Abstract
In Drosophila, PcG complexes provide heritable transcriptional silencing of target genes. Among them, the ESC/E(Z) complex is thought to play a role in the initiation of silencing whereas other complexes such as the PRC1 complex are thought to maintain it. PcG complexes are thought to be recruited to DNA through interaction with DNA binding proteins such as the GAGA factor, but no direct interactions between the constituents of PcG complexes and the GAGA factor have been reported so far. The Drosophila corto gene interacts with E(z) as well as with genes encoding members of maintenance complexes, suggesting that it could play a role in the transition between the initiation and maintenance of PcG silencing. Moreover, corto also interacts genetically with Trl, which encodes the GAGA factor, suggesting that it may serve as a mediator in recruiting PcG complexes. Here, we show that Corto bears a chromo domain and we provide evidence for in vivo association of Corto with ESC and with PC in embryos. Moreover, we show by GST pull-down and two-hybrid experiments that Corto binds to E(Z), ESC, PH, SCM and GAGA and co-localizes with these proteins on a few sites on polytene chromosomes. These results reinforce the idea that Corto plays a role in PcG silencing, perhaps by confering target specificity.
INTRODUCTION
The power of Drosophila genetics has permitted the discovery of genes encoding proteins involved in epigenetic mechanisms before knowing the molecular structure of chromatin. A striking example is given by the analysis of mutants that led Ed Lewis to propose that Polycomb (Pc) encodes a general repressor of the Bithorax Complex (1). Later on, genetic screens identified two classes of regulatory factors responsible for maintaining homeotic (Hox) gene expression throughout development. These factors are encoded by two groups of genes: the Polycomb group (PcG) and the trithorax group (trxG). Genetic data demonstrated an antagonistic role for PcG and trxG genes: mutations in trxG genes suppress phenotypes of PcG mutants. The PcG and TrxG proteins were later on shown to function as multimeric complexes that regulate Hox gene expression through modulation of the chromatin structure. In Drosophila, the domains of Hox gene expression are established early during development by the gap and pair rule gene products. Then, these domains are maintained throughout development by the PcG and TrxG complexes; the TrxG complexes are involved in maintaining the activation of Hox genes whereas the PcG complexes are involved in maintaining their silencing outside their expression domains. The pleiotropic phenotypes of PcG and trxG Drosophila mutants and the localization of the proteins they encode at multiple sites on polytene chromosomes were the first indications that the function of these regulators was not restricted to the Hox genes. Several non-Hox targets were further discovered and key genes, such as segmentation genes or dorso–ventral polarity genes, were shown to be targets for PcG (2,3) or TrxG (4,5) factors.
The PcG and the TrxG complexes are targeted to appropriate genes by regulatory DNA sequences called Polycomb Response Elements (PREs) and Trithorax Response Elements. The PREs exhibit autonomous silencing activity and are able to recruit PcG proteins in vivo (6,7). The first evidence that the GAGA factor, initially classified as a TrxG factor, was involved in PRE silencing came from the analysis of the Fab-7 PRE, the silencing activity of which requires the GAGA binding sites (8). This factor was further shown to be involved in targeting PcG complexes (9–11). There seems to be a multiplicity of distinct PcG complexes, yet only two PcG complexes have been purified from embryonic extracts so far. The first one, called the ESC/E(Z) complex, contains not only the PcG proteins ESC and E(Z) but also the histone deacetylase dRPD3 and the histone binding protein p55. It is involved in the early establishment of Hox gene silencing at the end of segmentation (12–14). A second complex, termed PRC1 for Polycomb Repressive Complex 1, has been shown to contain the PcG proteins PC, PH, PSC, dRING1 and SCM as well as several TAFs (TATA-binding protein Associated Factors) (15,16).
The mechanisms leading to silencing by PcG complexes have not been elucidated as yet. PcG complexes are supposed to render chromatin more compact and have been demonstrated to reduce DNA accessibility (17,18). However, silencing by PcG proteins does not exclude general transcription factors from promoters (19), and the presence of TAFs in the PRC1 complex led to the proposal that silencing may be due to a direct interaction between PcG complexes and the transcription machinery. Furthermore, there is some evidence of a direct connection between PcG silencing and chromatin structure. PC interacts with nucleosomal core particles in vitro (20) and the PRC1 complex inhibits chromatin remodeling of nucleosomal arrays (21). One hypothesis is that complexes would freeze the position of nucleosomes in a structure that prevents gene activity. The presence of the histone binding protein p55 together with the histone deacetylase dRPD3 in the ESC/E(Z) complex suggests that nucleosome deacetylation may underlie PcG silencing (14).
The corto gene of Drosophila melanogaster, also known as ccf, has been shown to play a role in chromosome condensation and Hox gene regulation (22,23). Loss-of-function males exhibit an ectopic sex comb on the second tarsal segment of the first leg that corresponds to the homeotic transformation of the first tarsal segment into the second. This phenotype can be allocated to misregulation of the Hox gene Sex comb reduced (Scr). Furthermore, corto mutations enhance the extra sex comb phenotype of some mutants of the PcG genes Enhancer of zeste [E(z)], multi sex-combs (mxc), Pc, Polycomb-like (Pcl), polyhomeotic (ph) and Sex-comb on midleg (Scm), strongly suggesting that corto and these PcG genes act synergistically in regulating Scr (22,23). Loss-of-function corto mutants also exhibit post-pronotum defects that exist in mutants for Trithorax-like (Trl), the gene that encodes the GAGA factor. This phenotype is enhanced in double heterozygotes for corto and Trl, suggesting a relationship between corto and Trl in Antennapedia (Antp) regulation (23). These genetic observations led us to address the question of Corto belonging to PcG complexes like ESC/E(Z) or PRC1, as well as its role in the direct connection between these complexes and the GAGA factor. In the present report, we show that Corto is a chromo domain protein. Using independent co-immunoprecipitation experiments, we provide evidence that, in embryos, Corto is associated with ESC, on the one hand, and with PC, on the other hand. Moreover, by in vitro binding assays and yeast two-hybrid analysis, we provide evidence that Corto directly interacts with ESC and E(Z), directly interacts with PH and SCM, but not with PC, and also directly interacts with the GAGA factor. Lastly, we demonstrate, by immunostaining experiments, that Corto partially co-localizes with ESC, E(Z), PC, PH, SCM and GAGA on third instar larvae polytene chromosomes. Taken together, these results strongly suggest that Corto belongs to PcG complexes that regulate the chromatin structure.
MATERIALS AND METHODS
cDNA and plasmids
The cDNAs corresponding to esc, E(z), Scm and Trl (isoform 519) were purchased from Invitrogen (EST numbers SD03549, LD30505, LD09463 and LD41963, respectively). The corto cDNA came from clone NB67 (22), the Pc cDNA came from plasmid pUHE/PcM-D1.5 (24) and the ph proximal cDNA (ph-p) came from clone C4-6 (25). Plasmid constructs are described in the Supplementary Material. Plasmids pJG-PH, pJG-PHΔN and pJG-PHΔS, derived from the ph-p cDNA, were kindly provided by Dr H. Brock (26).
Embryonic extracts and immunoprecipitation assays
Embryos from the hs-LexA-ESC strain (7) were collected every 4 h, heat shocked for 1 h at 37°C and allowed to recover for 1 h at 25°C. Embryos from the the α1T-LexA-PC strain (7) were collected every 12 h. Nuclear extracts were performed as described in Soeller et al. (27) except that complete protease inhibitor cocktail tablets (Roche) were used. The final proteic solution was dialyzed against incubation buffer (25 mM HEPES pH 7.6, 100 mM KCl, 0.1 mM EDTA, 10 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 10% glycerol) for 3 h at 4°C. Anti-LexA antibodies (Upstate Biotechnology) were covalently linked to protein A–agarose as described in Harlow and Lane (28). Protein A beads (mock) or protein A-anti-LexA beads (assay) were washed with 1 ml of incubation buffer then incubated with embryonic nuclear extract containing 150 µg of protein in 300 µl of incubation buffer for 3 h at 4°C. The beads were then washed three times with 1 ml of interaction buffer for 10 min at 4°C and analyzed by western blot.
GST pull-down assays
In vitro transcription/translation assays were performed using the appropriate RNA polymerase, the TNT® coupled reticulocyte lysate system (Promega) and [35S]methionine as indicated by the manufacturer. As PC contains only two methionines, the protein obtained by the transcription/translation reaction showed very low specific activity. Then, PC was produced in bacteria from plasmid pUHE/PcM-D1.5 and GST pull-down results were analyzed by western blot.
Aliquots of 50 µl of beads coated with GST or GST-Corto fusion proteins containing ∼1 µg of protein were incubated in 1 ml of interaction buffer [40 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.5% Igepal, 1 mM AEBSF, 0.5% protease inhibitors for bacterial extracts (Sigma), 3% BSA] with 10 µl of the translation mixtures. The beads were then washed four times in 1 ml of incubation buffer without BSA while progressively increasing the KCl concentration to 400 mM and submitted to PAGE.
Two-hybrid methods
Interaction mating assays were performed by mating, on yeast extract/peptone/dextrose plates, bait and prey carrier haploid strains that had been previously grown on selective medium (29). The mated diploids were then replicated on selective medium containing either dextrose or galactose/raffinose with or without leucine. After 24 h, the plates with leucine were overlaid with 10 ml of X-gal agarose (0.5 M Na2HPO4 pH 7.5, 0.2% SDS, 10% dimethylformamide, 0.1% X-gal, 1% agarose) to check the activity of the LacZ reporter gene. X-gal and leucine tests both gave the same results except for yeasts containing pEG-Corto, which never grow on medium without leucine. Thus, only the X-gal assays are presented here. Plasmids pJG4-5, which encodes the B42 activation domain, and pEG202, which encodes the LexA DNA binding domain, were used as negative controls. Plasmids pB42AD-T, which encodes a B42 activation domain/SV40 large T-antigen fusion protein, and pLexA53, which encodes a LexA/murine p53 fusion protein, were used as positive controls (30).
Antibodies and immunolocalization on polytene chromosomes
Squashes of third instar larvae salivary glands and immunostaining were performed as described by Cavalli (http:// www.igh.cnrs.fr/equip/cavalli/link.labgoodies.html). The w1118 strain was used for these assays except for immunolocalization of ESC, where the hs-LexA-ESC strain was used. In that case, larvae were repetitively heat shocked for 2 h at 37°C for the 3 days preceding the experiment. Polyclonal antibodies against the GST-C127/203 fusion protein were raised in the rat and stored at –20°C. They were used at a 1:40 dilution. Anti-PC (24), anti-PH (31), anti-SCM (32) and anti-E(Z) (33) were used at a 1:40 dilution and anti-GAGA (a generous gift from Dr P. B. Becker) at a 1:150 dilution. Anti-LexA was used at a 1:200 dilution. The secondary antibodies Alexa Fluor® 594 goat anti-rabbit IgG (H+L) conjugate and Alexa Fluor® 488 goat anti-rat IgG (H+L) conjugate ‘highly cross adsorb’ (Molecular Probes) were used at a 1:1000 dilution.
RESULTS
Structural analysis of Corto
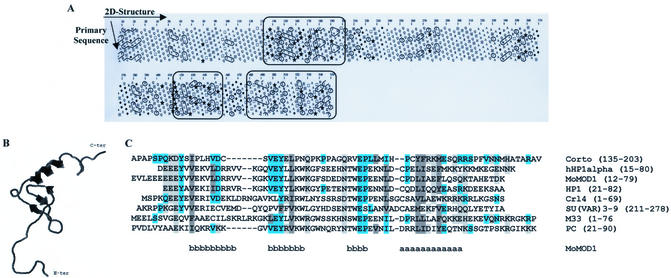
No significant similarity between Corto and known proteins could be detected by BLAST or PSI-BLAST. We performed a two-dimensional hydrophobic cluster analysis to reveal globular domains that might represent functional domains (34). Three main hydrophobic cores were detected, located at positions 127–203, 418–455 and 480–550, respectively (Fig. 1A). Structural comparison of these domains with protein folds from the Brookhaven Protein Databank were carried out using the Bioinformatics Tools Server (35). While no obvious similarities were observed between the two C-terminal globular domains and known proteins, the most N-terminal domain (amino acids 127–203) exhibited similarities with the chromo (chromatin organization modifier) domains of the mouse modifier protein 1 (MoMOD1) (PDB accession code 1ap0) (36) and the Schizosaccharomyces pombe Clr4 protein (PDB accession code 1g6z) (37). MoMOD1 and Clr4 are homologous to the heterochromatin associated protein 1 (HP1) and the SU(VAR)3-9 protein of Drosophila, respectively. Both proteins have been shown to be involved in repression of gene expression in heterochromatin (38,39). A putative three-dimensional structure of the Corto domain was obtained with MODELLER, a program for comparative modeling (40), using the MoMOD1 chromo domain as a template (Fig. 1B). This suggests that this domain exhibits a typical chromo domain structure composed of three β-sheets followed by one α-helix. Alignment of the primary sequences of the Corto 127/203 domain and the chromo domains of human HP1α, MoMOD1, Drosophila HP1, Crl4, Drosophila SU(VAR)3-9, mouse Polycomb protein M33 and Drosophila Polycomb protein PC supports this hypothesis (Fig. 1C).
Figure 1.
Structure of the Corto protein. (A) Hydrophobic cluster analysis plot (reviewed in 34) of Corto (Swiss-Prot identity code P41046). The clusters of hydrophobic residues are shown. The symbols used in the plot are: open square, threonine; dotted square, serine; diamond, glycine; star, proline. The three main globular domains, located at positions 127–203, 418–455 and 480–550, are framed. (B) Putative Corto chromo domain three-dimensional structure as obtained with MODELLER using the MoMOD1 chromo domain (PDB identity code 1ap0) as a template. (C) Alignment of the N-terminal globular domain of Corto and the chromo domains of human HP1α (P45973), mouse modifier protein 1 (MoMOD1) (PDB identity code 1ap0), D.melanogaster HP1 (P05205), S.pombe Clr4 protein (O60016), D.melanogaster SU(VAR)3-9 (P45975), mouse M33 (P30658) and D.melanogaster PC (P26017). Identical amino acids between Corto and the other sequences are on a blue blackground, similar ones on a gray background, hydrophobic residues in dark gray and charged residues in light gray. Positions of the secondary structures (b for β-sheets and a for α-helix), as experimentally determined for MoMOD1 (36), are indicated below the alignment.
The chromo domain and the C-terminal domain of Corto mediate self-association
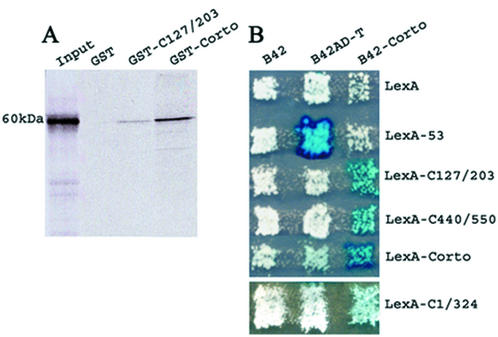
Self-association of Corto was first checked by GST pull-down assay. We observed that the labeled full-length Corto protein binds weakly to the chromo domain (amino acids 127–203) and more strongly to the whole protein (Fig. 2A). Under the same experimental conditions, the GST control beads did not retain any radioactivity.
Figure 2.
Self-association of Corto. (A) GST pull-down experiments: binding of 35S-labeled Corto to GST, GST-C127/203 and GST-Corto as revealed by autoradiography. (B) Two-hybrid experiments. Fusion proteins indicated horizontally (the negative control B42, B42AD-T, which binds to LexA-53, and B42-Corto) were expressed in the RFY206 strain (MATa). Fusion proteins indicated vertically (the negative control LexA, LexA-53, LexA-C127/203, LexA-C440/550 and LexA-Corto) were expressed in the RFY231 (MATα) strain that contains the LacZ reporter plasmid pSH18-34 (top). LexA-C1/324 was expressed in the EGY48SHΔSpe (MATα) strain which contains an endogenous LacZ gene (bottom). Strains were mated on appropriate medium and the activity of LacZ in the diploid yeasts was checked as described in Materials and Methods.
To test further if Corto is able to make multimers, we carried out two-hybrid experiments in yeast. We observed that the LacZ reporter gene was strongly activated in diploids expressing both B42-Corto and LexA-Corto on galactose/raffinose medium (Fig. 2B). This result leads us to conclude that Corto is able to make multimers in yeast. To clarify the position of the contacts between Corto molecules, we subcloned the regions corresponding to amino acids 1–324, to the chromo domain (amino acids 127–203) and to the C-terminal globular domain (amino acids 440–550) in pEG202 in-frame with LexA. A weak activation of the LacZ reporter was observed when B42-Corto was co-expressed with LexA-C1/324. Moreover, we observed a strong activation of the LacZ reporter gene in diploids co-expressing B42-Corto and LexA-C127/203 or B42-Corto and LexA-C440/550. These results show that at least two domains, i.e. the chromo domain and the C-terminal domain, are involved in Corto dimerization.
Corto co-immunoprecipitates with ESC and with PC
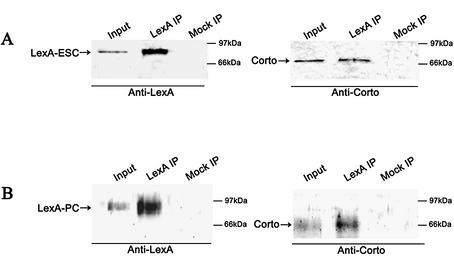
The fact that Corto belongs to PcG complexes was checked by co-immunoprecipitation experiments. As the rat anti-Corto antibodies did not immunoprecipitate Corto, we used the hs-LexA-ESC and LexA-PC strains. We prepared nuclear extracts from 0–4 h heat-shocked embryos of the hs-LexA-ESC strain and performed co-immunoprecipitation experiments using anti-LexA antibodies (Fig. 3A). We observed that the anti-LexA antibodies precipitated Corto indicating that, in early embryos, Corto is included in a complex containing ESC. We further tested the fact that Corto belongs to a PC-containing complex using overnight nuclear extract from embryos from the α1T-LexA-PC strain by immunoprecipitation (Fig. 3B). We observed that the anti-LexA antibodies precipitated Corto, indicating that in these embryos there is a complex containing both PC and Corto.
Figure 3.
Co-immunoprecipitation of LexA-ESC or LexA-PC and Corto. Nuclear extracts of heat-shocked 0–4 h embryos from the hs-LexA-ESC strain (A) or overnight embryos of the α1T-LexA-PC strain (B) were immunoprecipitated with anti-LexA beads (LexA IP) or with beads containing no antibody (Mock IP). Western blots were revealed with anti-LexA or with anti-Corto antibodies as indicated.
These results suggest that in the embryo, Corto belongs at least transiently to an ESC complex that may be the PcG initiation complex and also to a PC complex that may be the PRC1 complex. This is in agreement with our previous observations of genetic interactions between corto and some members of these complexes (23). Hence, we performed assays for direct interaction between Corto and these proteins.
Corto binds to ESC and E(Z), two members of the PcG initiation complex
We first performed GST pull-down assays to check the interactions between Corto and ESC and between Corto and E(Z). As shown in Figure 4B, both full-length Corto and the Corto chromo domain (amino acids 127–203) retained 35S-labeled ESC and E(Z). We concluded that both proteins were able to bind to Corto in vitro via its chromo domain.
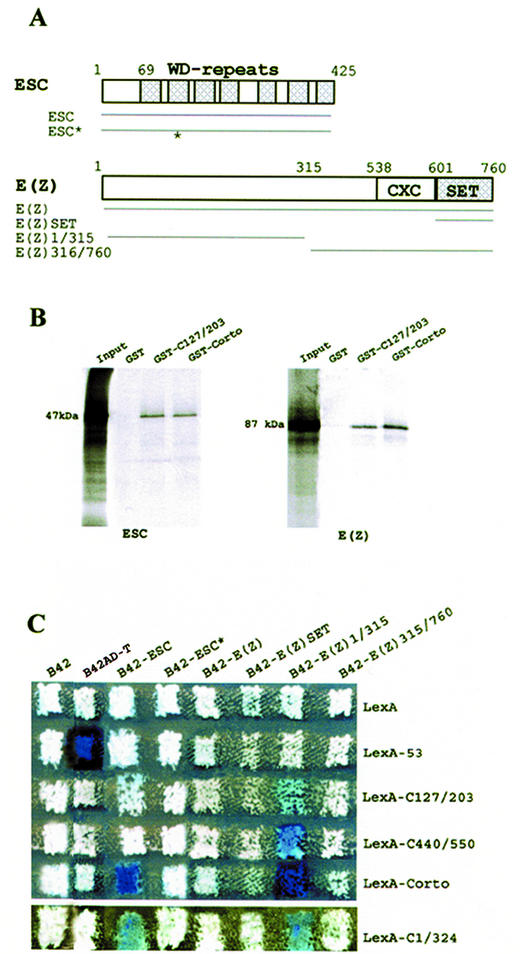
Figure 4.
Corto binds to ESC and E(Z). (A) Schematic representations of the ESC and E(Z) proteins and of the areas subcloned in-frame with B42 in plasmid pJG4-5. The ESC protein consists of seven WD repeats spread all along the protein. ESC* exhibits two point mutations in the second WD repeat (indicated by an asterisk). The E(Z) protein contains a SET domain at the C-terminal end preceded by a cystein-rich region, CXC. (B) GST pull-down experiments: binding of 35S-labeled ESC and E(Z) to GST, GST-C127/203 and GST-Corto as revealed by autoradiography. (C) Two-hybrid experiments. Mating was performed as described in Figure 2.
To extend these results, we subcloned the full-length ESC coding region in pJG4-5 and tested its ability to bind to Corto in yeast. The ESC protein (425 amino acids) is composed of seven WD repeats spanning the C-terminal 355 residues (12). We observed that co-expression of B42-ESC and LexA-Corto strongly activates the LacZ reporter gene (Fig. 4C). We also observed activation of LacZ when co-expressing B42-ESC and LexA-C1/324, but no activation could be detected when co-expressing B42-ESC and LexA-C107/203 or LexA-C440/550. These results confirmed that ESC contacts Corto in its N-terminal half. Nevertheless, in contrast to GST pull-down experiments, the chromo domain by itself is not sufficient to provide interaction. To further confirm the binding of ESC to Corto, we used the pJG-ESC* construct, in which esc displays two point mutations leading to the substitution of two amino acids in the second WD repeat (E125G and L142F). We observed that B42-ESC* did not interact with any of the LexA-Corto proteins. This result clearly shows that the structure adopted by the WD repeat motif is necessary for the binding of ESC to Corto. Interestingly, point mutations in the second WD repeat of the human ESC homolog EED also disrupts its interaction with the human E(Z) homolog EZH2 (41).
We also checked the interactions between Corto and E(Z) by yeast two-hybrid experiments (Fig. 4C). E(Z) is a 760 amino acid protein characterized by the presence of a highly conserved domain called the SET domain [for SU(VAR)3-9, E(Z), Trithorax] at the C-terminal end (Fig. 4A). This domain, initially identified at the C-terminal ends of these three proteins in Drosophila, mediates protein–protein interactions (reviewed in 42). The full E(Z) coding region as well as fragments corresponding to the N-terminal half of E(Z) (amino acids 1–315), to the C-terminal half of E(Z) (amino acids 315–760) and to the SET domain alone were subcloned in pJG4-5Y. No activation of the LacZ reporter gene was observed when B42-E(Z) and LexA-Corto were co-expressed. Moreover, no activation of LacZ was observed when co-expressing B42-E(Z)SET or B42-E(Z)315-760 with any of the LexA-Corto fusions. In contrast, co-expression of B42-E(Z)1/315 and LexA-Corto strongly activated the LacZ reporter gene, showing that the N-terminal half of E(Z) strongly interacts with Corto. Furthermore, co-expression of B42-E(Z)1/315 and LexA-C1/324, LexA-C127/203 or LexA-C440/550 also activated the LacZ reporter gene. These results corroborate the GST pull-down experiment results and strongly suggest that the N-terminal half of E(Z) directly interacts with Corto not only at the level of the chromo domain but also at the level of its C-terminal end.
Corto binds to some members of the PRC1 maintenance complex
We also tested for direct binding of Corto to some members of the PRC1 maintenance complex, i.e. PC, PH and SCM, by GST pull-down experiments. As shown in Figure 5B, PC was not retained either on the GST-C127/203 or on the GST-Corto beads, PH was strongly retained on the GST-C127/203 and GST-Corto beads, and SCM was weakly retained on the GST-C127/203 beads and strongly retained on the GST-Corto beads. These results suggest that Corto does not directly interact with PC but directly interacts with PH and SCM.
Figure 5.
Corto directly binds to PH and SCM but does not bind to PC. (A) Schematic representations of PC, PH and SCM and of the areas subcloned in-frame with B42 in pJG4-5. The PC chromo domain (CH) is located at the N-terminal end of the protein and the SAM motifs of PH and SCM are located at the C-terminal ends of these proteins. (B) GST pull-down experiments: binding of PC, PH and SCM to GST, GST-C127/203 and GST-Corto. PC was produced in bacteria and the GST pull-down assays were revealed with anti-PC antibodies; 35S-labeled PH or SCM were produced by in vitro transcription/translation and the GST pull-down assays were revealed by autoradiography. (C) Two-hybrid experiments. Mating was performed as described in Figure 2.
Two-hybrid experiments were then performed to confirm these results. PC is a 390 amino acid protein with a chromo domain located between positions 26 and 84 (Fig. 5A). The full coding region was subcloned in pJG4-5 and introduced in yeast. The co-expression of B42-PC and LexA-Corto, LexA-C1/324, LexA-C127/203 or LexA-C440/550 never led to activation of the LacZ reporter. These results corroborate the GST pull-down experiments and suggest that Corto does not directly interact with PC.
Interactions between PH and Corto were also examined by two-hybrid experiments (Fig. 5C). PH is a large protein (1589 amino acids) characterized by a SAM domain (Sterile Alpha Motif), a protein–protein interaction motif present in a wide variety of proteins and involved in many biological processes (26). This 165 amino acid domain is located at the C-terminal end of the protein (Fig. 5A). B42-PH is a fusion of the full-length PH protein with B42 whereas B42-PHΔN is deleted from the SAM domain and B42-PHΔS retains only amino acids 1–522. B42-PH and B42-PHΔN never led to activation of LacZ in the presence of any of the LexA-Corto fusion proteins. In contrast, co-expression of B42-PHΔS and LexA-Corto or LexA-C1/324 led to activation of the reporter. No activation was observed when co-expressing B42-PHΔS and LexA-C127/203 or LexA-C440/550. Then, two-hybrid assays did not reveal any contact between PH and the chromo domain of the Corto protein. From two-hybrid and GST pull-down assays, we can deduce that the N-terminal half of Corto binds to the N-terminal half of PH.
Interactions between Corto and SCM were also examined by two-hybrid experiments (Fig. 5C). Like PH, SCM possesses a SAM domain at its C-terminal end. The full Scm coding region was subcloned in pJG4-5Y and introduced into yeast. The LacZ reporter was strongly activated when co-expressing B42-SCM and LexA-Corto, LexA-C1/324, LexA-C127/203 or LexA-C440/550. These results corroborate the GST pull-down experiments and clearly allow us to conclude that Corto contacts SCM via two regions, i.e. the chromo domain and the C-terminal domain.
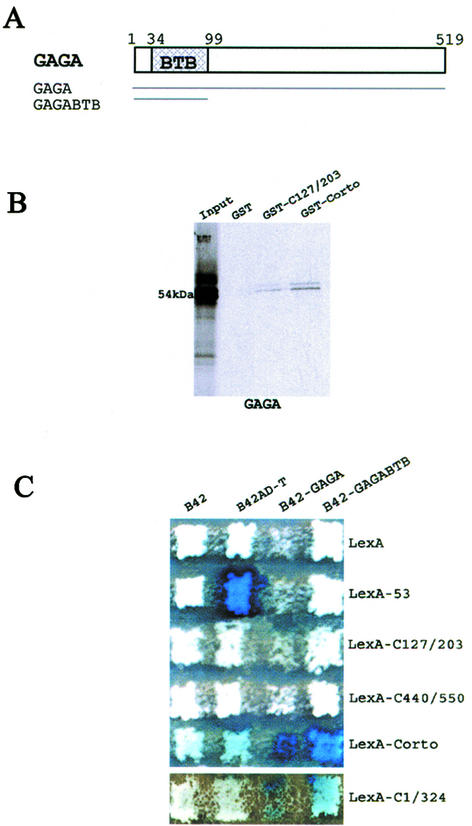
Corto binds to the GAGA factor
Since we had previously observed genetic interactions between corto and Trl (Trl encodes the GAGA factor), we then tested direct interactions between Corto and GAGA. GST pull-down experiments showed that GAGA was retained on GST-C127/203 and GST-Corto beads while it was not retained on control GST beads, suggesting that Corto interacts with GAGA at the level of the chromo domain (Fig. 6B).
Figure 6.
Corto directly binds to the BTB/POZ domain of the GAGA factor. (A) Schematic representation of the GAGA 519 amino acid isoform. The BTB/POZ domain is located in the N-terminal end of the protein. (B) GST pull-down experiments showing the binding of 35S-labeled GAGA to GST, GST-C127/203 and GST-Corto. (C) Two-hybrid experiments. Mating was performed as described in Figure 2.
Interactions between Corto and GAGA were also analyzed by two-hybrid experiments (Fig. 6C). The B42-GAGA fusion protein inhibits yeast growth. Nevertheless, we could observe an activation of LacZ when co-expressing B42-GAGA and LexA-Corto or LexA-C1/324. No activation was observed while co-expressing B42-GAGA and LexA-C127/203 or LexA-C440/550. Binding of GAGA to the chromo domain, although suggested by GST pull-down assays, could not be shown by two-hybrid assays. This may be due to the need for a larger domain. The GAGA protein contains a BTB/POZ domain (BR-C, Tramtrack, Bab, Pox virus, Zinc finger), a characteristic domain that mediates homomeric or heteromeric dimerization (Fig. 6A) (43,44). We further subcloned this domain in pJG4-5Y to test its binding to Corto. We observed that the co-expression of B42-GAGABTB with LexA-Corto or LexA-C1/324 activated LacZ whereas its co-expression with LexA-C127/203 or LexA-C440/550 did not activate LacZ. These results lead us to conclude that the N-terminal half of Corto directly interacts with the BTB/POZ domain of GAGA.
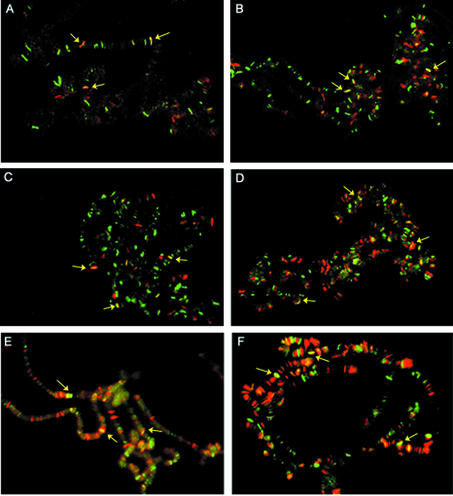
Corto co-localizes with PcG proteins and GAGA at a few sites on polytene chromosomes
The Corto protein was previously shown to bind to polytene chromosome arms at many loci and to co-localize partially with PSC (22). Although PSC shares many sites with PH and PC, which completely co-localize, about 50 sites are not shared by PSC and PC/PH (25,45). To extend the binding experiments in vivo, we wanted to determine whether Corto co-localizes with some other PcG proteins and with the GAGA factor. We immunodetected Corto and these proteins simultaneously on third instar larvae polytene chromosomes. As ESC is not expressed in larvae, we used the transgenic strain hs-LexA-ESC to artificially produce the protein in salivary glands. As shown in Figure 7, Corto binding sites coincide with a fraction of the ESC, E(Z), PC/PH, SCM and GAGA binding sites.
Figure 7.
Simultaneous immunofluorescence localizations of Corto and ESC (A), Corto and E(Z) (B), Corto and PC (C), Corto and PH (D), Corto and SCM (E) and Corto and GAGA (F) on salivary gland polytene chromosomes. The anti-Corto antibodies were detected with Alexa Fluor® 488 anti-rat IgG antibodies (green) and the anti-LexA, E(Z), PC, PH, SCM and GAGA antibodies were detected with Alexa Fluor® 594 anti-rabbit IgG antibodies (red). Yellow arrows indicate some co-localizations.
DISCUSSION
Previous studies have suggested that Corto acts as a chromatin structure modulator involved in PcG silencing (22,23). The present results provide molecular evidence supporting the fact that this protein exhibits a chromo domain shared with some chromatin binding proteins, participates in PcG complexes and directly contacts some PcG proteins as well as the GAGA factor.
Structural comparison with protein folds from the Brookhaven Protein Databank and primary sequence alignments help us to propose that the region of the Corto protein located between residues 127 and 203 is a chromo domain. A previous analysis of the whole D.melanogaster genome by primary sequence comparison had listed 13 chromo domain proteins, which did not include Corto (46). Our result illustrates the power of structural comparisons and lets us foresee that the chromo domain may be more widely represented among Drosophila proteins than previously thought. Chromo domains are conserved domains found in a variety of proteins associated with gene silencing, such as the heterochromatic protein 1 (HP1), SU(VAR)3-9 or the PcG protein PC. These domains are supposed to mediate protein– protein interactions. Consistent with this are our findings that some of the interactions reported here involve the Corto chromo domain. Furthermore, in vitro binding analysis and two-hybrid experiments show that this domain of the Corto protein self-associates, another general property of chromo domains (47).
To gain insight into the molecular mechanism responsible for genetic interactions between corto and some PcG genes, we first tested whether Corto is involved in PcG complexes and showed by co-immunoprecipitation that, in the embryo, Corto is a constituent of both a complex containing ESC and a complex containing PC. Then, we tested Corto for direct interaction with members of the PcG initiation complex [i.e. ESC and E(Z)], with members of the PRC1 maintenance complex (i.e. PC, PH and SCM) and with the GAGA factor. The whole Corto protein directly interacts with all these proteins except PC. The chromo domain is clearly involved in the interaction with E(Z) and SCM. For the two latter proteins, a strong interaction was also observed with the C-terminal domain of Corto. These results and previous genetic data (23) strongly suggest that Corto is functionally associated in multimeric complexes with these proteins.
Although ESC is expressed at high levels during early embryogenesis and at only very low levels in imaginal discs, E(Z), like Corto, is expressed all through development. E(Z) has indeed been shown to fulfill two functions in Drosophila: an early function linked to ESC in the initiation of PcG silencing and a late function in maintaining PcG repression. This observation led Ng et al. (48) to propose that E(Z) would have an as yet undiscovered proteic partner that would replace ESC in the ESC-E(Z) complex at late developmental stages. As Corto binds to the N-terminal end of E(Z) as does ESC (49), competition between Corto and ESC for binding to E(Z) may occur to favor the transition between initiation and maintenance complexes. Another PcG protein, PCL, has been shown to associate with E(Z) and ESC in embryos, indicating PCL as another candidate for this function (50). Genetic interactions between corto and Pcl (23) as well as GST pull-down results indicating that PCL binds to Corto (data not shown) are in favor of a ESC-E(Z)-PCL-Corto complex that plays a transient role in PcG silencing.
A maintenance complex, PRC1, containing PC, PH, SCM, PSC and other proteins was identified in embryos (15,16), and genetic interactions, co-localizations on polytene chromosomes and in vivo recruitment to transgenes suggest that these proteins are also functionally associated during the larval and pupal stages. The present study shows that Corto directly interacts with two members of the PRC1 maintenance complex, PH and SCM. It also shows that Corto, although exhibiting about 200 binding sites on polytene chromosomes, shares only a few of them with these PcG protein binding sites. Salivary gland cells are terminally differentiated and may be extreme examples of the maintenance stage in chromatin structure. Thus, it is possible that Corto and these proteins share more targets in other tissues. However, this supports the hypothesis that multiple PcG complexes of different compositions regulate different target genes (51) and leads us to propose that Corto joins transiently to the PRC1 complex, perhaps confering target specificity.
In agreement with genetic interactions between corto and Trl, the gene encoding the GAGA factor, we have also demonstrated a direct interaction of Corto with GAGA. This interaction was further confirmed by co-localization on a few sites on polytene chromosomes. Although originally described as a transcriptional activator, GAGA is also involved in PcG silencing: it has been shown to bind to multiple sites in PREs and to be involved in a PcG complex containing PC, PH and PSC in embryos (7). However, direct interactions between GAGA and these proteins have never been reported. Our results suggest that Corto might be a transient mediator in recruiting such PcG complexes at PREs. SAP18, a polypeptide associated with the Sin3-HDAC co-repressor complex, was also shown to directly interact with GAGA via a region that encompasses the BTB domain (52). SAP18, like Corto, co-localizes with GAGA at only a few sites on polytene chromosomes (52). This suggests that recruitment of specific repressor complexes at GAGA binding sites through BTB domain binding proteins like SAP18 or Corto might be a general mechanism in PcG silencing. The transcriptional activity of GAGA was assigned to its C-terminal glutamine-rich domain (53), but its BTB domain, which mediates self-oligomerization, has been shown to be necessary for efficient binding to natural target promoters (54). A speculative model in which DNA-bound GAGA molecules form oligomers through association of their BTB domain and bent DNA has been proposed (54). Considering this model, association of other proteins, such as Corto, to the BTB domain might result in partial disruption of the GAGA oligomers and might drive the promoter into a repressed rather than into an activated state.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Drs C. Antoniewski, H. Brock, S. Netter, R. Paro and J. -Y. Roignant for the vectors, Drs P. B. Becker, R. Jones, R. Paro, F. Maschat and A. Peterson for the antibodies, Dr V. Pirrotta for the hs-LexA-ESC and the α1T-LexA-PC strains, Dr M. Gho for help with fluorescence microscopy, Dr E. Mouchel-Vielh for helpful discussions and V. Ribeiro and N. Salmon for excellent technical assistance. This work was supported by the Centre National de la Recherche Scientifique and the Université Pierre et Marie Curie. F.P. is a recipient of Association pour la Recherche sur le Cancer grant 4492. J.S. is supported by a scholarship from the Ministère de la Recherche and A.L. by a scholarship from the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Lewis E.B. (1978) A gene complex controlling segmentation in Drosophila. Nature, 276, 565–570. [DOI] [PubMed] [Google Scholar]
- 2.McKeon J., Slade,E., Sinclair,D.A., Cheng,N., Couling,M. and Brock,H.W. (1994) Mutations in some Polycomb group genes of Drosophila interfere with regulation of segmentation genes. Mol. Gen. Genet., 244, 474–483. [DOI] [PubMed] [Google Scholar]
- 3.Netter S., Fauvarque,M.-O., Diez del Corral,R., Dura,J.-M. and Coen,D. (1998) white+ transgene insertions presenting a dorsal/ventral pattern define a single cluster of homeobox genes that is silenced by the polycomb-group proteins in Drosophila melanogaster. Genetics, 149, 257–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daubresse G., Deuring,R., Moore,L., Papoulas,O., Zakrajsek,I., Waldrip,W.R., Scott,M.P., Kennison,J.A. and Tamkun,J.W. (1999) The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development, 126, 1175–1187. [DOI] [PubMed] [Google Scholar]
- 5.Treisman J.E., Luk,A., Rubin,G.M. and Heberlein,U. (1997) eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev., 11, 1949–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poux S., McCabe,D. and Pirrotta,V. (2001) Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development, 128, 75–85. [DOI] [PubMed] [Google Scholar]
- 7.Poux S., Melfi,R. and Pirrotta,V. (2001) Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev., 15, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagstrom K., Muller,M. and Schedl,P. (1997) A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics, 146, 1365–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busturia A., Lloyd,A., Bejarano,F., Zavortink,M., Xin,H. and Sakonju,S. (2001) The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development, 128, 2163–2173. [DOI] [PubMed] [Google Scholar]
- 10.Horard B., Tatout,C., Poux,S. and Pirrotta,V. (2000) Structure of a Polycomb Response Element and in vitro binding of Polycomb Group complexes containing GAGA factor. Mol. Cell. Biol., 20, 3187–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra R.K., Mihaly,J., Barges,S., Spierer,A., Karch,F., Hagstrom,K., Schweinsberg,S.E. and Schedl,P. (2001) The iab-7 Polycomb Response Element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol., 21, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng J., Li,R., Morgan,K. and Simon,J. (1997) Evolutionary conservation and predicted structure of the Drosophila extra sex combs repressor protein. Mol. Cell. Biol., 17, 6663–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tie F., Furuyama,T. and Harte,P.J. (1998) The Drosophila Polycomb Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development, 125, 3483–3496. [DOI] [PubMed] [Google Scholar]
- 14.Tie F., Furuyama,T., Prasad-Sinha,J., Jane,E. and Harte,P.J. (2001) The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development, 128, 275–286. [DOI] [PubMed] [Google Scholar]
- 15.Saurin A.J., Shao,Z., Erdjument-Bromage,H., Tempst,P. and Kingston,R.E. (2001) A Drosophila Polycomb Group complex includes Zeste and dTAFII proteins. Nature, 412, 655–660. [DOI] [PubMed] [Google Scholar]
- 16.Shao Z., Raible,F., Mollaaghababa,R., Guyon,J.R., Wu,C.T., Bender,W. and Kingston,R.E. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell, 98, 37–46. [DOI] [PubMed] [Google Scholar]
- 17.Boivin A. and Dura,J.-M. (1998) In vivo chromatin accessibility correlates with gene silencing in Drosophila. Genetics, 150, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald D.P. and Bender,W. (2001) Polycomb Group repression reduces DNA accessibility. Mol. Cell. Biol., 21, 6585–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Breiling A., Turner,B.M., Bianchi,M.E. and Orlando,V. (2001) General transcription factors bind promoters repressed by Polycomb Group proteins. Nature, 412, 651–655. [DOI] [PubMed] [Google Scholar]
- 20.Breiling A., Bonte,E., Ferrari,S., Becker,P.B. and Paro,R. (1999) The Drosophila Polycomb protein interacts with nucleosomal core particles in vitro via its repression domain. Mol. Cell. Biol., 19, 8451–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francis N.J., Saurin,A.J., Shao,Z. and Kingston,R.E. (2001) Reconstitution of a functional core Polycomb repressive complex. Mol. Cell, 8, 545–556. [DOI] [PubMed] [Google Scholar]
- 22.Kodjabachian L., Delaage,M., Maurel,C., Miassod,R., Jacq,B. and Rosset,R. (1998) Mutations in ccf, a novel Drosophila gene encoding a chromosomal factor, affect progression through mitosis and interact with Pc-G mutations. EMBO J., 17, 1063–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez A., Higuet,D., Rosset,R., Deutsch,J. and Peronnet,F. (2001) corto genetically interacts with Pc-G and trx-G genes and maintains the anterior boundary of Ultrabithorax expression in Drosophila larvae. Mol. Genet. Genomics, 266, 572–583. [DOI] [PubMed] [Google Scholar]
- 24.Zink B. and Paro,R. (1989) In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature, 337, 468–471. [DOI] [PubMed] [Google Scholar]
- 25.DeCamillis M., Cheng,N.S., Pierre,D. and Brock,H.W. (1992) The polyhomeotic gene of Drosophila encodes a chromatin protein that shares polytene chromosome-binding sites with Polycomb. Genes Dev., 6, 223–232. [DOI] [PubMed] [Google Scholar]
- 26.Kyba M. and Brock,H.W. (1998) The Drosophila Polycomb Group protein Psc contacts Ph and Pc through specific conserved domains. Mol. Cell. Biol., 18, 2712–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soeller W.C., Poole,S.J. and Kornberg,T. (1988) In vitro transcription of the Drosophila engrailed gene. Genes Dev., 2, 68–81. [DOI] [PubMed] [Google Scholar]
- 28.Harlow E. and Lane,D. (1988) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 522–523.
- 29.Finley R.L. Jr and Brent,R. (1994) Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl Acad. Sci. USA, 91, 12980–12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwabuchi K., Li,B., Bartel,P. and Fields,S. (1993) Use of the two-hybrid system to identify the domain of p53 involved in oligomerization. Oncogene, 8, 1693–1696. [PubMed] [Google Scholar]
- 31.Serrano N., Brock,H.W., Demeret,C., Dura,J.M., Randsholt,N.B., Kornberg,T.B. and Maschat,F. (1995) polyhomeotic appears to be a target of engrailed regulation in Drosophila. Development, 121, 1691–1703. [DOI] [PubMed] [Google Scholar]
- 32.Bornemann D., Miller,E. and Simon,J. (1998) Expression and properties of wild-type and mutant forms of the Drosophila sex comb on midleg (SCM) repressor protein. Genetics, 150, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrington E.A. and Jones,R.S. (1996) The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development, 122, 4073–4083. [DOI] [PubMed] [Google Scholar]
- 34.Callebaut I., Labesse,G., Durand,P., Poupon,A., Canard,L., Chomilier,J., Henrissat,B. and Mornon,J.-P. (1997) Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci., 53, 621–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douguet D. and Labesse,G. (2001) Easier threading through web-based comparisons and cross-validations. Bioinformatics, 17, 752–753. [DOI] [PubMed] [Google Scholar]
- 36.Ball L.J., Murzina,N.V., Broadhurst,R.W., Raine,A.R., Archer,S.J., Stott,F.J., Murzin,A.G., Singh,P.B., Domaille,P.J. and Laue,E.D. (1997) Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J., 16, 2473–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horita D.A., Ivanova,A.V., Altieri,A.S., Klar,A.J. and Byrd,R.A. (2001) Solution structure, domain features and structural implications of mutants of the chromo domain from the fission yeast histone methyltransferase Clr4. J. Mol. Biol., 307, 861–870. [DOI] [PubMed] [Google Scholar]
- 38.Paro R. and Hogness,D.S. (1991) The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tschiersch B., Hofmann,A., Krauss,V., Dorn,R., Korge,G. and Reuter,G. (1994) The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J., 13, 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sali A., Potterton,L., Yuan,F., van Vlijmen,H. and Karplus,M. (1995) Evaluation of comparative protein modelling by MODELLER. Proteins, 23, 318–326. [DOI] [PubMed] [Google Scholar]
- 41.Sewalt R.G., van der Vlag,J., Gunster,M.J., Hamer,K.M., den Blaauwen,J.L., Satijn,D.P., Hendrix,T., van Driel,R. and Otte,A.P. (1998) Characterization of interactions between the mammalian Polycomb-Group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-Group protein complexes. Mol. Cell. Biol., 118, 3586–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez-Venegas R. and Avramova,Z. (2002) SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene, 285, 25–37. [DOI] [PubMed] [Google Scholar]
- 43.Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein-protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- 44.Zollman S., Godt,D., Prive,G.G., Couderc,J.-L. and Laski,F.A. (1994) The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl Acad. Sci. USA, 91, 10717–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rastelli L., Chan,C.S. and Pirrotta,V. (1993) Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J., 12, 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eissenberg J.C. (2001) Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene, 275, 19–29. [DOI] [PubMed] [Google Scholar]
- 47.Smothers J.F. and Henikoff,S. (2001) The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol., 10, 27–30. [DOI] [PubMed] [Google Scholar]
- 48.Ng J., Hart,C.M., Morgan,K. and Simon,J.A. (2000) A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb Group complexes and contains covalently modified ESC. Mol. Cell. Biol., 20, 3069–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones C.A., Ng,J., Peterson,A.J., Morgan,K., Simon,J. and Jones,R.S. (1998) The Drosophila esc and E(z) proteins are direct partners in Polycomb Group-mediated repression. Mol. Cell. Biol., 18, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Connell S., Wang,L., Robert,S., Jones,C., Saint,R. and Jones,R.S. (2001) Polycomblike PHD fingers mediate conserved interaction with Enhancer of zeste protein. J. Biol. Chem., 276, 43065–43073. [DOI] [PubMed] [Google Scholar]
- 51.Strutt H. and Paro,R. (1997) The Polycomb Group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol. Cell. Biol., 17, 6773–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Espinas M.L., Canudas,S., Fanti,L., Pimpinelli,S., Casanova,J. and Azorin,F. (2000) The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep., 1, 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaquero A., Espinas,M.L., Azorin,F. and Bernues,J. (2000) Functional mapping of the GAGA factor assigns its transcriptional activity to the C-terminal glutamine-rich domain. J. Biol. Chem., 275, 19461–19468. [DOI] [PubMed] [Google Scholar]
- 54.Katsani K.R., Hajibagheri,M.A. and Verrijzer,C.P. (1999) Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J., 18, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.