Abstract
The improvement of non-viral-based gene delivery systems is of prime importance for the future of gene and antisense therapies. We have previously described a peptide-based gene delivery system, MPG, derived from the fusion peptide domain of HIV-1 gp41 protein and the nuclear localisation sequence (NLS) of SV40 large T antigen. MPG forms stable non-covalent complexes with nucleic acids and improves their delivery. In the present work, we have investigated the mechanism through which MPG promotes gene delivery. We demonstrate that cell entry is independent of the endosomal pathway and that the NLS of MPG is involved in both electrostatic interactions with DNA and nuclear targeting. MPG/DNA particles interact with the nuclear import machinery, however, a mutation which affects the NLS of MPG disrupts these interactions and prevents nuclear delivery of DNA. Nevertheless, we show that this mutation yields a variant of MPG which is a powerful tool for delivery of siRNA into mammalian cells, enabling rapid release of the siRNA into the cytoplasm and promoting robust down-regulation of target mRNA. Taken together, these results support the potential of MPG-like peptides for therapeutic applications and suggest that specific variations in the sequence may yield carriers with distinct targeting features.
INTRODUCTION
The development of non-viral-based gene delivery systems constitutes an essential challenge in therapeutics. Although they exhibit several advantages over viral systems, the interest of non-viral synthetic gene delivery systems for therapeutic applications remains limited by their poor ability to escape from the endosomal compartment and to translocate DNA into the nucleus (1). During the past 10 years, several peptide-based gene delivery systems that can overcome both extracellular and intracellular limitations have been proposed (2–4). Poor release from the endosomal compartment after cellular uptake constitutes one of the major limitations of non-viral gene delivery systems. Research has therefore been focused on the design of strategies to either facilitate release from the endosome or to bypass the endosomal pathway (2–4). Peptide carriers that combine DNA binding and membrane destabilising properties have been demonstrated to promote gene transfer into cultured cells (5,6) and living animals (7).
Another major limitation of non-viral gene delivery systems is the poor nuclear delivery of DNA, which is however essential for transfection of non-dividing cells as well as for in vivo applications. In order to improve nuclear delivery of DNA, synthetic peptides containing nuclear localisation sequences (NLS) have been extensively used (8,9). Protein transduction domains cross the cell membrane independently of the endosomal pathway and have been used to improve the delivery of DNA (10,11). The third helix of the Antennapedia homeodomain has been shown to form stable non-covalent complexes with small oligonucleotides and to facilitate their internalisation in a non-endosomal fashion (12). The NLS domain of Tat has been shown to promote nuclear targeting of proteins and DNA (10,11,13). Tat peptide covalently attached to liposomes promotes rapid delivery of DNA, independently of the endosomal pathway (14,15). In contrast, oligomers of the arginine-rich motif of HIV-1 Tat protein have been reported to form stable particles with DNA through non-covalent interactions, but promote their delivery into cells through the endosomal pathway (16).
We have recently described a new peptide-based gene delivery system, MPG, which forms stable non-covalent complexes with nucleic acids and promotes their delivery into a large panel of cell lines (17–19). MPG is a bipartite amphipathic peptide derived from both the fusion peptide domain of HIV-1 gp41 protein and the NLS of SV40 large T antigen. In the present work, we have investigated the mechanism through which MPG promotes gene delivery into cells and have demonstrated that it is independent of the endosomal pathway. We show that the NLS of MPG is required for both electrostatic interactions with DNA and nuclear targeting, and that MPG/DNA particles interact with the nuclear import machinery. Moreover we have used MPG and a variant characterised by a mutation in the NLS to deliver siRNA into mammalian cells. Our results demonstrate that delivery of siRNA by both MPG and its NLS mutant enable robust down-regulation of target mRNA. Nevertheless, the NLS mutant induces rapid release of the siRNA into the cytoplasm, and this correlates with a more significant biological response. Taken together, these results support the potential of MPG for therapeutic applications.
MATERIALS AND METHODS
Peptide synthesis and analysis
All peptides were synthesised by solid phase peptide synthesis using AEDI-expansin resin with a 9050 Pep Synthesizer (Millipore, Watford, UK) according to the Fmoc/tBoc method and purified as described previously (18). MPG and MPGΔNLS were acetylated at their N-terminus and synthesised with a cysteamide group at their C-terminus. MPG, GALFLGFLGAAGSTMGAWSQPKKKRKV; MPGΔNLS, GALFLGFLGAAGSTMGAWSQPKSKRKV.
Cell culture and MPG-mediated transfection
HS-68, Cos-7 and HeLa cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM glutamine, 1% antibiotics (10 000 µg/ml streptomycin, 10 000 IU/ml penicillin) and 10% (w/v) foetal calf serum (FCS) at 37°C in a humidified atmosphere containing 5% CO2 as described previously (18). For MPG- or MPGΔNLS-mediated gene delivery, peptide carrier/DNA complexes were formed in DMEM or phosphate-buffered saline (500 µl of DMEM containing 100 ng of DNA complexed with MPG at a charge ratio of 5:1) and incubated for 30 min at 37°C. Cells grown to 60% confluence were then overlaid with these preformed complexes. After 30 min incubation at 37°C, 1 ml of fresh DMEM supplemented with 10% FCS was added to the cells, without removing the overlay of carrier/DNA, and cells were returned to the incubator. For cell cycle-dependent studies, HS-68 fibroblasts were synchronised by serum deprivation for 40 h, then restimulated to enter the cycle and grow into early G1 for 4 h by addition of fresh DMEM supplemented with 20% FCS, and incubated in the presence of MPG/DNA complexes as described above. Luciferase activity was monitored as described previously (19) and control transfection experiments were performed using Lipofectamine™. When transfected at 4°C, cells were incubated for 1 h at 4°C prior to transfection, then incubated with MPG/DNA complexes for another 2 h at 4°C, extensively washed and finally returned to incubation at 37°C in fresh medium supplemented with serum. When transfections were performed in the presence of bafilomycin A (175 nM), cytochalasin B (50 µg/ml) or chloroquine (100 µM), cells were preincubated with these inhibitors of endocytosis for 1 h prior to transfection. Transfections were performed for 1 h, after which cells were extensively washed and either analysed by fluorescence microscopy in order to determine the cellular localisation of the fluorescently labelled oligonucleotide or returned to the incubator at 37°C in the presence of fresh medium supplemented with serum.
MPG-mediated siRNA delivery
siRNA targeting the 3′-untranslated region of GAPDH were from the Silencer™ GAPDH siRNA kit (Ambion). Fluorescent labelling of siRNA was performed using either the Fam or the Cy3 Silencer™ labeling kit (Ambion) and modified as described in the manufacturer’s protocol. The pRL-Luc reporter gene was from Promega and siRNA targeting luciferase sense (5′-CUUACGCUGAGUACUUCGATT-3′) and antisense (3′-TTGAAUGCGACUCAUGAAGCU-5′) and mismatch sense (5′-CGUACGCGGAAUACUUCGATT-3′) and antisense (3′-TTGCAUGCGCCUUAUGAAGCU-5′) siRNAs were obtained from Genset Oligos. For siRNA delivery, peptide carrier/siRNA complexes were formed in DMEM at a charge ratio of 10:1 and incubated for 30 min at 37°C. Cells grown to 60% confluence were then overlaid with these preformed complexes. After 30 min incubation at 37°C, 1 ml of fresh DMEM supplemented with 10% FCS was added to the cells, without removing the overlay of carrier/siRNA.
Northern blotting
HS-68 or HeLa cells (5 × 106 cells) incubated with complexes were collected and total RNAs were isolated from cells using TriReagent™ (Sigma, St Louis, MO) according to the manufacturer’s recommendations. RNAs were then purified by phenol extraction followed by ethanol precipitation. RNA samples (10 µg) were separated by electrophoresis in formaldehyde agarose gels (1.2%), transferred onto nylon membranes (Hybond N+; Amersham Pharmacia Biotech) and hybridized with 32P-labelled GAPDH and actin probes (the latter used to normalise RNA loading). Signals were detected by Phosphorimaging (Molecular Dynamics) and quantified using ImageQuant software.
Protein preparation
Plasmids for the expression of recombinant GST–importin α and GST–importin β fusion proteins were kindly provided by B. Cullen. Proteins were purified as described previously (13). For affinity chromatography experiments, GST–importin α and GST–importin β were coupled onto GST beads in 150 mM NaCl, 1 mM EDTA and 10 mM HEPES pH 7.5. Importin-coupled beads were then incubated in batch with MPG/DNA complexes for 1 h, extensively washed and incubated with 50 nM glutathione to elute the complexes. DNA was then extracted from the complexes with phenol/chloroform, followed by ethanol precipitation, and finally analysed by agarose gel electrophoresis (0.8%).
RESULTS
MPG-mediated gene delivery is independent of the endosomal pathway
Transport of DNA into cells involves a number of selective steps, therefore understanding the mechanisms underlying each of these steps as well as the behaviour of carrier/DNA complexes within the cell is essential to develop more efficient carriers. We have already reported that MPG can deliver oligonucleotides into mammalian cells at low temperature with high efficiency (17). In order to understand the mechanism through which MPG mediates gene delivery, transfection experiments were performed in the presence of several inhibitors that interfere with the endosomal pathway, including cytochalasin B, bafilomycin A and chloroquine. Cytochalasin B induces depolymerisation of the microfilaments involved in phagocytosis and macropinocytosis without affecting other endocytotic processes (20). Bafilomycin A specifically prevents the acidification of early endosomes, by inhibiting a proton pump know as vacuolar ATPase (21), and chloroquine is a weak base, which inhibits the maturation of the transport vesicles into late endosomes and neutralises the pH of the latter (22).
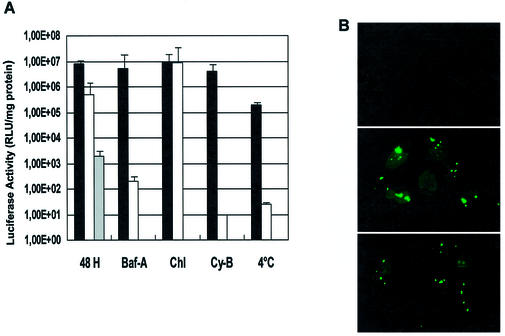
A plasmid encoding the reporter gene luciferase, pRL-SV40, was associated with the MPG carrier at a charge ratio of 5:1 (MPG/DNA) and overlaid onto cultured human fibroblasts (HS-68) or HeLa cells in the presence of either bafilomycin A (150 nM), chloroquine (100 µM) or cytochalasin B (50 µg/ml). Transfection efficiency was monitored 30 h after transfection and Lipofectamine™ was used as a standard control method for transfection. As shown in Figure 1, the transfection efficiency of MPG was not affected by bafilomycin A or cytochalasin B, suggesting that the cellular uptake of MPG/DNA complexes is independent of the classical endosomal pathway. In contrast, the efficiency of the lipid-based carrier was dramatically reduced by these inhibitors of the endosomal pathway. Chloroquine barely increased the efficiency of MPG (by only 10%), whereas it enhanced delivery by Lipofectamine™ more than 20-fold, as already reported for different liposome-based gene delivery systems (23,24). When transfection experiments were performed at 4°C, the efficiency of MPG was reduced 12-fold, compared to that of Lipofectamine™ by more than 1000-fold. Based on these results, we believe that the decrease in the efficiency of MPG is associated with cellular stress induced by low temperature. To exclude the possibility that the MPG/DNA complexes remain associated with the cell membrane in the presence of inhibitors and only enter the cell upon dilution of the inhibitor, transfection experiments were performed in the presence of inhibitors, using a fluorescently labelled oligonucleotide (17). As reported in Figure 1B, after 1 h most of the fluorescently labelled oligonucleotides localised to the nucleus and cytoplasm of the cells, suggesting that MPG/DNA complexes are not associated with the membrane and indeed enter the cells in the presence of inhibitors. Interestingly, a small fraction of these complexes are associated with the membrane, which may correspond to large aggregates unable to enter the cells. Taken together, these data suggest that the cellular uptake mechanism of MPG/DNA complexes is essentially independent of the endosomal pathway.
Figure 1.
MPG-mediated gene delivery in the presence of inhibitors of the endosomal pathway. (A) MPG/DNA complexes formed at a charge ratio of 5:1 with 100 ng pRL-SV40 plasmid encoding the reporter gene luciferase were overlaid onto human fibroblasts (HS-68) in the presence of either bafilomycin A (Baf-A, 150 nM), cytochalasin B (Cy-B, 50 µg/ml) or chloroquine (Chl, 100 µM). After 48 h cell extracts were prepared and luciferase activity was measured and reported as a function of total protein (black). Similar experiments were performed using a lipid-based delivery system, Lipofectamine™, as a control (white). Experiments in the absence of inhibitors were also performed using an MPG peptide lacking a C-terminal cysteamide group (grey). (B) A fluorescein-labelled 36mer oligonucleotide was complexed with MPG at a charge ratio of 5:1. Complexes were then overlaid onto cultured HeLa cells in the presence of bafilomycin A (middle panel) or cytochalasin B (bottom panel) and cellular localisation of the oligonucleotide was monitored by fluorescence microscopy on living cells 1 h after transfection. A control experiment with free oligonucleotide is shown in the top panel.
Role of the NLS of MPG in nuclear targeting of plasmids
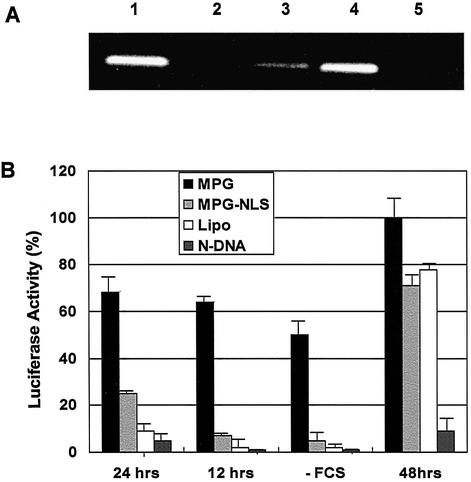
MPG peptide is derived from the NLS of the SV40 large T antigen and from the fusion peptide domain of HIV-1 gp41 protein (17). We have demonstrated that lysine residues are required for the formation of stable non-covalent interactions with DNA. We have investigated to what extent the NLS sequence was involved in nuclear targeting of plasmids. Nuclear import of proteins is generally mediated by the interaction of the NLS with importin, which in turn favours the interaction with importin β (25,26). Two peptides, wild-type MPG and MPGΔNLS, a variant characterised by a single mutation of the second lysine residue in the NLS motif to serine (KSKRKV), were used. This mutation has previously been reported to dramatically reduce import of NLS-containing proteins into the nucleus (25,26). To determine to what extent the NLS moiety of MPG was required for nuclear import of plasmids, we first performed in vitro binding experiments using GST–importin fusion proteins. Importin α and β immobilized on beads were incubated with MPG/DNA or MPGΔNLS/DNA complexes for 1 h at 25°C. Then, both bound and unbound fractions were analysed by agarose gel electrophoresis. As shown in Figure 2A, MPG/DNA complexes significantly interacted specifically with importin α (lane 4) but not with importin β (lane 5). Moreover, binding of the complex to importin α was dramatically reduced when the plasmid was associated with MPGΔNLS (lane 3), suggesting that mutations in the NLS sequence significantly affect the interaction with importin α. As a control, we verified that the plasmid alone did not interact with either importin α (lane 2) or β (data not shown).
Figure 2.
The NLS of MPG is essential for nuclear translocation. (A) Purified GST–importin α and GST–importin β were coupled onto GST-Sepharose resin and incubated in batch with MPG/DNA or MPGΔNLS/DNA complexes for 1 h, extensively washed and incubated with 50 nM glutathione to elute the complexes. DNA was then extracted and analysed by agarose gel electrophoresis (0.8%). Lane 1, control DNA; lane 2, free plasmid does not associate with importin α; lane 3, binding of MPG/DNA to importin α; lane 4, binding of MPG/DNA to importin β; lane 5, MPG/DNA does not associate with importin β. (B) Delivery of pRL-SV40 plasmid. MPG/DNA (black boxes) or MPGΔNLS/DNA (light grey boxes) complexes formed at a charge ratio of 5:1 with 100 ng pRL-SV40 plasmid encoding the reporter gene luciferase were overlaid onto synchronised or arrested (–FCS) human fibroblasts (HS-68). Luciferase activity was monitored 12 and 24 h after release from synchrony or after 24 h for arrested cells and 48 h for asynchronous cells. Control transfections were performed with Lipofectamine™ (white boxes) or naked plasmid (dark grey boxes).
One of the major limitations of most synthetic gene delivery systems, which significantly hampers gene delivery in vivo and into non-dividing cells, is their poor ability to translocate plasmid DNA into the nucleus of cells (1). In order to study the impact of the NLS of MPG on nuclear translocation of plasmids, we performed transfection experiments on synchronised or growth-arrested cells with either MPG or its NLS mutant. Cells were synchronised in G0 by serum starvation then released back into the cell cycle by addition of serum. Four hours after release, cells were transfected with MPG/DNA or MPGΔNLS/DNA complexes and transgene expression was monitored 12 h later, when most of the cells were in late G1, and 20 h later, in late G2 prior to mitosis. Data were normalised with luciferase activity obtained 48 h after transfection and compared with asynchronous cells. As reported in Figure 2B, high levels of luciferase expression were observed with MPG after only 12 h, which corresponds to 65% of expression observed after 48 h. After 24 h, transfection efficiency reached 70%. In contrast, when transfection was performed with MPGΔNLS, luciferase activity was negligible after 12 h (7%) and only reached 26% after 24 h. Nevertheless, after 48 h, the efficiency of MPGΔNLS was 72% that of MPG. In comparison, lipid-based systems induced basal levels of luciferase activity after 12 h (10%), which may be explained by a fraction of unsynchronised, mitotic cells in the initial population. Similar experiments were performed on serum-starved, arrested cells and transgene expression monitored 24 h after transfection revealed that when transfected with MPG, these cells exhibited 50% luciferase activity in comparison with dividing cells (Fig. 2B). In contrast, no transfection was observed with either MPGΔNLS or lipid-based formulations. These data demonstrate that MPG facilitates translocation of DNA into the nucleus of arrested cells and that the NLS plays a crucial role in its nuclear uptake. The low efficiency of transgene expression observed with MPGΔNLS reveals that nuclear translocation of MPG/DNA particles is directly mediated by the import machinery through interaction with importin α.
Role of the cysteamide group in the mechanism of MPG-mediated gene delivery
Cysteamide was originally attached to the C-terminus of MPG, so as to enable covalent attachment of probes or drugs. We previously demonstrated that this modification did not affect the ability of MPG to form stable complexes with DNA. In order to investigate the impact of this C-terminal cysteamide group on gene delivery, comparative experiments were performed with an MPG peptide lacking a C-terminal cysteamide group. As shown in Figure 1A (grey bars), luciferase expression levels measured following delivery of pRL-SV40 by MPG lacking a cysteamide group were 1000-fold lower than those observed with MPG modified with a cysteamide group. These data suggest that the cysteamide moiety is required to promote efficient transfection. One explanation for this observation is that the cysteamide stabilises the carrier/DNA particle, as similarly reported for gene delivery mediated by cysteine-containing peptides (27).
MPG-mediated siRNA delivery
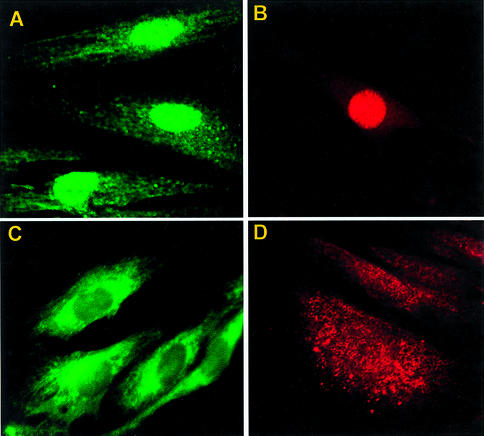
RNA interference (siRNA) constitutes a powerful tool to silence gene expression post-transcriptionally (28,29). In order to validate the potential of MPG-like carriers, we investigated the ability of MPG and MPGΔNLS to promote cellular uptake of siRNA. Fluorescently labelled siRNA designed to silence GAPDH was complexed with both MPG and MPGΔNLS at a molar ratio of 10:1, incubated on cultured HS-68 human fibroblasts in the presence of serum for 30 min, following which internalisation and subcellular localisation of siRNA were examined by fluorescence microscopy. Two different fluorescent probes (fluorescein and Cy3) were used in order to avoid any artefacts which might have been associated with the nature of the probe. As fixation procedures have sometimes been reported to cause artifactual uptake (30–32), experiments were performed on living cells. As shown in Figure 3, both peptide carriers were able to deliver siRNA into cultured cells with high efficiency (∼90% cells). Interestingly, the final subcellular localisation of the siRNA was dependent on the MPG carrier used. Fluorescently labelled siRNA localised rapidly to the nucleus when transfected with MPG, but remained mostly in the cytoplasm when transfected with MPGΔNLS. Moreover, these experiments demonstrate that MPG-mediated siRNA delivery is independent of the presence of serum.
Figure 3.
MPG-mediated delivery of siRNA. siRNA designed to target GAPDH was fluorescently labelled using Silence™ kits with fluorescein or Cy3. siRNA/MPG (A and B) and siRNA/MPGΔNLS (C and D) complexes were overlaid onto cultured HS-68 and cellular localisation of the siRNA was monitored by fluorescence microscopy on living cells 1 h after transfection.
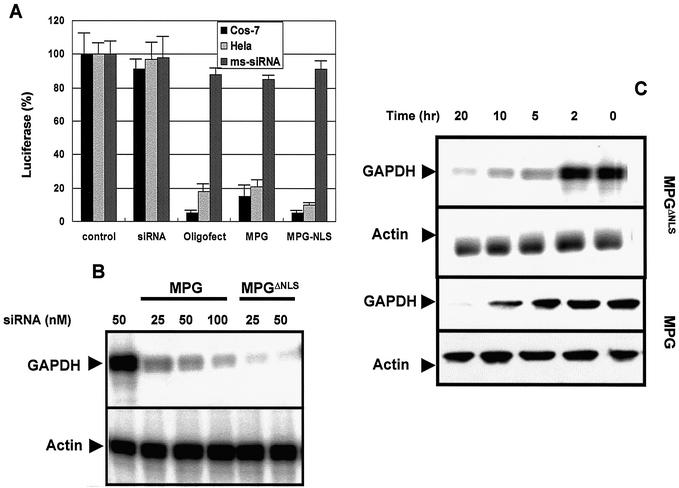
We next investigated the biological response of MPG-mediated siRNA delivery. Two series of experiments were performed. First we used the protocol described by Elbashir et al. (28): HeLa and Cos-7 cells transfected with the plasmid encoding the reporter gene luciferase were incubated with siRNA/MPG or siRNA/MPGΔNLS complexes in which the siRNA was designed to silence luciferase and luciferase activity was measured 48 h later. siRNA (50 nM) was associated with either MPG or MPGΔNLS at a charge ratio of 10:1. As a control, siRNA transfection was performed with the commercially available lipid-based delivery system Oligofectamine™. MPG-mediated delivery of siRNA yielded decreases of 78 and 85% in luciferase activity in Cos-7 and HeLa cells, respectively (Fig. 4A). This effect was further enhanced to 90 and 95% by MPGΔNLS. Similar experiments performed with a mismatch siRNA complexed with MPG or MPGΔNLS did not result in any change in levels of luciferase activity, suggesting that there is no side-effect associated with the presence of MPG. These experiments revealed that the efficiency of MPG-like peptides is similar to that of Oligofectamine™, suggesting that MPG is able to release the siRNA rapidly and does not affect its biological effect upon cellular internalisation.
Figure 4.
MPG-mediated delivery of siRNA induces a robust biological response. (A) MPG-mediated delivery of a siRNA targeting the luciferase gene. MPG/siRNA or MPGΔNLS/DNA complexes were formed in DMEM at a charge ratio of 10:1 and incubated for 30 min at 37°C, then overlaid onto Cos-7 or HeLa cells grown to 60% confluence and previously transfected with the pRL-SV40 plasmid encoding the reporter gene luciferase. After 30 min incubation at 37°C, 1 ml of fresh DMEM supplemented with 10% FCS was added to the cells. Luciferase activity was measured 48 h after transfection. Transfections were also performed with Oligofectamine™ as a standard. Control experiments were performed with either naked siRNA or using a mismatch siRNA (dark grey bars). (B) MPG-mediated delivery of siRNA targeting the GAPDH gene: western blot analysis. Different concentrations of siRNA (25, 50 and 100 nM) were transfected with either MPG or MPGΔNLS and the levels of GAPDH protein were analysed by western blotting 30 h post-transfection. Actin was used as a control to normalise protein loading. (C) MPG-mediated delivery of siRNA targeting GAPDH gene: northern blot analysis. The kinetics of siRNA (50 nM)- induced degradation of GAPDH mRNA following transfection with either MPG or MPGΔNLS were analysed by northern blotting. Actin was used as a control to normalise mRNA levels in each sample.
Implication of the NLS sequence of MPG in the kinetics of the response to siRNA
To further understand the impact of the NLS in MPG, we examined the biological response of a siRNA targeting the GAPDH gene following transfection into human fibroblasts (HS-68). Data found with HS-68 human fibroblasts were essentially the same as with HeLa and Cos-7 cells (data not shown). Different concentrations of siRNA (25, 50 and 100 nM) were transfected with either MPG or MPGΔNLS and the levels of GAPDH protein were analysed by western blotting 30 h post-transfection. As shown in Figure 4B, GAPDH protein levels were dramatically reduced when siRNA was transfected with either MPG carrier. Quantification of the signals revealed that a concentration of 25 nM siRNA transfected with MPGΔNLS reduced GAPDH protein levels by 80%, whereas 100 nM siRNA was required to reduce GAPDH levels by 60% when delivered by wild-type MPG, demonstrating that MPGΔNLS is at least 2-fold more efficient. The kinetics of siRNA-induced degradation of GAPDH mRNA following transfection with either MPG or MPGΔNLS were analysed by northern blotting. As shown in Figure 4C, MPG promoted a significant reduction in GAPDH mRNA after 20 h, which was initiated after 10 h. In comparison, MPGΔNLS induced a robust down-regulation of GAPDH mRNA after only 5 h. These data are comparable to those already reported for lipid-mediated siRNA delivery and suggest that the rapid release of siRNA into the cytoplasm significantly improves the silencing response. Taken together, this set of experiments has led us to conclude that mutations which affect the NLS of MPG are essential to design a potent vector for specific delivery of siRNA.
DISCUSSION
The two major barriers to the development of efficient non-viral gene delivery systems are the cell membrane and the nuclear membrane. In order to overcome these limitations we have designed a single peptide-based gene delivery system, MPG, which combines a hydrophobic domain derived for the fusion sequence of the HIV-1 gp41 protein with the NLS of the large T antigen of SV40 (17–19). MPG can cross cell membranes in a receptor-independent fashion and can therefore be associated with the class of peptides termed cell-penetrating peptides (for a review see 33). In the present work, we have investigated the mechanism through which MPG promotes gene delivery and have demonstrated that it is independent of the endosomal pathway. In this respect the mechanism through which MPG promotes gene delivery differs from that of other peptide-based gene delivery systems described so far (2,3) and is more related to that proposed for cell-penetrating peptides such as penetratin, transportan and TAT (9–11). However, recently some controversy concerning the mechanism of cell-penetrating peptides has arisen and divergent interpretations have been ascribed to differences in experimental procedures or in cell lines used (12,30,32). Recent progress in live cell imaging microscopy should help to avoid misinterpretations. Several studies have revealed that in the case of the TAT-based gene delivery system, the size of the peptide/DNA complexes plays an essential role in the uptake mechanism. Particles smaller than 300 nm do not enter the cell through the endosomal pathway (14,15), in contrast to particles of 500–700 nm, which are taken up by endocytosis (16). Along these lines, an explanation for the cellular uptake mechanism of MPG may therefore be found within the size of the MPG/DNA particles. Indeed, we previously determined by dynamic light scattering that the size of MPG/DNA particles was ∼200–300 nm (18). In this study, we also report that small MPG/DNA particles are able to enter cells in the presence of inhibitors of the endosomal pathway, in contrast to larger MPG/DNA aggregates. One part of the controversy concerning the mechanism of cell-penetrating peptides may be due to the size of the particles, which plays a crucial role in the internalisation pathway.
Most transfection systems described so far require the nuclear membrane to break down during mitosis for DNA to get into the nucleus, and are therefore dependent on active cell cycle progression (1). As the nucleus constitutes one of the major cellular compartments to be targeted, several strategies have been proposed to improve nuclear uptake of DNA. NLSs have been associated with gene delivery formulations, either covalently attached to DNA (34,35), fused with a peptide nucleic acid (36) or incorporated into lipid-based formulations (37). The nuclear translocation property of TAT has been used to promote nuclear targeting of plasmids (13,16). Here we have demonstrated that the NLS of MPG facilitates nuclear translocation of DNA and that MPG/DNA complexes interact directly with the nuclear import machinery though importin α. Hence, the NLS constitutes an essential and unique feature of MPG in comparison with other gene delivery systems. Moreover, we have shown that a specific mutation in the NLS impairs translocation of DNA into the nucleus and have used this property to promote delivery of siRNA into the cytoplasm, where it was more efficient in its silencing function than in the nucleus, consistent with the recently reported active site of siRNA (38).
Our investigation of the mechanism underlying the ability of MPG to deliver nucleic acids into cells has revealed that MPG-mediated delivery is independent of the endosomal pathway, and in part dependent on the NLS, which is involved in electrostatic interactions with DNA as well as specific interactions with the nuclear import machinery. Moreover, we have shown that the NLS is essential to promote delivery of nucleic acids to the nucleus, but that it is not required for cytoplasmic targeting. This latter feature suggests that specific variations in the sequence of MPG may yield carriers with distinct targeting features. In conclusion, the efficiency together with the targeting diversity of MPG-like peptides supports their potential for a wide range of therapeutic applications.
Acknowledgments
ACKNOWLEDGEMENTS
We express our gratitude to B. Cullen for importin constructs. We thank M. Dorée for continuous support, M. Billaud for helpful discussions and P. Travo, head of the IFR 24 Integrated Imaging Facility, for technical advice on microscopy. This work was supported in part by the Centre National de la Recherche Scientifique (CNRS) and by grants from the Agence Nationale de Recherche sur le SIDA (ANRS), the European Community (QLK2-CT-2001-01451) and the Association pour la Recherche sur le Cancer to M.C.M. (ARC-4326) and to G.D. (ARC-5271).
REFERENCES
- 1.Luo D. and Saltzman,M. (2000) Synthetic DNA delivery systems. Nat. Biotechnol., 18, 33–37. [DOI] [PubMed] [Google Scholar]
- 2.Mahato R.I. (1997) Non-viral peptide-based approaches to gene delivery. J. Drug Targeting, 7, 249–268. [DOI] [PubMed] [Google Scholar]
- 3.Morris M.C., Chaloin,L., Heitz,F. and Divita,G. (2000) Translocating peptides and proteins and their use for gene delivery. Curr. Opin. Biotechnol., 11, 461–466. [DOI] [PubMed] [Google Scholar]
- 4.Gariepy J. and Kawamura,K. (2000) Vectorial delivery of macromolecules into cells using peptide-based vehicles. Trends Biotechnol., 19, 21–26. [DOI] [PubMed] [Google Scholar]
- 5.Wyman T.B., Nicol,F., Zelphati,O., Scaria,P.V., Plank,C. and Szoka,F.C.,Jr (1997) Design, synthesis and characterization of a cationic peptide that binds to nucleic acids and permeabilizes bilayers. Biochemistry, 36, 3008–3017. [DOI] [PubMed] [Google Scholar]
- 6.Gottschalk S., Sparrow,J.T., Hauer,J., Mims,M.P., Leland,F.E., Woo,S.L. and Smith,L.C. (1996) A novel DNA-peptide complex for efficient gene transfer and expression in mammalian cells. Gene Ther., 3, 48–57. [PubMed] [Google Scholar]
- 7.Rittner K., Benavente,A., Bompart-Sorlet,A., Heitz,F., Divita,G., Brasseur,R. and Jacob,E. (2002) New basic membrane-destabilizing peptides for plasmid-based gene delivery in vitro and in vivo. Mol. Ther., 5, 104–114. [DOI] [PubMed] [Google Scholar]
- 8.Cartier R. and Reszka,R. (2002) Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther., 9, 157–167. [DOI] [PubMed] [Google Scholar]
- 9.Morris M.C., Chaloin,L., Heitz,F. and Divita,G. (2002) Signal sequence-based cell-penetrating peptides and their applications for gene delivery. In Langel,U. (ed.), Cell Penetrating Peptides: Processes and Application. CRC Press, Boca Raton, FL.
- 10.Lindgren M., Hällbrink,M., Prochiantz,A. and Langel,U. (2000) Cell-penetrating peptides. Trends Pharmacol. Sci., 21, 99–103. [DOI] [PubMed] [Google Scholar]
- 11.Wadia J.S. and Dowdy,S.F. (2002) Protein transduction technology. Curr. Opin. Biotechnol., 13, 52–56. [DOI] [PubMed] [Google Scholar]
- 12.Dom G., Shaw-Jackson,C., Matis,C., Bouffioux,O., Picard,J.J., Prochiantz,A., Mingeot-Leclercq,M.P., Brasseur,R. and Rezsohazy,R. (2003) Cellular uptake of Antennapedia Penetratin peptides is a two-step process in which phase transfer precedes a tryptophan-dependent translocation. Nucleic Acids Res., 31, 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truant R. and Cullen,B.R. (1999) The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol., 19, 1210–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torchilin V.P., Rammohan,R., Weissig,V. and Levchenko,T. (2001) TAT peptide on the surface of liposomes affords their efficient intracellular delivery even at low temperature and in the presence of metabolic inhibitors. Proc. Natl Acad. Sci. USA, 98, 8786–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torchilin V.P., Levchenko,T.S., Rammohan,R., Volodina,N., Papahadjopoulos-Sternberg,B. and D’Souza,G.G. (2003) Cell transfection in vitro and in vivo with nontoxic TAT peptide-liposome-DNA complexes. Proc. Natl Acad. Sci. USA, 100, 1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudolph C., Plank,C., Lausier,J., Schillinger,U., Muller,R.H. and Rosenecker,J. (2003) Oligomers of the arginine-rich motif of the HIV-1 TAT protein are capable of transferring plasmid DNA into cells. J. Biol. Chem., 278, 11411–11418. [DOI] [PubMed] [Google Scholar]
- 17.Morris M.C., Vidal,P., Chaloin,L., Heitz,F. and Divita,G. (1997) A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res., 25, 2730–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris M.C., Chaloin,L., Mery,J., Heitz,F. and Divita,G. (1999) A novel potent strategy for gene delivery using a single peptide vector as a carrier. Nucleic Acids Res., 27, 3510–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidal P., Morris,M.C., Chaloin,L., Heitz,F. and Divita,G. (1997) New strategy for RNA vectorization in mammalian cells. Use of a peptide vector. C. R. Acad. Sci. III, 320, 279–287. [DOI] [PubMed] [Google Scholar]
- 20.Silverstein S.C., Steinman,R.M. and Cohn,Z.A. (1977) Endocytosis. Annu. Rev. Biochem., 46, 669–710. [DOI] [PubMed] [Google Scholar]
- 21.Bowman E.J., Siebers,A. and Altendorf,K. (1988) Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells and plant cells. Proc. Natl Acad. Sci. USA, 85, 7972–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxfield F.R. (1982) Weak bases and ionophores rapidly and reversibly raise the pH of endocytic vesicles in cultured mouse fibroblasts. J. Cell Biol., 95, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brisson M., Tseng,W.C., Almonte,C., Watkins,S. and Huang,L. (1999) Subcellular trafficking of the cytoplasmic expression system. Hum. Gene Ther., 10, 2601–2611. [DOI] [PubMed] [Google Scholar]
- 24.Zelphati O. and Szoka,F.C.,Jr (1996) Mechanism of oligonucleotide releases from cationic liposomes. Proc. Natl Acad. Sci. USA, 93, 11493–11498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- 26.Cullen B.R. (2001) Journey to the center of the cell. Cell, 105, 697–700. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie D.L., Kwok,K.Y. and Rice,K.G. (2000) A potent new class of reductively activated peptide gene delivery agents. J. Biol. Chem., 275, 9970–9977. [DOI] [PubMed] [Google Scholar]
- 28.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 29.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 30.Richard J.P., Melikov,K., Vives,E., Ramos,C., Verbeure,B., Gait,M.J., Chernomordik,L.V. and Lebleu,B. (2003) Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem., 278, 585–590. [DOI] [PubMed] [Google Scholar]
- 31.Pichon C., Monsigny,M. and Roche,A.C. (1999) Intracellular localization of oligonucleotides: influence of fixative protocols. Antisense Nucleic Acid Drug Dev., 9, 89–93. [DOI] [PubMed] [Google Scholar]
- 32.Koppelhus U., Awasthi,S.K., Zachar,V., Holst,H.U., Ebbesen,P. and Nielsen,P.E. (2002) Cell-dependent differential cellular uptake of PNA, peptides and PNA-peptide conjugates. Antisense Nucleic Acid Drug Dev., 2, 51–63. [DOI] [PubMed] [Google Scholar]
- 33.Langel U. (ed.) (2002) Cell Penetrating Peptides: Processes and Applications. CRC Press, Boca Raton, FL.
- 34.Zanta M.A., Belguise-Valladier,P. and Behr,J.P. (1999) Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl Acad. Sci. USA, 96, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciolina C., Byk,G., Blanche,F., Thuillier,V., Scherman,D. and Wils,P. (1999) Coupling of nuclear localization signals to plasmid DNA and specific interaction of the conjugates with importin. Bioconjug. Chem., 10, 49–54. [DOI] [PubMed] [Google Scholar]
- 36.Branden L.J., Mohamed,A.J. and Smith,C.I. (1999) A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol., 17, 784–787. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian A., Ranganathan,P. and Diamond,S.L. (1999) Nuclear targeting peptide scaffolds for lipofection of non-dividing mammalian cells. Nat. Biotechnol., 17, 873–877. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Y. and Cullen,B.R. (2002) RNA interference in human cells is restricted to the cytoplasm. RNA, 8, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]