Abstract
The 10–23 RNA cleaving DNAzyme has been shown to cleave any purine–pyrimidine (RY) junction under simulated physiological conditions. In this study, we systematically examine the DNAzymes relative activity against different RY combinations in order to determine the hierarchy of substrate core dinucleotide sequence susceptibility. The reactivity of each substrate dinucleotide compared in the same background sequence with the appropriately matched DNAzyme was found to follow the scheme AU = GU ≥≥ GC >> AC. The relatively poor activity of the DNAzyme against AC and GC containing substrates was found to be improved substantially by modifications to the binding domain which subtly weaken its interaction with the substrate core. The most effective modification resulting in rate enhancement of up to 200-fold, was achieved by substitution of deoxyguanine with deoxyinosine such that the base pair interaction with the RNA substrates core C is reduced from three hydrogen bonds to two. The increased cleavage activity generated by this modification could be important for application of the 10–23 DNAzyme particularly when the target site core is an AC dinucleotide.
INTRODUCTION
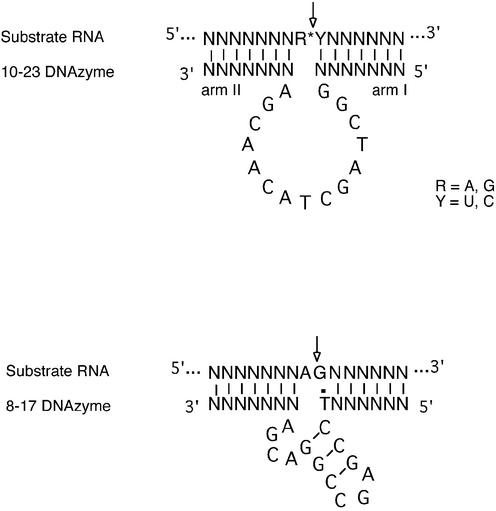
The 10–23 RNA cleaving DNAzyme is a catalytic nucleic acid composed entirely of DNA (Fig. 1) (1). It was derived from a combinatorial library of sequences by in vitro selection. The tremendous activity and sequence specificity against its target RNA under simulated physiological conditions (2,3) has generated the expectation that it may function in cells as a gene suppression agent. Indeed there are now many reports of specific gene suppression activity in a number of biological systems in vitro and in some animal models (4,5, reviewed in 6). In this respect, the DNAzyme may have an advantage over existing nucleic acid based approaches to gene suppression as it displays endogenous RNA cleavage activity more typical of ribozymes such as the ‘hammerhead’ or ‘hairpin’ (7,8) with the superior chemical and biological stability found in passive antisense reagents (6).
Figure 1.
Secondary structure of the 10–23 and 8–17 DNAzyme–substrate complexes. The 10–23 DNAzyme (top) consists of two variable binding domains, designated arm I and arm II, which flank a conserved 15 base unpaired motif that forms the catalytic domain. The only requirement of the RNA polynucleotide substrate is for a core sequence containing an RY junction. The 8–17 DNAzyme (bottom) has a similar configuration to the 10–23 DNAzyme except that it has a 12 base catalytic motif and a core substrate requirement for AG in which the A is unpaired and the G forms a wobble pair.
In addition to extraordinary cleavage activity and specificity, the 10–23 DNAzyme has a broad target range with its only substrate requirement being a purine–pyrimidine (RY) dinucleotide (Fig. 1) (1). This is ideal for target selection as the more sites tested in long folded mRNA the greater the likelihood of finding highly accessible and cleavable sites (9). It also means that DNAzymes can be targeted with greater flexibility to discrete sites such as those spanning polymorphisms or fusion transcripts produced as a consequence of chromosomal translocation. Despite the reported activity against any RY cleavage site, our early experiences with DNAzyme targeting RC dinucleotides indicated that these sequences were cleaved less efficiently than those containing RU. This led us to exclude these cytosine sites in our search for DNAzymes with efficient gene suppression activity (9). In the study reported here, we determine if there was a rational basis in our preference for RU cleavage sites over those consisting of RC. This was accomplished through systematic analysis of DNAzyme activity against various combinations of RY containing substrates. As each of these substrates were derivatives of a common ancestor, they all contained the same background sequence. From these experiments we are able to report that there was a hierarchy of RY reactivity to the 10–23 DNAzyme, which appeared to follow the general scheme AU = GU ≥ GC >> AC.
In view of the relatively poor activity of RC cleavage sites (particularly AC), we modified the DNAzyme in an attempt to improve its cleavage efficiency against these targets. One of the differences between RU and RC cleavage sites is the increased strength of the base pair interaction between the pyrimidine base in the substrate and its cognate purine in the binding domain of the DNAzyme. Interestingly, we found that various conservative substitutions which subtly disturb or weaken this interaction had a profoundly positive effect on the reaction rate and extent. The enhancement of activity generated by these modifications has the potential to significantly broaden the utility of the 10–23 DNAzyme, particularly against RNA target sites that contain an AC dinucleotide core.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were supplied by Sigma Genosys (Sydney) and Trilink (San Diego, CA). The length and integrity of each oligonucleotide was confirmed by 5′-end labelling and denaturing polyacrylamide gel electrophoresis. The sequence of template oligos for transcription and the names of the corresponding transcripts were as follows: S-AU1, TCCA CAAAGATATCCCAGCCTCTCCCTATAGTGAGTCGTA TTAAATT; S-GU1, TCCACAAAGACATCCCAGCCTC TCCCTATAGTGAGTCGTATTAAATT; S-AC1, TCCAC AAAGGTATCCCAGCCTCTCCCTATAGTGAGTCGTAT TAAATT; S-GC1, TCCACAAAGGCATCCCAGCCTCTC CCTATAGTGAGTCGTATTAAATT; S-AU2, TTCTCC CAATGTGCGCCATCCCTATAGTGAGTCGTATTAAAT T; S-GU2, TTCTCCCAACGTGCGCCATCCCTATAGTGAGTCGTATTAAATT; S-AC2, TTCTCCCAGTGTGCGCC ATCCCTATAGTGAGTCGTATTAAATT; S-GC2, TTCTC CCAGCGTGCGCCATCCCTATAGTGAGTCGTATTAAA TT; S-AC3, TCCACAAAAGTATCCCAGCCGCTCCCT ATAGTGAGTCGTATTAAATT; S-GC3, TCCACAAAAG CATCCCAGCCGCTCCCTATAGTGAGTCGTATTAAATT.
In vitro transcription
Labelled RNA substrates were prepared by in vitro transcription from template oligonucleotides using a method adapted from Milligan et al. (10). Briefly, template oligonucleotides with the antisense of the desired RNA, plus the antisense of a conserved +1 to +4 sequence (GGGC) and the T7 promoter (–) were annealed overnight with an oligonucleotide containing the conserved T7 promoter (+) sequence. The hybrid template (2 µmol) was then transcribed in a 10 µl volume in the presence 7.5 µCi of [α-32P]UTP (GeneWorks, Adelaide, SA) with the Ampliscribe transcription kit (Epicentre, Madison, WI). To enhance the specific activity of label and maintain a high transcript yield, the concentration of unlabelled NTPs including UTP was lowered slightly (to 6 mM) from the maximum (10 mM) recommended by the kits manufacturer. After 2 h of transcription at 37°C the RNA substrates were combined with an equal volume of formamide dyes and purified by electrophoresis on a 16% denaturing polyacrylamide gel. RNA oligos were eluted from the gel overnight in 300 mM sodium acetate pH 5.4 and precipitated with 2 vol of ethanol. After washing in 70% ethanol, each transcript was redissolved in water and the concentration determined by measuring the optical density at λ = 260 nm. The extinction coefficient for each transcript sequence was estimated using the Oligo™ 6.0 software package.
Cleavage reactions
The activity of various DNAzymes in vitro was determined by measuring the extent of labelled transcript cleavage over a 1 h time course. Each reaction was set up with the DNAzyme in excess of the substrate in order to establish single turnover conditions. In each reaction vessel a DNAzyme (usually 5 µM) and substrate (0.6 µM) was combined in 50 mM Tris–HCl pH 7.5 and preheated to 85°C for 30 s before equilibrating at 37°C for 5 min. Reactions were initiated by the addition of MgCl2 (at 37°C in 50 mM Tris–HCl pH 7.5) to give a final concentration of 10 mM. Incubation was carried out at 37°C and terminated at various time intervals by the transfer of reaction aliquots to an equal volume of ice-cold stop buffer (90% formamide, 20 mM EDTA and loading dye). At completion, the uncleaved substrate and products were resolved by electrophoresis on a 16% denaturing polyacrylamide gel.
Kinetic analysis
The extent of reaction at each time point was quantified using a PhosphorImager (Molecular Dynamics). For the analysis of DNAzyme kinetics, the cleavage band intensity volume for each sample as a percentage of total intensity volume was transformed graphically in a plot against time. To derive the first-order rate constant from single turnover experiments, a curve was fitted to the data (least squares) using the equation %P = %P∞ – C · exp[–kt]. In this expression %P is the percentage product, %P∞ is the percentage product at t = ∞, C is the difference in %P between t = ∞ and t = 0, and k is the first-order rate constant (11).
RESULTS AND DISCUSSION
The 10–23 DNAzyme has been shown to be capable of cleaving the RY junction of RNA substrates. In order to determine systematically the relative efficiency of the DNAzymes against all four combinations of RY pairs, we designed RNA substrates that each contained one of the four core RY dinucleotides. As each of these substrates shared the same background sequence, the respective difference in cleavage susceptibility could be attributed exclusively to the difference in the core sequence. To monitor the activity of various DNAzyme–substrate combinations, their cleavage reaction kinetics were followed under single turnover conditions through a 1 h time course. Quantitative data on the progress of each reaction (produced from gel images using Imagequant software) was used to generate the first-order rate constant kobs. In this reaction system it was generally assumed that all substrate was bound at initiation such that the kobs value was indicative of the rate of the cleavage step. This assumption was tested by comparison of observed rates achieved under different DNAzyme:substrate ratios, in which no significant difference was seen between an 8- and 60-fold excess of DNAzyme (data not shown).
Hierarchy of substrate core sequence sensitivity
As expected, the RU containing substrates were the most susceptible to cleavage by their matched DNAzyme with consistently high rate constants (Table 1). In accordance with previous work, the identity of the unpaired purine (R) made no significant difference to the cleavage rate (12). However, when the pyrimidine component (Y) of the RY core was changed to cytosine the difference between unpaired purines became substantial. For example, in the first substrate set (Fig. 2), while the GC substrate S-GC1 (kobs = 0.056 min–1) was cleaved with a rate ∼7-fold less than that observed for the corresponding GU core S-GU1 (kobs = 0.378 min–1), the AC substrate S-AC1 (kobs = 5.2 × 10–4 min–1) was cleaved very poorly with an average rate ∼600-fold less than the corresponding AU (kobs = 0.311 min–1). This general pattern was supported by experiments in substrate series 2 with the exception that the GC containing substrate S-GC2 was cleaved as efficiently as the RU substrates (Table 2), allowing the formulation of a scheme for the reactivity of RY substrate core sequences as follows with AU = GU ≥ GC >> AC.
Table 1. Summary of the substrate S1 reaction series, corresponding DNAzymes and reaction rates.
aThe sequence of four RNA substrates, each containing a different RY junction, are at the top of the list. These are followed by the respective DNAzyme sequences written as the bottom strand of the heteroduplex (3′–5′) with substitutions in bold and the catalytic motif shown only for the last in the list.
*The average reaction rate constants for the respective DNAzyme–substrate pairs are shown in a matrix where each column relates to a particular substrate in the list indicated by the corresponding asterisk. The variance between replicates was determined by the standard deviation. In reactions where no cleavage was detected or the rate constant was <0.0001 min–1, the kobs was denoted ND.
Figure 2.
Reaction progress for each RY substrate (S1 series) and their matched DNAzymes. Reaction progress at each time point was determined by densitometry of cleavage band intensity. Data points represented by squares, circles, triangles and rhombi correspond to the GU, AU, GC and AC substrates, respectively. Curves were fitted to the data by regression using the formula described in Materials and Methods. The average rates of these reactions were also recorded in Table 1.
Table 2. Summary of the substrate S2 reaction series, corresponding DNAzymes and reaction rates.
First-order reaction rates for substrate series S2. See Table 1 for explanation of symbols.
DNAzyme base substitution
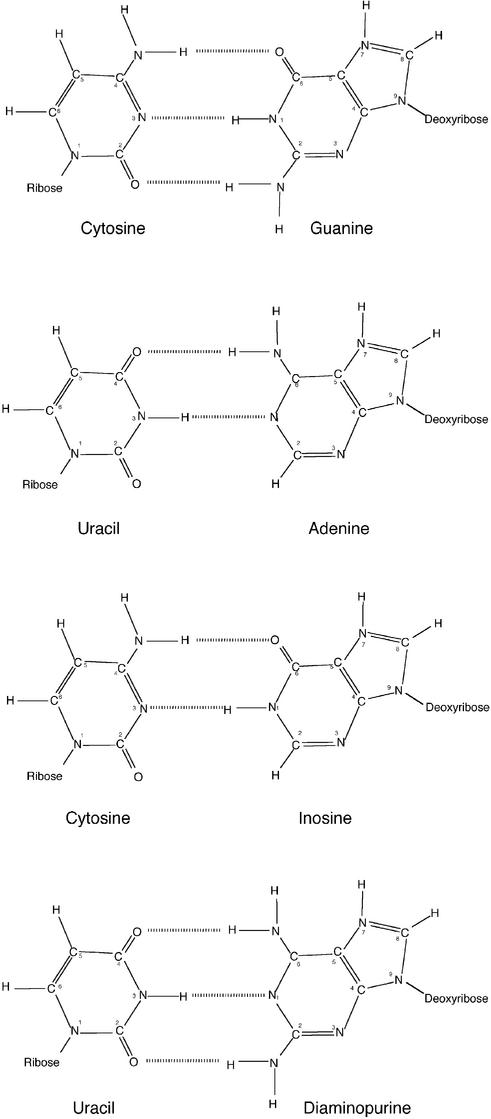
One possible explanation for the poor activity of the 10–23 DNAzyme against RNA cleavage sites consisting of RC (particularly AC) compared to those with RU, is the difference in pairing strength between the core pyrimidine ribonucleotide of the substrate and the corresponding deoxynucleotide in the DNAzyme binding domain. In the case of the rU-dA pairing there is a relatively weak interaction consisting of two hydrogen bonds (13). In contrast, the rC-dG pairing is substantially stronger consisting of a tridentate or three hydrogen bond interaction (Fig. 3). To test the hypothesis that this stronger bonding between the DNAzyme and substrate at this position is antagonistic to catalysis, a range of DNAzymes with various nucleotide substitutions corresponding to this position and to the analogous site in the other binding domain (Table 1) were produced and used to challenge each of the respective RNA substrates. Interestingly, the DNAzymes with substitutions that were predicted to weaken its interaction with the substrate core produced a large enhancement of cleavage activity in RY substrates compared to the matched counterparts. The most significant improvement was produced by substitution of dG with dI such that an rC-dI pairing interaction was established at this critical core position in the DNAzyme–substrate complex. This was exemplified in substrate S-GC1 and S-AC1 by a 2.7-fold and at least 20-fold enhancement of the observed rate compared with the matched DNAzyme (Fig. 4). This pattern was also repeated in the other substrates tested, particularly those with an AC core such as S-AC2 and S-AC3, which were cleaved by the inosine substituted DNAzyme at rates enhanced by a factor of 8.6- and 193-fold, respectively, over their conventionally matched counterparts (Tables 2 and 3). The dramatic improvement in cleavage activity elicited by the G to I substitution in DNAzymes targeting RC substrates (particularly AC) appeared to support the suggestion that a weaker bidentate hydrogen bond interaction more similar to that of the AU pairing was more efficient than a stronger tridentate interaction provided by the matched GC pair (Fig. 3).
Figure 3.
Base pair structures showing hydrogen bond interactions.
Figure 4.
Reaction progress curves comparing the activity of rC-dG and rC-dI core DNAzyme–substrate pairs for the substrate series S1. The average rates of these reactions were also recorded in Table 1.
Table 3. Summary of the substrate S3 reaction series, DNAzymes and reaction rates.
See Table 1 for explanation of symbols.
Diaminopurine substitution
To further examine the hypothesis that strong tridentate pairing is counter productive to the 10–23 DNAzyme mechanism, a diaminopurine (DAP) substitution was made to the RU targeting DNAzyme (Dz3) such that the weaker bidentate rU-dA pair became a stronger tridentate rU-dDAP pair (Fig. 3) (14). As expected, this modification was detrimental to cleavage of the substrate S-AU1, producing a 6-fold reduction in rate (kobs value of 0.311–0.050 min–1) compared to the normal pairing (Table 1). While the impact of this substitution was substantial in reactions with the substrate containing an AU core sequence, there was essentially none on substrates with a GU core (0.378–0.328 min–1). This situation closely reflected the observations in the RC substrates in that the reaction rate could only be extensively modulated by pairing strength when the unpaired purine was adenine. This seemed to indicate that in addition to being unpaired, adenine cleavage sites also require some extra conformational freedom on the 3′ side in order to be cleaved efficiently. While this is provided by standard nucleotides when followed by uracil (dA-rU pair), a paired guanine needed to be replaced by a wobble paired inosine.
Arm II modification
This requirement for additional flexibility extending beyond the cleavage site zone into the base pair interactions of the binding domain, while conditional for unpaired A in the 10–23 DNAzyme, was found to be an absolute requirement in another DNAzyme model designated 8–17 (Fig. 1). The 8–17 DNAzyme was evolved in the same selection experiment as the 10–23 DNAzyme and can only cleave RNA sequences with the core AG in which the rG forms a wobble pair with a dT in the binding domain (1). This preference for weaker interaction near the cleavage site was also observed in a recent study of the 10–23 DNAzyme (12). In this instance the activity of the DNAzyme against some target sequences was also improved by base substitutions which reduced the strength of interaction between the DNAzyme and the substrate. In these experiments, various substitutions in arm II of the DNAzyme (Fig. 1) which produced wobble and mispairs immediately 5′ rather than 3′ of the cleavage site, were found to be responsible for the enhancement of activity. In the present study, we revisited this substitution series with respect to the current RY substrate analogues. Again, as seen with inosine substitution, substrates which contained RC cleavage sites were more amenable to hydrolysis by a subset of DNAzymes that were not fully matched at the site immediately 5′ to the cleavage site. This included some wobble pairs and mispairs with conventional bases which often performed better than those with inosine. This was exemplified in reactions with the substrate S-GC1 where the DNAzyme Dz7 with a rate of 0.102 min–1 was significantly faster than the matched DNAzyme Dz1 (0.056 min–1). In this DNAzyme–substrate complex the rU immediately 5′ to the unpaired G is mispaired to a cognate T in the DNAzyme. The activity of this DNAzyme was followed closely by Dz6 (0.078 min–1) and Dz9 (0.062 min–1) which produce rU-dI and rU-dG wobble pairs, respectively (Table 1). Interestingly, when the unpaired G was changed to an unpaired A (S-AC1), the activity of the rU-dT mispaired DNAzymes was lost, whereas the relative activity of the two wobble paired DNAzymes was maintained. Further investigation of this substitution series was carried out in a another two substrate sets S2 and S3. S-AC2, despite presenting an rC base before unpaired purine rather than rU, was still cleaved more efficiently by the mispaired and wobble paired DNAzymes Dz19 (rC-dC, kobs = 0.031) and Dz17 (rC-dI, kobs = 0.022 min–1) compared to the other paired and mispaired DNAzymes in this series (Table 2). The rate of cleavage of the unpaired G version of S2, substrate S-GC2, however, was not enhanced by DNAzyme modification (Dz17, kobs = 0.444 min–1 and Dz19, kobs = 0.171 min–1) compared to their matched counterpart (Dz12, kobs = 0.451 min–1) which already possessed relatively high activity. Substrate series S3 had the same core sequence (URY) as S1 and also displayed a very similar response to the respective modifications with the reaction rates of wobble paired and rU-dT mispaired DNAzymes exceeding that of the normally paired analogue (Table 3).
Comparison with ribozyme
The relationship between the intensity of base pairing around the cleavage site and cleavage rate observed for the 10–23 and 8–17 DNAzymes has also been seen in the hammerhead ribozyme (15). In this study, ribozyme cleavage activity was shown to be very poor against RNA substrates containing an NCH core triplet, compared with the conventional NUH triplet that is generally used as a rule to for hammer ribozyme targeting. The difference was potentially associated with the strength of pairing between the ribozyme and substrate, where the stronger GC (NCH) was at a disadvantage compared to the weaker AU (NUH) pair. Interestingly, the cleavage activity against an NCH triplet containing substrate was restored in the matched ribozyme when the corresponding G (giving a GC pair) was substituted with inosine such that an IC pair is formed (15). At other positions proximal to the cleavage site, any disturbance of helix integrity between ribozyme and substrate was found to be detrimental to cleavage activity (16).
Combination modification
In view of the reaction rate enhancement achieved with substitutions in each binding domain, it was possible that there may be an advantage for the DNAzyme to incorporate a combination of modifications which affect interactions on both the 5′ and 3′ side of an RC cleavage site. To investigate this prospect a DNAzyme with two inosine substitutions (Dz30) was generated and tested against its cognate substrates. Against the GC containing substrate S-GC1 this DNAzyme with a kobs = 0.071 min–1, did not perform significantly better than the fully matched version (0.056 min–1) with a rate that was less than half that produced by the single inosine modified DNAzyme Dz2. However, against the AC substrate S-AC1, it produced an additional 2-fold enhancement of cleavage rate compared to the single inosine version Dz2 which was already enjoying a rate at least 21-fold that of the matched counterpart. While this increase in performance was remarkable, it is possible that the benefit of the combined modification including dI or some other wobble base on the 3′ side of the cleavage site may be even greater against the substrate triplets GAC and CAC in which the unpaired A is flanked by strong pairs with the DNAzyme on both sides. To test this we altered the substrate S-AC1 core triplet from UAC to CAC and compared the activity of its matched DNAzyme with analogues that contained inosine corresponding to C on either the 5′ or 3′ side of the cleavage site and a combination of both. As expected, the combination modification against this target sequence was substantially greater than either single modification alone with a 5-fold faster rate (data not shown).
Summary
From our investigation of DNAzyme cleavage activity in various substrates containing each of the four RY core sequences, we have found that a hierarchy of susceptibility follows the general scheme AU = GU ≥ GC >> AC. While the performance of the 10–23 DNAzyme against substrates containing an RC core was generally lower than that of those with an RU core, it could be enhanced substantially by reducing the strength of Watson–Crick pairing between them at bases flanking the cleavage site. This was achieved with the greatest effect by substituting dG with dI such that the tridentate bond between rC-dG was converted to a bidentate rC-dI. As the enhancement was strongest in substrates with an AC core, it suggested that the unpaired A was more sensitive to the strength of interaction between its adjacent pairs. This was supported by the substantial loss of comparative activity against the AU substrate core produced by the substitution of dA with dDAP (a tridentate bond forming analogue), which had no significant impact on the corresponding GU substrate cleavage rate. From these observations it would appear that the DNAzyme–substrate geometry necessary to catalyse the transesterification of the linkage following an unpaired A, requires the additional flexibility that can be provided in the subsequent pair by the relative relaxation of an rU-dA or rC-dI interaction rather than the less forgiving rU-dDAP or rC-dG pairs.
These experiments, while yielding further insight to the 10–23 DNAzyme mechanism, have also provided a practical solution for enhancing its activity against substrates containing an RC core sequence, particularly in regard to those with an AC. Inosine could be used instead of guanine at the site described to improve both the chemical and biological activity of DNAzymes that target RC junctions and therefore greatly expand their utility.
Acknowledgments
ACKNOWLEDGEMENT
We would like to thank Dr Phillip Hendry for his critical reading of this manuscript.
REFERENCES
- 1.Santoro S.W. and Joyce,G.F. (1997) A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santoro S.W. and Joyce,G.F. (1998) Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- 3.Cairns M.J., King,A. and Sun,L.Q. (2000) Nucleic acid mutation analysis using catalytic DNA. Nucleic Acids Res., 28, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L.Q., Cairns,M.J., Gerlach,W., Witherington,C., Wang,L. and King,A. (1999) Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J. Biol. Chem., 274, 17236–17241. [DOI] [PubMed] [Google Scholar]
- 5.Santiago F.S., Kavurma,M.M., Lowe,H.C., Chesterman,C.N., Baker,A., Atkins,D.G. and Khachigian,L.M. (1999) New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nature Med., 5, 1264–1269. [DOI] [PubMed] [Google Scholar]
- 6.Sun L.Q., Cairns,M.J., Saravolac,E.G., Baker,A. and Gerlach,W.L. (2000) Catalytic nucleic acids: from lab to applications. Pharmacol. Rev., 52, 325–347. [PubMed] [Google Scholar]
- 7.Haseloff J. and Gerlach,W.L. (1988) Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature, 334, 585–591. [DOI] [PubMed] [Google Scholar]
- 8.Hampel A. and Tritz,R. (1989) RNA catalytic properties of the minimum (–) sTRSV sequence. Biochemistry, 28, 4929–4933. [DOI] [PubMed] [Google Scholar]
- 9.Cairns M.J., Hopkins,T.M., Witherington,C., Wang,L. and Sun,L.Q. (1999) Target site selection for an RNA-cleaving catalytic DNA. Nat. Biotechnol., 17, 480–486. [DOI] [PubMed] [Google Scholar]
- 10.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendry P. and McCall,M.J. (1996) Unexpected anisotropy in substrate cleavage rates by asymmetric hammerhead ribozymes. Nucleic Acids Res., 24, 2679–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairns M.J., Hopkins,T.M., Whitherington,C. and Sun,L.Q. (2000) The influence of arm length asymmetry and base substitution on the activity of the 10–23 DNA enzyme. Antisense Nucleic Acid Drug Dev., 10, 323–332. [DOI] [PubMed] [Google Scholar]
- 13.Sugimoto N., Nakano,S., Katoh,M., Matsumura,A., Nakamuta,H., Ohmichi,T., Yoneyama,M. and Sasaki,M. (1995) Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry, 34, 11211–11216. [DOI] [PubMed] [Google Scholar]
- 14.Bailly C. and Waring,M.J. (1998) The use of diaminopurine to investigate structural properties of nucleic acids and molecular recognition between ligands and DNA. Nucleic Acids Res., 26, 4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig J., Blaschke,M. and Sproat,B.S. (1998) Extending the cleavage rules for the hammerhead ribozyme: mutating adenosine15.1 to inosine15.1 changes the cleavage site specificity from N16.2U16.1H17 to N16.2C16.1H17. Nucleic Acids Res., 26, 2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werner M. and Uhlenbeck,O.C. (1995) The effect of base mismatches in the substrate recognition helices of hammerhead ribozymes on binding and catalysis. Nucleic Acids Res., 23, 2092–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]