Abstract
To ensure the fidelity of protein biosynthesis, aminoacyl-tRNA synthetases (aaRSs) must recognize the tRNA identity elements of their cognate tRNAs and discriminate their cognate amino acids from structurally similar ones through a proofreading (editing) reaction. For a better understanding of these processes, we investigated the role of tRNALeu tertiary structure in the aminoacylation and editing reactions catalyzed by leucyl-tRNA synthetase (LeuRS). We constructed a series of Escherichia coli tRNALeu mutated transcripts with alterations of the nucleotides involved in tertiary interactions. Our results revealed that any disturbance of the tertiary interaction between the tRNALeu D- and TψC-loops affected both its aminoacylation ability and its ability to stimulate the editing reaction. Moreover, we found that the various tertiary interactions between the D- and TψC-loops (G18:U55, G19:C56 and U54:A58) functioned differently within the aminoacylation and editing reactions. In these two reactions, the role of base pair 19:56 was closely correlated and dependent on the hydrogen bond number. In contrast, U54:A58 was more important in aminoacylation than in editing. Taken together, our results suggest that the elbow region of tRNA formed by the tertiary interactions between the D- and TψC-loops affects the interactions between tRNA and aaRS effectively both in aminoacylation and in editing.
INTRODUCTION
The fidelity of protein biosynthesis depends on the specific recognition of tRNAs and amino acids by their cognate aminoacyl-tRNA synthetases (aaRSs), which charge the correct amino acid to the 3′-end of tRNAs (1,2). However, most tRNAs have a similar L-shaped tertiary structure, which complicates the ability of aaRSs to discriminate their cognate tRNAs from other tRNA species. A variety of identity determinants and anti-determinants of tRNA ensure the specificity of each binding to its corresponding synthetase (3,4). On the other hand, some small amino acids are so similar in structure that aaRSs have great difficulty in discriminating them from non-cognate ones [e.g. leucyl-tRNA synthetase (LeuRS) has difficulty in discriminating leucine from norvaline, homocysteine or isoleucine], resulting in misactivation of non-cognate amino acids and potential misacylation of tRNAs (5). To correct such errors, aaRSs catalyze proofreading (editing) reactions to hydrolyze misactivated (pre-transfer editing) and/or mischarged amino acids (post-transfer editing) (5). In many instances, such a proofreading reaction requires cognate tRNA (5). Thus, these aaRSs must recognize their cognate tRNA both in aminoacylation and in editing.
tRNAs are grouped into two classes based on the structure of their variable loop. In Escherichia coli, tRNALeu, tRNASer and tRNATyr contain stem–loop structures consisting of more than five nucleotides between the anticodon and the TψC stem, which is identified as the variable arm. These three tRNAs are classified as type II, while those without the variable arm are classified as type I. Various studies have begun to elucidate the important elements of tRNALeu for recognition by LeuRS (6–8). The important tertiary base pair A15:U48 and the location of G18 and G19 play important roles for LeuRS to discriminate tRNALeu from the other type II tRNA molecules, tRNASer and tRNATyr (6,7). According to the crystal structure of yeast tRNAPhe, G18 and G19 are involved in forming tertiary structures with the bases of the TψC-loop (G18:ψ55 and G19:C56) (9). Cysteine-tRNA synthetase (CysRS), which belongs to the same subgroup of aaRSs along with LeuRS (10), relies on tertiary interactions of G15:G48 and A13:A22:A46 as crucial identity elements for aminoacylation (11,12). Thus, it is intriguing to test if the tertiary base pairs between tRNALeu G18 G19 and the nucleotides of the TψC-loop are actual identity elements. Larkin et al. (8) used in vitro transcribed truncated suppressor tRNALeu with deletion of the anticodon stem–loop and variable arm to study the function of two tertiary base pairs in aminoacylation, and found that any replacement of G18:U55 or G19:C56 abolished the leucylation activity. However, it has been shown that the context sequence of tRNALeu also contributes to the recognition by LeuRS (7,13). Thus, additional study of the role of the nucleotides G18 and G19 in aminoacylation by LeuRS is warranted in the context of the full-length tRNALeu.
At the same time, we are interested in the function of tRNALeu in the editing reaction. Escherichia coli LeuRS binds tRNALeu in the editing domain (connecting peptide 1, CP1), which is distinct from the aminoacylation active site, to edit misaminoacylated products hydrolytically (8,14–17). Editing reactions have been best characterized for the closely related valyl-tRNA synthetase (ValRS) and isoleucyl-tRNA synthetase (IleRS) (VlaRS and IleRS also belong to the same subgroup of aaRSs along with LeuRS and CysRS), and studies have shown that the editing reactions are dependent on their cognate tRNAs (18–22). The tRNAIle determinants of editing are localized to three nucleotides (G16, D20 and D20a) in the D-loop at the elbow region of the L-shape, though these three bases are not required for the aminoacylation reaction (21,22). In the elbow region of the two-domain L-shaped tRNA molecule, the tertiary base pairs between nucleotides 18 and 55, 19 and 56, and 54 and 58 stack together to sustain the tertiary structure of tRNA (9,23,24), leading us to question whether these three tertiary base pairs of tRNALeu are also involved in editing.
Nucleotides at positions 16, 17, 20a, 59 and 60 in the D- and TψC-loops form a ‘variable pocket’ contiguous with the tertiary base pairs of G18:U55, G19:C56 and U54:A58 (9,23–26), which serves as a critical identity element in various tRNA species (3,4). Asahara et al. showed that U16, A20a and G59 of tRNALeu were not responsible for the leucine-specific aminoacylation (6). However, it is unknown whether these tertiary component nucleotides contribute to tRNA-dependent editing.
In the present work, we examined the role of tRNALeu tertiary structural components in aminoacylation and editing by creating substitution and deletion mutations at U17, G18, G19, U54, U55, C56 and G59 of E.coli tRNALeu transcripts. Our results revealed that any disturbance of the tertiary interaction between the D- and TψC-loops of tRNALeu not only greatly affected its ability to bind with E.coli LeuRS in aminoacylation, but also affected its ability to stimulate the editing reaction. Moreover, each of these tertiary interactions between nucleotides 18 and 55, 19 and 56, and 54 and 58 played different roles in aminoacylation and editing. The role of 19:56 in aminoacylation and editing was closely correlated and dependent on the hydrogen bond number, while the role of 18:55 in the two reactions was not so important as that of 19:56. The tertiary base pair U54:A58 played more important roles in the aminoacylation reaction. However, the nucleotides of the ‘variable pocket’ did not play significant roles in either the aminoacylation or editing reaction. Our results imply that the elbow region of tRNA effectively impacts its interactions with cognate aaRS in both aminoacylation and amino acid editing reactions.
MATERIALS AND METHODS
Materials
l-Leucine, l-isoleucine, l-norvaline, ATP, GTP, CTP, UTP, inorganic pyrophosphatase (PPase) and dithiothreitol (DTT) were purchased from Sigma (USA). l-[14C]Leucine (50 µCi/ml), l-[14C]isoleucine (50 µCi/ml) and [γ-32P]ATP (10 mCi/ml, 3000 Ci/mmol) were products of Amersham Life Science. Enzymes were products of MBI Fermentas or Promega. Ribonuclease (RNase) inhibitor was obtained from Takara Biotechnology Company. T7 RNA polymerase was purified from an E.coli overproducing strain in our laboratory (27). The E.coli LeuRS (2100 U/mg) was purified in our laboratory as described previously (28). Oligonucleotides for tRNALeu mutation were synthesized on an Applied Biosystems Oligonucleotide Synthesizer.
Preparation of tRNALeu transcripts
The deletion and substitution mutants of tRNALeu (the anticodon is GAG) were constructed as previously reported (29) and confirmed by DNA sequencing. Escherichia coli tRNALeu was transcribed in vitro by T7 RNA polymerase (17,30). tRNALeu transcripts were purified by 7 M urea–20% PAGE and resolved in diethyl pyrocarbonate-treated Milli-Q water (containing 5 mM MgCl2). All tRNALeu transcripts were annealed by heating for 5 min at 80°C and slowly cooled to room temperature. The tRNALeu concentrations were determined by UV absorbance at 260 nm. Extinction coefficients were calculated from the sequences of each tRNA (31).
Assay of aminoacylation and misaminoacylation of tRNALeu
The time course of aminoacylation was determined at 37°C in an 80 µl reaction mix consisting of 100 mM Tris–HCl pH 7.8, 30 mM KCl, 12 mM MgCl2, 4 mM ATP, 0.1 mM EDTA, 5 mM DTT, 1 U/µl RNase inhibitor, 20 µM [14C]leucine (50 µCi/ml), 3 µM tRNALeu transcripts and 1 µM E.coli LeuRS. The reaction was initiated by addition of LeuRS. At various time intervals (5, 10, 15, 25, 40 and 60 min), 10 µl aliquots were applied to Whatman Grade 3 qualitative filter paper, precipitated with 5% trichloroacetic acid (TCA), washed with 5% TCA and ethanol, and determined by scintillation counting. The aminoacylation kinetic constants for tRNALeu were measured at 37°C as described above, except that the concentrations of tRNALeu were varied from 0.5 to 20 µM or from 2 to 50 µM and the concentration of LeuRS was varied from 10 to 100 nM, depending on the accepting activities of tRNALeu transcripts. The kcat and Km values of tRNALeu and its mutants were the average of three independent determinations.
Misaminoacylation of tRNALeu with isoleucine was determined by the same procedure as used in the aminoacylation assay, except that 1 mM [14C]isoleucine (40 µCi/ml) was added instead of 20 µM [14C]leucine (50 µCi/ml), along with 10 µM tRNALeu transcripts and 4 µM LeuRS.
Measurement of ATP consumption in editing
Assays measuring the overall tRNA-dependent editing were carried out essentially as reported (22) and were performed at 37°C in 100 µl reaction mixes containing 100 mM Tris–HCl pH 7.8, 30 mM KCl, 12 mM MgCl2, 0.1 mM EDTA, 5 mM DTT, 1 U/µl RNase inhibitor, 2 U/ml PPase, 3 mM [γ-32P]ATP (3–5 c.p.m./pmol), 25 mM norvaline, 5 µM tRNALeu transcripts and 0.2 µM LeuRS. Aliquots (15 µl) of the editing reaction were taken and mixed with 350 µl of quenching liquid containing 6% activated charcoal, 7% HClO4 and 10 mM tetrasodium pyrophosphate. After centrifugation, the amount of inorganic phosphate [32P] in 50 µl of supernatant was quantified by scintillation counting. The rate of ATP hydrolysis is the average of three determinants. The background ATP hydrolysis in the absence of tRNALeu and LeuRS was assayed within each determinant as control.
RESULTS
Construction of E.coli tRNALeu mutants
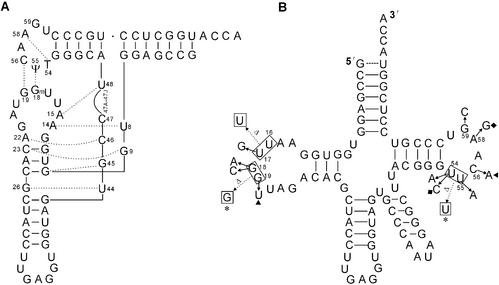
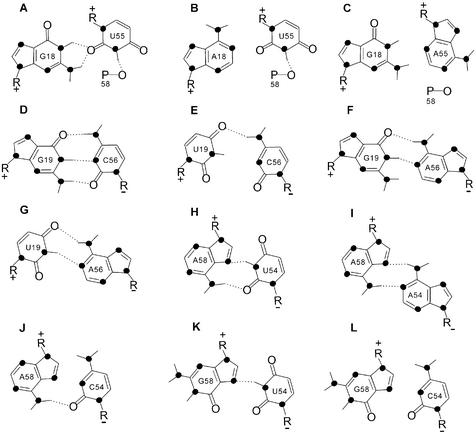
According to the crystal structure of yeast tRNAPhe (9,23,24), the hydrogen bondings between Gm18:ψ55, G19:C56 and T54:A58 of natural tRNALeu are important tertiary interactions (Fig. 1A). Though the modification of nucleotides Gm18, T54 and ψ55 may strengthen the tertiary interactions, they are not involved in the tertiary hydrogen bonding directly. Thus, G18:U55, G19:C56 and U54:A58 in in vitro transcribed unmodified tRNA will result in little change at the elbow region in comparison with those in natural modified tRNA (32). In this study, we created different substitution and deletion mutations located at the elbow region of in vitro transcribed tRNALeu (Fig. 1B). At these sites, eight single substitutions (A18 or C18 for G18, A55 for U55, U19 for G19, A56 for C56, A54 or C54 for U54 and G58 for A58) and two double substitutions (U19:A56 for G19:C56 and C54:G58 for U54:A58) mutants of tRNALeu were constructed. Based on previously reported work (32–34), the modeled interactions between these nucleotides (18:55, 19:56 and 54:58) are shown in Figure 2. The G18:U55 base pair is of the ‘GU imino: amino-2-carbonyl bifurcated’ type (32), and an additional intraloop hydrogen bond occurs between the imino proton of U55 and an oxygen atom of the nearby phosphate 58 (33) (Fig. 2A). A18:U55 (Fig. 2B) retains only the intraloop hydrogen bond. However, G18:A55 (Fig. 2C) cannot be fitted in without serious distortion of the backbone, so it has no hydrogen bond (34). Because a pyrimidine at 18 would cause a large loss in stacking energy (34), C18:U55 is not included in Figure 2. G19 and C56 form a Watson–Crick base pair of three hydrogen bonds (Fig. 2D). U19:C56 (Fig. 2E) retains one hydrogen bond, while G19:A56 (Fig. 2F) and U19:A56 (Fig. 2G) retain two hydrogen bonds each. U54 and A58 in wild-type tRNALeu (Fig. 2H) form a reverse Hoogsteen base pair consisting of two hydrogen bonds, while A54 and A58 (Fig. 2I) form a similar reverse Hoogsteen base pair with a minimal alteration of the phosphodiester backbone. Model building suggests that only one hydrogen bond is formed between C54:A58 (Fig. 2J) and U54:G58 (Fig. 2K). In addition, the double substitution mutation of U54 and A58 (C54:G58; Fig. 2L) would eliminate the hydrogen bonds and therefore disrupt the tertiary interaction.
Figure 1.
The structure of tRNALeu. (A) The L-shaped tertiary structure based on tRNAPhe. Dotted lines indicate the tertiary interactions. (B) The cloverleaf structure of transcripts and the mutants used in this study. Dotted arrows indicate the deletion mutations, Δ16U/17U, Δ18G/19G and Δ54U/55U. Asterisks indicate the double deletion mutation (Δ18G/19G–Δ54U/55U), triangles indicate the double substitution mutation (U19:A56) and diamonds indicate the double substitution mutation (C54:G58).
Figure 2.
Proposed hydrogen bonding in the tertiary base pairs. (A) G18:U55. (B) A18:U55. (C) G18:A55. (D) G19:C56. (E) U19:C56. (F) G19:A56. (G) U19:A56. (H) U54:A58. (I) A54:A58. (J) C54:A58. (K) U54:G58. (L) C54:G58. Predicted hydrogen bonds are shown as dotted lines. Nitrogen atoms are shown as solid circles. R+ indicates that the ribose–phosphate chain is coming toward the reader, while R– indicates that it is facing away from the reader.
In order to disrupt the tertiary interaction at the elbow region of tRNALeu more severely, two deletion mutants (tRNALeu Δ18G/19G and tRNALeu Δ54U/55U) and one double deletion mutant (tRNALeu Δ18G/19G–Δ54U/55U) were constructed. Two single substitution mutants (tRNALeu 17G and tRNALeu 59C) and one deletion mutant (tRNALeu Δ16U/17U) in the ‘variable pocket’ were constructed to identify the role of the ‘variable pocket’ nucleotides in tRNA-dependent editing. It was thought that the deletion mutation Δ16U/17U could affect tRNALeu in two ways—by directly reducing the number of nucleotides in the ‘variable pocket’, which may result in differential recognition by LeuRS, or by changing the tertiary base pairs between the invariant nucleotides G18 G19 and U55 C56 by reducing the number of nucleotides in the α region of the D-loop (which precedes G18 G19) from four to three.
Effect of mutations on aminoacylation of tRNALeu
The effect of the above mutations on the accepting activity by LeuRS was measured (Fig. 3). For tRNALeu with mutations at the base pairs between D- and TψC-loops, tRNALeu 18A (one intraloop hydrogen bond, Fig. 2B), tRNALeu 19U (one hydrogen bond, Fig. 2E) and tRNALeu U19:A56 (two hydrogen bonds, Fig. 2G) had high accepting activity (98, 86 and 91%, respectively; Table 1). Notably, deletion mutant tRNALeu Δ18G/19G also had a high level of aminoacylation (87%, Table 1), though this mutant is predicted to have impaired tertiary interaction between D- and TψC-loops. The plateau levels of aminoacylation of other tRNALeu mutants with impaired tertiary base pairs at 18:55, 19:56 and 54:58 were much lower (Table 1). The double substitution mutant tRNALeu C54:G58, the single deletion mutant tRNALeu Δ54U/55U and the double deletion mutant, tRNALeu Δ18G/19G–Δ54U/55U, had the lowest accepting activity (39, 49 and 41%, respectively; Table 1).
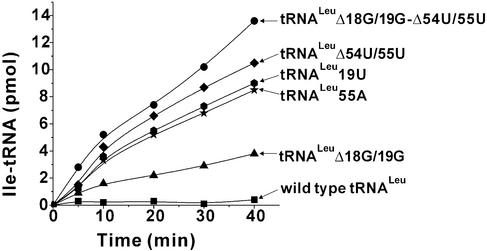
Figure 3.
The misaminoacylation of tRNA transcripts by E.coli LeuRS with isoleucine.
Table 1. Leucylation of the wild-type and mutants of E.coli tRNALeu.
| tRNA transcripts | kcat (s–1) | Km (µM) | kcat/Km (µM–1 s–1) | kcat/Km (relative) | Relative plateau of aminoacylation (%) |
|---|---|---|---|---|---|
| Wild-type tRNALeu | 3.2 | 1.8 | 1.8 | 1.0 | 100 |
| tRNALeu Δ18G/19G | 3.1 | 45 | 0.069 | 0.038 | 87 |
| tRNALeu Δ54U/55U | 0.06 | 40 | 1.5 × 10–3 | 8.3 × 10–4 | 49 |
| tRNALeu Δ18G/19G–Δ54U/55U | 0.04 | 50 | 8.0 × 10–4 | 4.4 × 10–4 | 41 |
| tRNALeu 18A | 3.5 | 2.6 | 1.3 | 0.72 | 98 |
| tRNALeu 18C | 1.8 | 18 | 0.10 | 0.056 | 60 |
| tRNALeu 55A | 1.6 | 19 | 0.084 | 0.047 | 65 |
| tRNALeu 19U | 3.0 | 24 | 0.13 | 0.072 | 86 |
| tRNALeu 56A | 1.8 | 6.4 | 0.28 | 0.17 | 69 |
| tRNALeu U19:A56 | 3.0 | 7.2 | 0.42 | 0.24 | 91 |
| tRNALeu 54A | 2.0 | 21 | 0.095 | 0.053 | 65 |
| tRNALeu 54C | 2.9 | 36 | 0.08 | 0.044 | 68 |
| tRNALeu 58G | 3.0 | 38 | 0.079 | 0.044 | 75 |
| tRNALeu C54:G58 | 2.6 | 150 | 0.017 | 9.6 × 10–3 | 39 |
| tRNALeu Δ16U/17U | 2.9 | 3.1 | 0.94 | 0.52 | 89 |
| tRNALeu 17G | 3.4 | 1.1 | 3.1 | 1.7 | 90 |
| tRNALeu 59C | 3.5 | 1.0 | 3.5 | 1.9 | 93 |
The Km values of LeuRS for tRNALeu with alterations between the D- and TψC-loops increased 1.7- to 83-fold (Table 1), indicating that the binding of these tRNA mutants with LeuRS decreased overall, but to different extents depending on the mutation type and location. The kcat values of tRNALeu mutants were quite consistent with their accepting activities. For these tRNALeu mutants, the aminoacylation efficiency (kcat/Km) of LeuRS decreased 1.4- to 2273-fold (Table 1). Two deletion mutants, tRNALeu Δ54U/55U and tRNALeu Δ18G/19G–Δ54U/55U, were the only two tRNALeu mutants that had a significant change both in Km and in kcat (kcat/Km decreased 1200- and 2273-fold, respectively), indicating that they are of the worst tertiary structures and are the worst substrate of LeuRS. However, another single deletion mutant, tRNALeu Δ18G/19G, only had an increase in Km, indicating that it was a better substrate than tRNALeu Δ54U/55U and tRNALeu Δ18G/19G–Δ54U/55U. As to the tRNALeu mutants involved in tertiary interaction between nucleotides 18 and 55, tRNALeu 18A with just one intraloop hydrogen bond (Fig. 2B) showed kcat and Km values similar to wild-type tRNALeu (Table 1). However, the two mutants lacking a hydrogen bond, tRNALeu 18C and tRNALeu 55A (Fig. 2C), showed a drastic increase in Km. Among the mutants involved in the 19:56 interaction, tRNALeu 56A and tRNALeu U19:A56 (both with two hydrogen bonds, Fig. 2F and G) had much lower Km (Table 1) than tRNALeu 19U (one hydrogen bond, Fig. 2E), suggesting that tRNALeu with two hydrogen bonds at this tertiary base pair is a better substrate than those with only one hydrogen bond. For mutants related to 54:58, any change of U54 and A58 resulted in an obvious increase in Km (Table 1), no matter whether the hydrogen bond number between 54 and 58 is two (tRNALeu 54A, Fig. 2I) or one (54C and 58G of tRNALeu, Fig. 2J and K). The mutant without a hydrogen bond between 54 and 58 (C54:G58, Fig. 2L) had the largest increase in Km (Table 1). These results suggest that U54:A58 plays a critical role for tRNALeu binding to LeuRS.
As regards the mutants involved in the ‘variable pocket’, the plateau level of tRNALeu Δ16U/17U, tRNALeu 17G and tRNALeu 59C was very high (89, 90 and 93% that of wild-type tRNALeu, respectively; Table 1). Kinetic analysis showed that the Km values of LeuRS for the two ‘variable pocket’ substitution mutants decreased while their catalytic efficiency increased (Table 1), indicating that they became better substrates for LeuRS. The deletion mutant, tRNALeu Δ16U/17U, showed a 50% decrease in kcat/Km, which was largely due to an increased Km.
Mischarging tRNALeu transcripts with isoleucine
In order to understand the role of the tertiary interaction in the discrimination of leucine from non-cognate amino acids, we examined the ability of LeuRS to misaminoacylate all tRNALeu variants (Fig. 3). Two deletion mutants, tRNALeu Δ54U/55U and tRNALeu Δ18G/19G–Δ54U/55U, which were the worst substrates for aminoacylation, yielded the largest mischarging products. In the two substitution mutants, the mischarging levels of tRNALeu 19U (one hydrogen bond) and tRNALeu 55A (no hydrogen bonds) were also high. The deletion mutant tRNALeu Δ18G/19G was mischarged weakly, and the other eight mutants, including the ‘variable pocket’ mutants, showed no significant mischarging under the experimental conditions.
Determination of consumption of ATP during the editing reaction
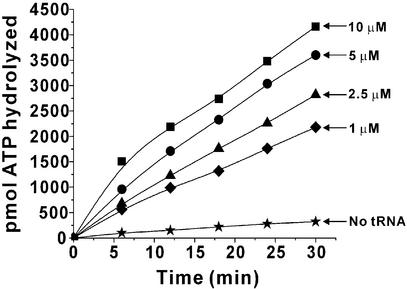
The net effect of the proofreading reaction is the consumption of ATP. Therefore, editing can be measured in another way by using the tRNA-dependent conversion of ATP to AMP and PPi in the presence of non-cognate amino acids. This ATPase assay measures the overall editing without discrimination between the pre- and post-transfer editing. Such an assay is widely used in ValRS and IleRS (20–22). In the present work, a definite stimulation of ATP hydrolysis by LeuRS was observed with in vitro transcribed wild-type tRNALeu in the presence of norvaline (Fig. 4).
Figure 4.
tRNALeu-dependent stimulation of ATP hydrolysis by E.coli LeuRS in the presence of norvaline. All the data were corrected against background ATP hydrolysis.
However, this ATP hydrolysis was attenuated by any change in the tertiary base pairs of tRNALeu (Table 2). When the worst substrates in aminoacylation, tRNALeu Δ54U/55U and tRNALeu Δ18G/19G–Δ54U/55U, were used, the rates of ATP hydrolysis decreased to 14 and 10% that of wild-type tRNALeu, respectively, indicating that these two mutants also had the worst ability to stimulate the LeuRS editing reaction. The ATP hydrolysis rate of tRNALeu 55A was only ∼20% that of wild type, in contrast to that of tRNALeu 18A and tRNALeu 18C, which were 63 and 46% respectively. Among the three mutants related to the tertiary interaction of 19:56, tRNALeu 56A and tRNALeu U19:A56 (both with two hydrogen bonds) had higher ATP hydrolysis rates (33 and 36%) than did tRNALeu 19U (one hydrogen bond) (18%). The ATP consumption stimulated by tRNALeu with mutations at 54 and 58 decreased moderately (33–60%, Table 2). Two tRNALeu substitution mutants at the ‘variable pocket’ consumed ATP at the same rate as wild-type tRNALeu, indicating that the two mutants had no defect in proofreading ability. However, the ATP hydrolysis of one ‘variable pocket’ deletion mutant, tRNALeu Δ16U/17U, decreased by 30%.
Table 2. tRNALeu-dependent ATP hydrolysis by E.coli LeuRS.
| tRNA transcripts | ATP hydrolysis rates (pmol/min)a | Relative ATP hydrolysis rates |
|---|---|---|
| Wild-type tRNALeu | 126 | 1.0 |
| tRNALeu Δ18G/19G | 34.6 | 0.27 |
| tRNALeu Δ54U/55U | 17.9 | 0.14 |
| tRNALeu Δ18G/19G–Δ54U/55U | 12.1 | 0.096 |
| tRNALeu 18A | 79.2 | 0.63 |
| tRNALeu 18C | 57.5 | 0.46 |
| tRNALeu 55A | 26.1 | 0.21 |
| tRNALeu 19U | 23.1 | 0.18 |
| tRNALeu 56A | 40.8 | 0.33 |
| tRNALeu U19:A56 | 45.3 | 0.36 |
| tRNALeu 54A | 65 | 0.51 |
| tRNALeu 54C | 60 | 0.48 |
| tRNALeu 58G | 76 | 0.60 |
| tRNALeu C54:G58 | 42 | 0.33 |
| tRNALeu Δ16U/17U | 88.7 | 0.70 |
| tRNALeu 17G | 126 | 1.0 |
| tRNALeu 59C | 125 | 0.99 |
aThe results were corrected by subtracting the background ATP hydrolysis.
DISCUSSION
Tertiary interactions within the tRNA molecule play an important role in tRNA aminoacylation (11,12,33,35), and have been similarly identified in tRNA-like structures in plant viral RNAs and tmRNA (34,36). Here, we show that the tertiary base pairs at the elbow region of L-shaped tRNALeu, G18:U55, G19:C56 and U54:A58, are important for both aminoacylation and editing reactions catalyzed by LeuRS. However, U17 and G59, which are involved in the ‘variable pocket’ of tRNALeu, contributed very little to aminoacylation and editing, which is consistent with previous reports (6). It suggests that the defect of tRNALeu Δ16U/17U in both aminoacylation and editing (Tables 1 and 2) was not because the deletion of U16 or U17 changed the ‘variable pocket’ itself, but because the deletion changed the position of the invariant nucleotides G18 G19, resulting in the disturbance of the tertiary interactions between D- and TψC-loops, which supported the importance of these interactions further.
Larkin et al. reported that the substitutions of G18:U55 and G19:C56 abolished the leucylation of truncated tRNALeu (8). In the present work, all tRNALeu mutants in the elbow region disrupted the tertiary interactions between the D- and TψC-loops. These mutants showed decreases in kcat/Km, but were still able to be aminoacylated by LeuRS relatively efficiently. This suggests that although tRNALeu tertiary interactions are important to aminoacylation, the overall tertiary structure of tRNALeu also relates to aminoacylation efficiency, as previously reported (7,13). The decrease of aminoacylation efficiency of tRNALeu mutants was due mainly to increased Km, suggesting that the affinity between tRNALeu mutants and LeuRS was weakened by perturbation of the tertiary interactions at the elbow region.
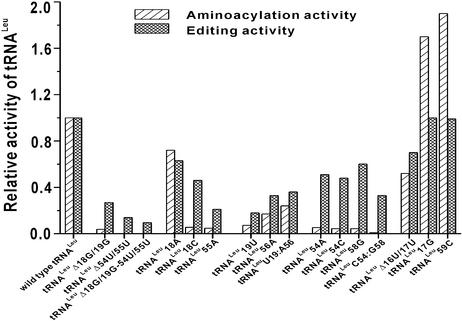
To gain insight into the function of G18:U55, G19:C56 and U54:A58 of tRNALeu in aminoacylation and editing reactions, we compared the activities of the related tRNALeu mutants in the two reactions (Fig. 5). Obviously, three deletion mutations (tRNALeu Δ18G/19G, tRNALeu Δ54U/55U and tRNALeu Δ18G/19G–Δ54U/55U) will greatly affect both aminoacylation and editing. Moreover, it seems that in the three mutants, the retained editing activity is correlated with the aminoacylation activity. The tertiary interaction between nucleotides 19 and 56 is important in both aminoacylation and editing. Although the single substitution mutant tRNALeu 19U and the double substitution mutant tRNALeu U19:A56 possessed the same 19U, the aminoacylation efficiencies and editing efficiencies between the two tRNALeu mutants were of 3.3- and 2-fold difference, respectively. However, the aminoacylation and editing efficiencies between tRNALeu 56A and tRNALeu U19:A56 (both having an A at position 56) were similar. We note that the hydrogen bond numbers between bases 19 and 56 in wild-type tRNALeu, tRNALeu U19:A56, tRNALeu 56A and tRNALeu 19U were three, two, two and one, respectively (Fig. 2D, G, F and E, respectively). This suggests that aminoacylation and editing efficiency might be correlated with the hydrogen bond number between nucleotides 19 and 56, not with the exact nucleotides themselves. This is supported by the report that tRNAPhe with C19:G56 (three hydrogen bonds) was equally efficient in the aminoacylation reaction as was wild-type tRNAPhe with G19:C56 (23).
Figure 5.
Combined comparison of the activities of tRNALeu mutants correlated with the substitution mutation at G18:U55, G19:C56 and U54:A58 in aminoacylation and editing reactions. The aminoacylation activity is the relative kcat/Km, and the editing activity is the relative ATP hydrolysis rate.
It has been reported that the replacement of G18:U55 with A18:G55, which does not change the hydrogen bond number between nucleotides 18 and 55, resulted in little change of tRNA-like structures in plant viral RNAs or tmRNA (34,36). In the present study, although there are no hydrogen bonds predicted between A18 and U55 in tRNALeu 18A, and only one intraloop hydrogen bond occurs between U55 and A58 in this mutant (Fig. 2B), tRNALeu 18A is recognized efficiently by LeuRS in both the aminoacylation and editing processes (Fig. 5). Only when the tertiary interaction between 18 and 55 is almost completely disrupted, such as in the case of tRNALeu 18C and tRNALeu 55A (Fig. 2C), would the recognition of these two mutants by LeuRS in aminoacylation be significantly decreased (relative kcat/Km = 0.056 and 0.047). Though LeuRS catalyzes the aminoacylation of tRNALeu 18C and tRNALeu 55A with similar low efficiencies, there is a 2-fold difference in editing ability (Fig. 5). While the reason for this divergence is obscure, it implies that the consequences of these two mutations in aminoacylation are different from those in editing. tRNALeu with mutations at 54 and 58 had very low aminoacylation ability, but retained high editing ability (Fig. 5), implying that the tertiary base pair U54:A58 plays a more significant role in aminoacylation than in editing. It seems that U54:A58 is a part of the leucine identity elements of tRNALeu, though the model of LeuRS complexed with tRNALeu shows no obvious direct interaction between the tertiary base pair 54:58 and LeuRS (37).
Based on the above data, we conclude that the elbow region of L-shaped tRNALeu affects its interaction with LeuRS in aminoacylation and editing. Further, the roles of the three tertiary base pairs (between nucleotides 19 and 56, 18 and 55, and 54 and 58) in these two reactions are different; only the tertiary interaction of 19:56 plays an equally important role in both aminoacylation and editing. The reason for this may be that this tertiary base pair is located in the outer side of the elbow region of two-domain L-shaped tRNA (Fig. 1B) and will affect the tRNA tertiary structure mostly in both the aminoacylation and editing reaction. Although there is still no evidence for direct contacts between the elbow region and the synthetase (19,37–39), we propose that the elbow region mediates the communications between the two domains of the L-shaped tRNA to prime the correct recognition by cognate aaRS in aminoacylation and editing reactions. The tertiary base pairs of G19:C56, G18:U55 and U54:A58 may contribute differently to adjust such communications for the different requirements in aminoacylation and editing.
Here, the nucleotides of tRNALeu involved in aminoacylation and editing are different from those of tRNAVal and tRNAIle (20,21), and their effects on aminoacylation and editing are summarized in Table 3. The data suggest that the roles of tRNAs in aminoacylation and editing are very complicated. In the case of tRNAVal (20), all mutants that actively promote editing can be aminoacylated, but not all aminoacylated tRNAs are able to trigger the editing response of E.coli ValRS. In the case of tRNAIle (21), aminoacylation and editing rely on distinct tRNAIle domains and nucleotide determinants to trigger both activities. However, the tertiary interaction between nucleotides 19 and 56 of tRNALeu is important for aminoacylation and editing reactions, and both activities are correlated. In tRNALeu, nucleotides important only for editing are not found.
Table 3. Effects of nucleotides in tRNALeu, tRNAVal and tRNAIle on aminoacylation and editing.
| Effects on aminoacylation and editing | Nucleotides in three systems | ||
|---|---|---|---|
| tRNALeu | tRNAVal,a | tRNAIle,b | |
| Important for neither aminoacylation nor editing | U17 and G59 | ||
| Important for aminoacylation and editing and both are correlated | G19:C56 | Most nucleotides studied in Tardif and Horowitz (20) | |
| More important for aminoacylation than for editing | U54:A58 | Anticodon | |
| Important only for editing | A76 | G16, D20 and D20a | |
| Mixed type | G18:U55 | ||
Acknowledgments
ACKNOWLEDGEMENTS
This work was funded by the Natural Science Foundation of China (Grant 30270310), the Chinese Academy of Sciences (Grant KSCX-2-2-04) and Shanghai Committee of Science and Technology (Grant 02DJ140567).
REFERENCES
- 1.Ibba M. and Söll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 2.Alexander R.W. and Schimmel,P. (2001) Domain–domain communication in aminoacyl-tRNA synthetases. Prog. Nucleic Acid Res. Mol. Biol., 69, 317–349. [DOI] [PubMed] [Google Scholar]
- 3.Giegé R., Sissler,M. and Florentz,C. (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res., 26, 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saks M.E., Sampson,J.R. and Abelson,J. (1994) The transfer RNA identity problem: a search for rules. Science, 263, 191–197. [DOI] [PubMed] [Google Scholar]
- 5.Jakubowski H. and Goldman,E. (1992) Editing of errors in selection of amino acids for protein synthesis. Microbiol. Rev., 56, 412–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asahara H., Himeno,H., Tamura,K., Hasegawa,T., Watanabe,K. and Shimizu,M. (1993) Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol., 231, 219–229. [DOI] [PubMed] [Google Scholar]
- 7.Tocchini-Valentini G., Saks,M.E. and Abelson,J. (2000) tRNA leucine identity and recognition sets. J. Mol. Biol., 298, 779–793. [DOI] [PubMed] [Google Scholar]
- 8.Larkin D.C., Williams,A.M., Martinis,S.A. and Fox,G.E. (2002) Identification of essential domains for Escherichia coli tRNALeu aminoacylation and amino acid editing using minimalist RNA molecules. Nucleic Acids Res., 30, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S.H., Suddath,F.L., Quigley,G.J., McPherson,A., Sussman,J.L., Wang,A.H., Seeman,N.C. and Rich,A. (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science, 185, 435–440. [DOI] [PubMed] [Google Scholar]
- 10.Cusack S. (1995) Eleven down and nine to go. Nature Struct. Biol., 2, 824–831. [DOI] [PubMed] [Google Scholar]
- 11.Hamann C.S. and Hou,Y.M. (1997) An RNA structural determinant for tRNA recognition. Biochemistry, 36, 7967–7972. [DOI] [PubMed] [Google Scholar]
- 12.Hou Y.M., Westhof,E. and Giegé,R. (1993) An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl Acad. Sci. USA, 90, 6776–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asahara H., Nameki,N. and Hasegawa,T. (1998) In vitro selection of RNAs aminoacylated by Escherichia coli leucyl-tRNA synthetase. J. Mol. Biol., 283, 605–618. [DOI] [PubMed] [Google Scholar]
- 14.Chen J.F., Guo,N.N., Li,T., Wang,E.D. and Wang,Y.L. (2000) CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry, 39, 6726–6731. [DOI] [PubMed] [Google Scholar]
- 15.Mursinna R.S., Lincecum,T.L.,Jr and Martinis,S.A. (2001) A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry, 40, 5376–5381. [DOI] [PubMed] [Google Scholar]
- 16.Englisch S., Englisch,U., von der Haar,F. and Cramer,F. (1986) The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E.coli and yeast. Nucleic Acids Res., 14, 7529–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du X. and Wang,E.D. (2002) Discrimination of tRNALeu isoacceptors by the mutants of Escherichia coli leucyl-tRNA synthetase in editing. Biochemistry, 41, 10623–10628. [DOI] [PubMed] [Google Scholar]
- 18.Schimmel P. and Schmidt,E. (1995) Making connections: RNA-dependent amino acid recognition. Trends Biochem. Sci., 20, 1–2. [DOI] [PubMed] [Google Scholar]
- 19.Fukai S., Nureki,O., Sekine,S., Shimada,A., Tao,J.S., Vassylyev,D.G. and Yokoyama,S. (2000) Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell, 103, 793–803. [DOI] [PubMed] [Google Scholar]
- 20.Tardif K.D. and Horowitz,J. (2002) Transfer RNA determinants for translational editing by Escherichia coli valyl-tRNA synthetase. Nucleic Acids Res., 30, 2538–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale S.P., Auld,D.S., Schmidt,E. and Schimmel,P. (1997) Discrete determinants in transfer RNA for editing and aminoacylation. Science, 276, 1250–1252. [DOI] [PubMed] [Google Scholar]
- 22.Farrow M.A., Nordin,B.E. and Schimmel,P. (1999) Nucleotide determinants for tRNA-dependent amino acid discrimination by a class I tRNA synthetase. Biochemistry, 38, 16898–16903. [DOI] [PubMed] [Google Scholar]
- 23.Robertus J.D., Ladner,J.E., Finch,J.T., Rhodes,D., Brown,R.S., Clark,B.F. and Klug,A. (1974) Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature, 250, 546–551. [DOI] [PubMed] [Google Scholar]
- 24.Ladner J.E., Jack,A., Robertus,J.D., Brown,R.S., Rhodes,D., Clark,B.F. and Klug,A. (1975) Structure of yeast phenylalanine transfer RNA at 2.5 Å resolution. Proc. Natl Acad. Sci. USA, 72, 4414–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moras D., Comarmond,M.B., Fischer,J., Weiss,R., Thierry,J.C., Ebel,J.P. and Giegé,R. (1980) Crystal structure of yeast tRNAAsp. Nature, 288, 669–674. [DOI] [PubMed] [Google Scholar]
- 26.Woo N.H., Roe,B.A. and Rich,A. (1980) Three-dimensional structure of Escherichia coli initiator tRNAMet f. Nature, 286, 346–351. [DOI] [PubMed] [Google Scholar]
- 27.Li Y., Wang,E.D. and Wang,Y.L. (1999) A modified procedure for fast purification of T7 RNA polymerase. Protein Expr. Purif., 16, 355–358. [DOI] [PubMed] [Google Scholar]
- 28.Li T., Wang,E.D. and Wang,Y.L. (1997) The overproduction and purification of leucyl-tRNA synthetase in E. coli. Acta Biochim. Biophys. Sin., 29, 591–596. [PubMed] [Google Scholar]
- 29.Li Y., Wang,E.D. and Wang,Y.L (1998) Overproduction and purification of Escherichia coli tRNALeu. Sci. China (Ser. C), 41, 225–231. [Google Scholar]
- 30.Li Y., Chen,J.F., Wang,E.D. and Wang,Y.L. (1999) T7 RNA polymerase transcription of Escherichia coli isoacceptors tRNALeu. Sci. China (Ser. C), 42, 185–190. [DOI] [PubMed] [Google Scholar]
- 31.Puglisi J.D. and Tinoco,I.,Jr (1989) Absorbance melting curves of RNA. Methods Enzymol., 180, 304–325. [DOI] [PubMed] [Google Scholar]
- 32.Sampson J.R., DiRenzo,A.B., Behlen,L.S. and Uhlenbeck,O.C. (1990) Role of the tertiary nucleotides in the interaction of yeast phenylalanine tRNA with its cognate synthetase. Biochemistry, 29, 2523–2532. [DOI] [PubMed] [Google Scholar]
- 33.Nagaswamy U., Larios-Sanz,M., Hury,J., Collins,S., Zhang,Z., Zhao,Q. and Fox,G.E. (2002) NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res., 30, 395–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Smit M.H., Gultyaev,A.P., Hilge,M., Bink,H.H., Barends,S., Kraal,B. and Pleij,C.W. (2002) Structural variation and functional importance of a D-loop–T-loop interaction in valine-accepting tRNA-like structures of plant viral RNAs. Nucleic Acids Res., 30, 4232–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puglisi J.D., Putz,J., Florentz,C. and Giege,R. (1993) Influence of tRNA tertiary structure and stability on aminoacylation by yeast aspartyl-tRNA synthetase. Nucleic Acids Res., 21, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barends S., Bjork,K., Gultyaev,A.P., de Smit,M.H., Pleij,C.W. and Kraal,B. (2002) Functional evidence for D- and T-loop interactions in tmRNA. FEBS Lett., 514, 78–83. [DOI] [PubMed] [Google Scholar]
- 37.Cusack S., Yaremchuk,A. and Tukalo,M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J., 19, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvian L.F., Wang,J. and Steitz,T.A. (1999) Insight into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science, 285, 1074–1077. [PubMed] [Google Scholar]
- 39.Hendrickson T.L. (2001) Recognizing the D-loop of transfer RNAs. Proc. Natl Acad. Sci. USA, 98, 13473–13475. [DOI] [PMC free article] [PubMed] [Google Scholar]