The protein expression was induced at OD600 = 0.6 by adding MgSO4 (10 mmol l–1) and phage λCE6 (Novagen) to a final concentration of 2 × 109 p.f.u. ml–1. Cells were harvested 3 h later and the RegB protein purified, at 4°C, by cobalt affinity chromatography (Clontech) with lysis buffer [50 mmol l–1 sodium phosphate pH 8.0, 300 mmol l–1 NaCl, 0.1 mmol l–1 aminoethylbenzenesulfonyl fluoride (AEBSF) protease inhibitor (Euromedex) and 5.0 mmol l–1 β-mercaptoethanol], wash buffer (lysis buffer supplemented with 10 mmol l–1 imidazole) and elution buffer (lysis buffer supplemented with 150 mmol l–1 imidazole).
Purification of 15N-labeled RegB samples for NMR analysis
An overnight preculture of transformed E.coli strain BL21(DE3) (Novagen) was inoculated to 1 l of the M9 minimal medium [12.8 g l–1 (Na2HPO4·2H2O), 3.0 g l–1 KH2PO4, 0.5 g l–1 NaCl, 1.0 g l–1 15NH4Cl, 0.1 mmol l–1 CaCl2, 1.0 mmol l–1 MgSO4 and 4.0 g l–1 glucose] containing 100 µg ml–1 ampicillin. Protein expression was induced at OD600 = 0.8 for 3 h using 1.0 mmol l–1 isopropyl-β-d-thiogalactopyranoside (IPTG) (Promega). The protein was purified as described above.
RNA synthesis
The Selex22tb RNA was synthesized on a Pharmacia LKB Gene Assembler Plus using phenoxyacetyl β-RNA phosphoramidites (Amersham Pharmacia Biotech). The protocol was adapted from that used for DNA synthesis, the main difference being the use of 5-ethylthio-1H-tetrazol as activator (22). The terminal 5′-O-dimethoxytrityl group was removed in the synthesizer at the end of the synthesis. The oligoribonucleotides were cleaved from the support and deprotected according to the procedure described in Snoussi and Leroy (22). The fragment purity was checked using a Q-Sepharose HPLC column running on a Beckman system and, due to the very low amount of the abortive forms, was not purified further.
In vitro assay of cleavage of Selex22tb RNA by different RegB forms
Cleavage reactions were performed at 37°C in 4 mmol l–1 sodium phosphate pH 8.0, 24 mmol l–1 NaCl and 0.4 mmol l–1 β-mercaptoethanol. The reaction mixture contained 3.6 µmol l–1 of the RNA substrate Selex22tb, 0.36 µmol l–1 RegB and 40 U of RNA Guard® RNase inhibitor (Pharmacia) to prevent cleavage by contaminating RNase A-like enzymes. The reaction was followed for 40 min. Aliquots were taken every 10 min and the reaction was stopped by adding formamide (40% final) and heating for 5 min at 95°C. The substrate and the products were separated by gel electrophoresis (20% acrylamide, 7 mol l–1 urea), stained with ethidium bromide (0.25 µg l–1) for 7 min, washed for 30 s with sterile water and detected with a UV light transilluminator coupled to a digital acquisition card (Vilbert Lourmat). The band intensities were quantified using Bio1D software (Vilbert Lourmat) and corrected for the RNA length (21 nt for the RNA substrate Selex22tb, and 15 and 6 nt for the products). The 6 nt product is too short to fix sufficient ethidium bromide, and hence we quantified the appearance of the 15 nt product.
NMR experiments
All nuclear magnetic resonance (NMR) experiments were carried out on a Bruker DRX600 spectrometer equipped with a TXI triple resonance probe [1H/15N heteronuclear single quantum correlation (HSQC) spectra] or a 1H-BB probe (phosphorus experiments). The spectra were processed off-line using GIFA software (23).
The formation of the 2′,3′-cyclic phosphodiester could be followed conveniently by 31P NMR. Two samples were prepared, containing 300 µmol l–1 of the Selex22tb substrate with 30 µmol l–1 of either the RegB enzyme or the R52L inactive mutant previously dialyzed against a Tris cleavage buffer (final concentration in the sample: Tris 5 mmol l–1 pH 8.0, NaCl 30 mmol l–1, β-mercaptoethanol 0.5 mmol l–1). The reaction was followed at 25°C by recording 175 successive 31P spectra (carrier: 2915.24 Hz, spectral width: 7300 Hz and transients: 1024). The pH of the negative control sample (R52L mutant) was slightly different from that of the reaction sample (wild-type RegB), resulting in a small shift of the peaks corresponding to the residual amount of inorganic phosphorus.
The 15N-labeled samples of the R52L, H48A and H68A RegB mutants were prepared in a sodium phosphate 50 mmol l–1 pH 6.0, 300 mmol l–1 NaCl, 5 mmol l–1 β-mercaptoethanol buffer at a final concentration of ∼500 µmol l–1. The HSQC spectra were recorded at 25°C using the fast HSQC sequence (24). Each experiment consisted of 264 rows of 512 points, each row being the sum of 256 transients. The spectral widths were set to 36 p.p.m. (15N) and 5.5 p.p.m. (1H), and the frequency offsets were to 7300 Hz (15N) and 4800 Hz (1H). The spectra were processed to obtain real matrices of 512 × 512 points.
Surface plasmon resonance experiments
The experiments were carried out using a BIAcore 2000 instrument (BIAcore) with a CM5 sensor chip maintained at 25°C. The protein was coupled to the surface of the sensor chip using CO2H/NH2 chemistry. Flow cell 1 was used as a blank surface. The running buffer consisted of 10 mmol l–1 HEPES pH 7.4, 150 mmol l–1 NaCl, 3 mmol l–1 EDTA and 0.005% polysorbate 20 (v/v). A 70 µl aliquot of RNA was injected, at 25 µl min–1, at different concentrations ranging from 75 to 140 µmol l–1. At least two independent injections with two different RNA concentrations were carried out for each RegB form. Regeneration of the sensor chip was achieved by two short 50 µl pulses of NaCl 1 mol l–1 and NaOH 50 mmol l–1. All sensorgrams were corrected for background and bulk refractive index by subtracting the reference flow cell signal. The 1:1 Langmuir binding interaction model was used to fit the data with the BIA evaluation 3.0 software (BIAcore).
RESULTS AND DISCUSSION
Characterization of the reaction product
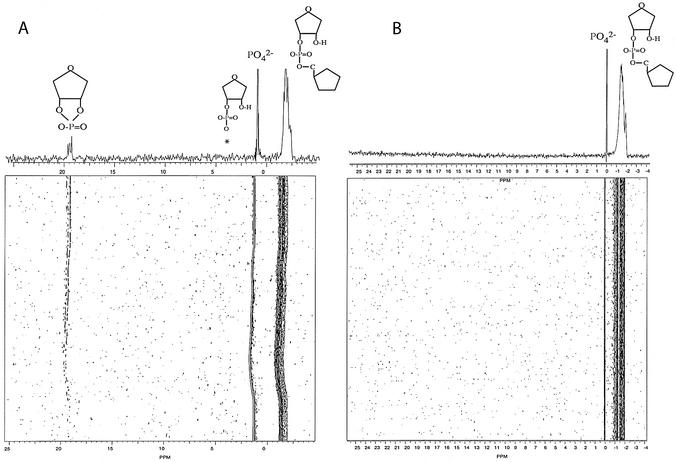
The cleavage of the GGAG tetranucleotide by RegB leads to the appearance of 3′-phosphate and 5′-OH extremities (1). This suggests that the mechanism of RegB is similar to that of barnase, RNase A, RNase T1 and RNase T2-related enzymes. The true product of these enzymes is a cyclic 2′,3′-phosphodiester (Fig. 1). We thus wondered whether RegB was also producing a cyclic phosphodiester or directly releasing a linear 3′-monoester. To check this, we analyzed the product of the reaction by 31P phosphorus NMR. Indeed, the chemical shift of the 31P nuclei involved in a linear 3′,5′-phosphodiester bond and in a 3′-monoester range from –2.0 to –0.5 and from 3.0 to 4.0 p.p.m., respectively while that of a cyclic 2′,3′-phosphodiester resonates near 19 p.p.m. (25,26). The nature of the product of the reaction (linear or cyclic) can thus be determined unambiguously on a simple one-dimensional spectrum.
Figure 1.
Proposed acid–base catalysis of phosphodiester bond cleavage by barnase, RNase A, RNase T1 and RNase T2. HB-Enz. and –B-Enz. (Enz-HA and Enz-A–) denote, respectively, the acidic and the basic forms of the residue acting as a general base catalyst (general acid catalyst).
As substrate, we chose the Selex22tb RNA (GGUGCGAGAAAACGG↓AGCACC), already studied in this laboratory (3). This substrate derives from the 22nd clone obtained in a study intended to isolate good RegB substrates by the SELEX method (Systematic Evolution of Ligands by EXponential enrichment) (27). We previously showed that Selex22tb is cleaved efficiently by RegB, in the absence of the ribosomal protein S1 (3). In addition, it is sufficiently short to be produced by chemical synthesis on the mg scale. The reaction was carried out at 25°C (instead of 37°C) because RegB is stable at this temperature for several days. We recorded 175 successive one-dimensional experiments. Each experiment corresponded to 1024 transients. We indeed anticipated a very faint signal for the product; about 40 times lower than that of the substrate. RegB indeed cleaves only one phosphodiester bond among the 20 present in the 21 nt Selex22tb. The cleavage, in addition, is not quantitative, the maximal amount of product obtained corresponding to 50–60% of the substrate (3). Finally, the reaction was performed twice, once in the presence of the wild-type RegB enzyme and once in the presence of the inactive R52L mutant (28). The mutant enzyme was produced and purified rigorously in the same conditions as the wild-type and used as a negative control, to ensure that the observed reaction was not due to a contaminant RNase.
Figure 2A represents the results obtained when we mix the wild-type RegB protein with Selex22tb at a ratio of 1:10. In this two-dimensional-like spectrum, the x-axis represents the 31P chemical shifts, and the y-axis the time course of the reaction. The one-dimensional spectrum above is the sum of the last 10 rows. The broad region centered at –1.5 p.p.m. represents the different phosphorus nuclei of the substrate Selex22tb. The narrow peak at 1.5 p.p.m. corresponds to residual phosphate molecules originating from the first dialysis buffer. At 19 p.p.m., we clearly identified a peak that resonates in the region of the 2′,3′-cyclic phosphorus. This peak is likely to be the cyclic compound produced by RegB. It is absent at time zero (beginning of the reaction) and gradually accumulates during the time course of the reaction. In addition, it is not seen when Selex22tb is mixed with the RegB inactive mutant R52L (Fig. 2B), indicating that it results from the catalytic activity of the enzyme and is, in particular, not due to an RNase A-like contaminant. We anticipated a sharp single peak corresponding to the cyclic product. This is not the case in Figure 2A where we can see a broad multiplet at 19 p.p.m. Multiple solution conformations of the cyclic 15 nt product could explain this observation.
Figure 2.
Detection, by 31P NMR, of a 2′,3′-cyclic phosphorus product during the cleavage of the substrate Selex22tb RNA by RegB. (A) Cleavage by the wild-type RegB protein. (B) Negative control experiment using the inactive point mutant RegB R52L. The asterisk indicates the position of the hydrolyzed form of the cyclic product. A peak at this position could be observed provided that the experiment was recorded for a much longer time.
Functional properties of the RegB histidine-to-alanine mutants
All the RNases described to date which produce a cyclic 2′,3′-phosphodiester possess a histidine in their active site (17–21). We thus decided to check whether this was also the case for RegB. The phage T4 RegB sequence possesses five histidines (H17, H42, H48, H68 and H73). We checked in GenBank all the sequences that have a detectable homology to RegB. We found only 16 sequences. All these sequences encode RegB proteins originating from 16 T4-related phages (L.Piesiniene, unpublished data). Among these 16 sequences, 15 were not of interest because they were almost identical (99.4–100%) to the T4 RegB protein. However, one sequence, originating from phage RB69, was informative (only 77.5% identity). Figure 3 presents the alignment of the RegB proteins from phage T4 and phage RB69. It shows that histidines H42, H48, H68 and H73 are conserved. They were thus mutated into alanines by a PCR-based site-directed protocol (see Materials and Methods). The occurrence of each mutation was controlled by DNA sequencing.
Figure 3.
Sequence alignment of the RegB proteins from phage T4 and phage RB69. Four histidines (H42, H48, H68 and H73) out of five are conserved.
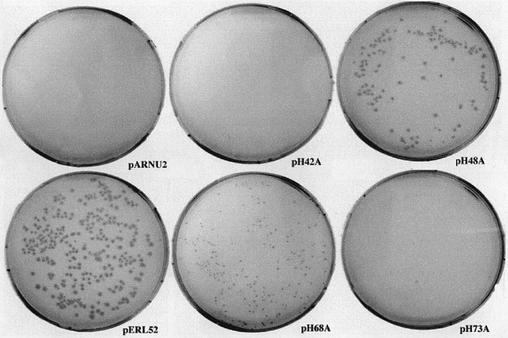
The template for the mutagenesis was the plasmid pARNU2 (2) which contains a copy of the wild-type regB gene under the control of the T7 promoter. This plasmid can be propagated in a BL21 E.coli strain (devoid of the T7 polymerase) but not in BL21(DE3) (possessing the T7 polymerase) even in the absence of inducer. Indeed, the leaky expression of the T7 polymerase in this latter strain is sufficient to produce enough of the highly toxic RegB protein to prevent bacterial growth (F.Saïda, unpublished data). Since the fully inactive mutant R52L is not toxic, we hypothesized that RegB toxicity is linked to its activity and thus speculated that a histidine-to-alanine mutant reducing the enzyme activity would also reduce its toxicity. Such a mutant would thus be easily isolated by checking the viability of a transformed BL21(DE3) strain. Figure 4 shows the result of the electro-transformation of BL21(DE3) cells with the four plasmids resulting from the mutation protocol (pH42A, pH48A, pH68A and pH73A). The plasmid pARNU2 was used as a negative control, and pERL52 (coding for the inactive R52L mutant) as a positive control. Only pH48A and pH68A are maintained in BL21(DE3), suggesting that the activity of the corresponding mutants, RegB H48A and RegB H68A, is reduced. However, other hypotheses, such as a folding default or the disappearance of a critical interaction with another partner, for example, could not be excluded at this stage.
Figure 4.
In vivo toxicity test of RegB mutants. The viability of E.coli strain BL21(DE3) was checked after transformation with plasmids pARNU2, pERL52, pH42A, pH48A, pH68A and pH73A expressing, respectively, the wild-type RegB protein, the inactive point mutant R52L (positive control) and the four histidine-to-alanine mutants. The only mutants that are maintained in BL21(DE3) are H48A and H68A.
Kinetics of cleavage of the Selex22tb RNA by RegB mutants
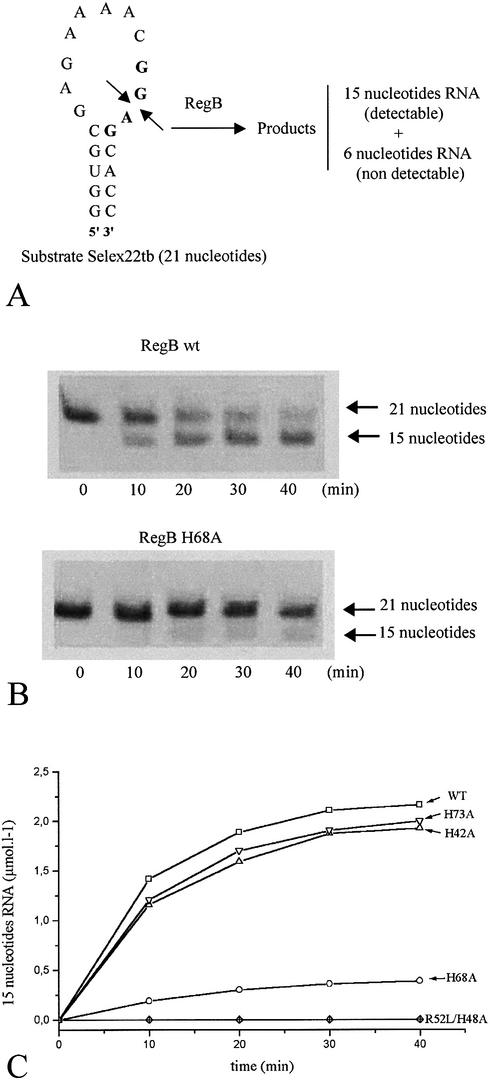
The wild-type RegB protein and the four histidine-to-alanine mutants were expressed in E.coli strain XL1-Blue using the phage λCE6 induction system (see Materials and Methods). The inactive point mutant RegB R52L was produced from a pET15b-derivative plasmid. The quick non-radioactive RNA cleavage assay described in Materials and Methods was used to compare the cleavage ability of the wild-type RegB protein and the H42A, H48A, H68A, H73A and R52L RegB mutants. All cleavage reactions were monitored for 40 min in the same conditions in the presence of an RNase A-like RNases inhibitor. The appearance of the 15 nt RNA product plotted as function of time is presented in Figure 5. As expected, the R52L mutant is devoid of any activity, whereas the wild-type RegB cleaves Selex22tb up to 58% in 40 min. The H42A and H73A mutants cleave the substrate with a rate similar to that of the wild-type. A slight difference is in fact observed (55% for the mutant instead of 58% for the wild-type) but does not seem to be significant because it is within the experimental error. In contrast, RegB activity is completely abolished by the H48A substitution and dramatically reduced by the H68A mutation. This result is in accordance with the colony phenotype observed in Figure 4 where we notice that colonies transformed with the partially active H68A mutant are smaller (the growth rate is lower) than those transformed with the H48A mutant (devoid of any activity). We hypothesized that these substitutions either disrupt the overall structure of RegB or highlight the significance of H48 and H68 in the binding of the substrate or in the direct catalysis of the cleavage reaction. To distinguish between these three hypotheses, we decided to probe by 1H/15N correlation spectroscopy the overall structure of RegB H48A and RegB H68A and to quantify the RNA-binding capabilities of these two mutants by surface plasmon resonance (SPR).
Figure 5.
Cleavage reaction kinetics of the Selex22tb RNA by different forms of RegB. (A) The 21 nt Selex22tb RNA is cleaved specificially by RegB in the middle of the GGAG motif, leaving 15 and 6 nt fragments. These fragments are separated on a 20% polyacrylamide gel and stained with ethidium bromide. The light emission of the UV-excited ethidium bromide, in RNA, was recorded using a Vilbert Lourmat Imager and quantified using Bio1D V99 software. (B) Example of two UV-excited gels showing the cleavage of the Selex22tb RNA by the wild-type and the H68A RegB forms. (C) Appearance of the 15 nt product during cleavage of Selex22tb RNA by different forms of RegB (wild type, H42A, H48A, R52L, H68A and H73A).
1H/15N correlation spectroscopy
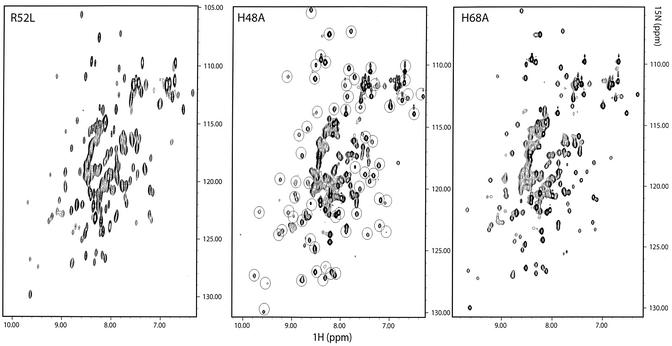
15N uniformly labeled samples of RegB H48A and RegB H68A were prepared using the BL21(DE3)/IPTG induction system (see Materials and Methods). The 1H/15N HSQC spectra (24) were recorded on a Bruker proton 600 MHz spectrometer. Each peak on the two-dimensional spectrum represents the backbone H/N correlation of one amino acid residue, and its special position reflects the effect of the structural environment. Hence, the HSQC spectrum can be considered as a two-dimensional fingerprint of the three-dimensional structure of the protein. Since the HSQC spectrum of the wild-type RegB protein is unavailable because RegB is too toxic to be produced on the NMR scale, we compared the HSQC spectra of the H48A and the H68A mutants with that of the independent inactive point mutant RegB R52L. The results are presented in Figure 6. We highlighted using circles the highly resolved widespread peaks of the RegB H48A HSQC spectrum. These peaks originate from residues in a well-structured environment and thus represent a significant probe of the protein scaffold (this probe is more relevant than circular dichroism which only describes the secondary structure composition of the protein). All circle-enclosed peaks are found in the same positions in the spectrum of RegB H68A and of the independent mutant RegB R52L. Since it is highly improbable that three independent and different substitutions cause the same structural alteration, we suggest that the amino acid substitutions H48A and H68A (as well as R52L) did not affect the overall structure of the protein.
Figure 6.
1H/15N HSQC spectra of the R52L, H48A and H68A RegB mutants recorded at 25°C on a Bruker DRX600 spectrometer. The encircled peaks in the spectrum of RegB H48A are found in the same positions in the spectra of the two independent mutants RegB R52L and RegB H68A.
Binding capabilities of RegB mutants
We used the SPR optical phenomenon to monitor the interactions between RegB mutants and the RNA Selex22tb (29). SPR provides both equilibrium and kinetic information about the RNA–protein interactions. We hypothesized that the formation and breakdown of the complex between RegB and the RNA Selex22tb are dictated by both association (ka) and dissociation (kd) rates, as described by
The dissociation constant can then be calculated according to
KD = kd/ka2
Equation 1 assumes that we neglect the breakdown of the RegB:RNA complex by enzymatic cleavage (in the case of the wild type and the mutants that have a residual activity). This assumption is justified by the fact that we recorded SPR sensorgrams over a short time period (2.8 min) and at 25°C. In these conditions, the percentage of cleavage of Selex22tb will be much lower than 10%. Indeed, Figure 5 shows that the percentage of cleavage, after 2.8 min, is 10% at 37°C, and it is known, according to the law of Arrhenius, that the rate of a catalyzed reaction is decreased by a significant factor (about 2) when the temperature drops from 37 to 25°C. The results of SPR measurements are presented in Table 1. We notice that the wild-type RegB protein and the mutants H42A, H48A and H73A have similar dissociation constants within the range of experimental error. This demonstrates that the introduced mutations, in particular H48A, do not change the binding capabilities of RegB. The affinity of the inactive mutant R52L is decreased by an order of magnitude compared with that of the wild type, suggesting that Arg52 may be important for the binding of the substrate RNA. The result for the H68A mutant seems to be slightly different from that of the wild-type: 2.5 µmol l–1 for H68A and 1.5 µmol l–1 for the wild type. This difference of 1.0 µmol l–1 in the KD may not be significant according to experimental errors (Table 1).
Table 1. Kinetics and affinity constants at 25°C for the interactions of wild-type RegB protein and the five RegB point mutants with the Selex22tb RNA substrate.
| Protein | ka (M–1 s–1) | kd (10–4 s–1) | KD (µM) |
|---|---|---|---|
| RegB wild type | 75 ± 3 | 1.1 ± 0.3 | 1.5 ± 0.5 |
| RegB R52L | 4.0 ± 0.5 | 0.60 ± 0.03 | 15 ± 2 |
| RegB H42A | 75 ±5 | 1.5 ± 0.2 | 2.0 ± 0.3 |
| RegB H48A | 53 ± 2 | 1.0 ± 0.1 | 1.9 ± 0.3 |
| RegB H68A | 64 ± 10 | 1.6 ± 0.4 | 2.5 ± 0.3 |
| RegB H73A | 64 ± 10 | 1.4 ± 0.5 | 2.2 ± 0.2 |
Values were recorded on a BIAcore 2000 SPR apparatus.
The fact that the in vitro cleavage kinetics and the HSQC NMR experiments show that the R52L, H68A and H48A mutations inactivate (completely or partially) the RegB enzyme without altering its structure supports the idea that these mutations result in a perturbation of the global catalytic process (binding of the substrate and/or direct chemical catalysis). The SPR results indicate that the R52L mutation induces a 10-fold decrease in the affinity between the enzyme and its substrate. Arg52 may be important for binding of the substrate, but we cannot exclude that it is also involved in the direct chemical catalysis because the decrease in the affinity for the RNA substrate (10-fold) seems too weak to abolish the activity of the enzyme completely. In contrast, the H48A and H68A mutations have no significant effect on the binding constant. In addition, the H48A mutant has no detectable activity, while H68A conserves a small (∼14%) but significant residual activity. These results suggest that H48 and H68 may be directly involved in the catalysis of the reaction mediated by RegB but in a different fashion. At this stage, several hypotheses could be presented. H48 may be a crucial residue involved directly in the acid–base catalysis (acceptor or donor of protons). H68 may polarize chemical bonds, enhancing the protons transfer, or interact with a particular intermediate.
The fact that the reaction product is a 2′,3′-cyclic phosphodiester and the involvement of at least one histidine in the catalytic mechanism strongly suggest that RegB shares the catalytic mechanism of barnase, RNase A, RNase T1 and RNase T2. However, RegB is structurally and functionally unrelated to all these RNases. First, RegB primary structure has no significant homology to any RNase from the barnase, RNase A and RNase T1 families, which have characteristic conserved residues encompassing their catalytic residues (9–12,15). Also, RegB does not possess the two typical cassettes of 9 and 12 residues that characterize the RNases from the RNase T2 family (13–15). Moreover, RegB is an intracellular enzyme (with two probably reduced cysteines, a reducing agent being required to avoid precipitation), whereas barnase, RNase A, RNase T1 and RNase T2 are secreted extracellular enzymes, the members of the three last families possessing several disulfide bridges. Finally, RegB has absolute sequence specificity, towards the GGAG sequence, which is not found in any described RNase. This suggests that RegB could form a new class of enzymes possessing a cyclizing mechanism.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Dr J. L. Leroy for helpful advice on RNA chemical synthesis and to Mrs N. Duvignaud for critical reading of the manuscript. This work was supported by a national grant from the French Department of Scientific Research (Ministere de la recherche).
REFERENCES
- 1.Uzan M., Favre,R. and Brody,E. (1988) A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc. Natl Acad. Sci. USA, 85, 8895–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanson B., Hu,R.M., Troitskaya,E., Mathy,N. and Uzan,M. (2000) Endoribonuclease RegB from bacteriophage T4 is necessary for the degradation of early but not middle or late mRNAs. J. Mol. Biol., 297, 1063–1074. [DOI] [PubMed] [Google Scholar]
- 3.Lebars I., Hu,R.M., Lallemand,J.Y., Uzan,M. and Bontems,F. (2001) Role of the substrate conformation and of the S1 protein in the cleavage efficiency of the T4 endoribonuclease RegB. J. Biol. Chem., 276, 13264–13272. [DOI] [PubMed] [Google Scholar]
- 4.Ruckman J., Ringquist,S., Brody,E. and Gold,L. (1994) The bacteriophage T4 regB ribonuclease. Stimulation of the purified enzyme by ribosomal protein S1. J. Biol. Chem., 269, 26655–26662. [PubMed] [Google Scholar]
- 5.Lima W.F., Wu,H. and Crooke,S.T. (2001) Human RNases H. Methods Enzymol., 341, 430–440. [DOI] [PubMed] [Google Scholar]
- 6.Kanaya S. (2001) Prokaryotic type 2 RNases H. Methods Enzymol., 341, 377–394. [DOI] [PubMed] [Google Scholar]
- 7.Nicholson A.W. (1997) Escherichia coli ribonucleases: paradigms for understanding cellular RNA metabolism and regulation. In D’Allessio,G. and Riordan,J.F. (eds), Ribonucleases: Structures and Functions. Academic Press, New York, pp. 1–49.
- 8.Ross J. (1997) RNA-processing RNases in mammalian cells. In D’Allessio,G. and Riordan,J.F. (eds), Ribonucleases: Structures and Functions. Academic Press, New York, pp. 553–587.
- 9.Mauguen Y., Hartley,R.W., Dodson,E.J., Dodson,G.G., Bricogne,G., Chothia,C. and Jack,A. (1982) Molecular structure of a new family of ribonucleases. Nature, 297, 162–164. [DOI] [PubMed] [Google Scholar]
- 10.Beintema J.J., Breukelman,H.J., Carsana,A. and Furia,A. (1997) Evolution of vertebrate ribonucleases: ribonuclease A superfamily. In D’Allessio,G. and Riordan,J.F. (eds), Ribonucleases: Structures and Function. Academic Press, New York, pp. 245–269.
- 11.Zhang J., Dyer,K.D. and Rosenberg,H.F. (2002) RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res., 30, 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beintema J.J. (1998) Introduction: the ribonuclease A superfamily. Cell. Mol. Life Sci., 54, 763–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida H. (2001) The ribonuclease T1 family. Methods Enzymol., 341, 28–41. [DOI] [PubMed] [Google Scholar]
- 14.Deshpande R.A. and Shankar,V. (2002) Ribonucleases from T2 family. Crit. Rev. Microbiol., 28, 79–122. [DOI] [PubMed] [Google Scholar]
- 15.Irie M. and Ohgi,K. (2001) Ribonuclease T2. Methods Enzymol., 341, 42–55. [DOI] [PubMed] [Google Scholar]
- 16.Cuchillo C.M., Pares,X., Guasch,A., Barman,T., Travers,F. and Nogues,M.V. (1993) The role of 2′,3′-cyclic phosphodiesters in the bovine pancreatic ribonuclease A catalysed cleavage of RNA: intermediates or products? FEBS Lett., 333, 207–210. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J.E. and Raines,R.T. (1994) Value of general acid–base catalysis to ribonuclease A. J. Am. Chem. Soc., 116, 5467–5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikawa S., Morioka,H., Kim,H.J., Fuchimura,K., Tanaka,T., Uesugi,S., Hakoshima,T., Tomita,K., Ohtsuka,E. and Ikehara,M. (1987) Two histidine residues are essential for ribonuclease T1 activity as is the case for ribonuclease A. Biochemistry, 26, 8620–8624. [DOI] [PubMed] [Google Scholar]
- 19.Steyaert J., Hallenga,K., Wyns,L. and Stanssens,P. (1990) Histidine-40 of ribonuclease T1 acts as base catalyst when the true catalytic base, glutamic acid-58, is replaced by alanine. Biochemistry, 29, 9064–9072. [DOI] [PubMed] [Google Scholar]
- 20.Irie M., Ohgi,K., Iwama,M., Koizumi,M., Sasayama,E., Harada,K., Yano,Y., Udagawa,J. and Kawasaki,M. (1997) Role of histidine 46 in the hydrolysis and the reverse transphosphorylation reaction of RNase Rh from Rhizopus niveus. J. Biochem., 121, 849–853. [DOI] [PubMed] [Google Scholar]
- 21.Irie M., Ohgi,K., Watanabe,H., Iwama,M., Nakamura,K.T., Kurihara,H., Nonaka,T., Mitsui,Y., Horiuchi,H. and Takagi,M. (1994) pH profile of kinetic constants of RNase Rh from Rhizopus niveus and its mutant enzymes towards UpU and possible mechanisms of RNase Rh. J. Biochem., 115, 1083–1087. [DOI] [PubMed] [Google Scholar]
- 22.Snoussi K. and Leroy,J.L. (2001) Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry, 40, 8898–8904. [DOI] [PubMed] [Google Scholar]
- 23.Pons J.L., Malliavin,T.E. and Delsuc,M.A. (1996) Gifa V.4: a complete package for NMR data set processing. J. Biomol. NMR, 8, 445–452. [DOI] [PubMed] [Google Scholar]
- 24.Mori S., Abeygunawardana,C., Johnson,M.O., Berg,J. and van Zijl,P.C.M. (1995) Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. Ser. B, 108, 94–98. [DOI] [PubMed] [Google Scholar]
- 25.Thompson J.E., Venegas,F.D. and Raines,R.T. (1994) Energetics of catalysis by ribonucleases: fate of the 2′,3′-cyclic phosphodiester intermediate. Biochemistry, 14, 7408–7414. [DOI] [PubMed] [Google Scholar]
- 26.Palmer H.R., Bedford,J.J., Leader,J.P. and Smith,R.A. (1990) 31P and 1H NMR studies of the effect of the counteracting osmolyte trimethylamine-N-oxide on interactions of urea with ribonuclease A. J. Biol. Chem., 275, 27708–27711. [DOI] [PubMed] [Google Scholar]
- 27.Jayasena V.K., Brown,D., Shtatland,T. and Gold,L. (1996) In vitro selection of RNA specifically cleaved by bacteriophage T4 RegB endonuclease. Biochemistry, 35, 2349–2356. [DOI] [PubMed] [Google Scholar]
- 28.Ruckman J., Parma,D., Tuerk,C., Hall,D.H. and Gold,L. (1989) Identification of a T4 gene required for bacteriophage mRNA processing. New Biol., 1, 54–65. [PubMed] [Google Scholar]
- 29.Katsamba P.S., Park,S. and Laird-Offringa,I.A. (2002) Kinetic studies of RNA–protein interactions using surface plasmon resonance. Methods, 26, 95–104. [DOI] [PubMed] [Google Scholar]