Abstract
Influenza A virus replication requires the interaction of viral RNA-dependent RNA polymerase (RdRp) with promoters in both the RNA genome (vRNA) and the full-length complementary RNA (cRNA) which serve as templates for the generation of new vRNAs. Although RdRp binds both promoters effectively, it must also discriminate between them because they serve different functional roles in the viral life cycle. Even though the inherent asymmetry between two RNA promoters is considered as a cause of the differential recognition by the RdRp, the structural basis for the ability of the RdRp to recognize the RNA promoters and discriminate effectively between them remains unsolved. Here we report the structure of the cRNA promoter of influenza A virus as determined by heteronuclear magnetic resonance spectroscopy. The terminal region is extremely unstable and does not have a rigid structure. The major groove of the internal loop is widened by the displacement of a novel A*(UU) motif toward the minor groove. These internal loop residues show distinguishable dynamic characters, with differing motional timescales for each residue. Comparison of the cRNA promoter structure with that of the vRNA promoter reveals common structural and dynamic elements in the internal loop, but also differences that provide insight into how the viral RdRp differentially recognizes the cRNA and vRNA promoters.
INTRODUCTION
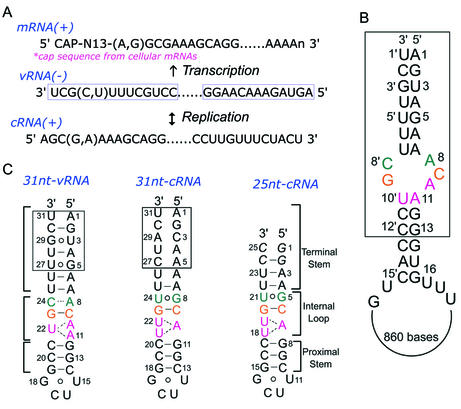
The influenza A virus genome consists of eight negative-sense, single-stranded RNA molecules. These genomic segments (vRNAs) act as templates for both messenger RNA (mRNA) and complementary RNA (cRNA) synthesis. The cRNAs, full-length copies of the vRNA molecules (1), serve as templates for viral genome replication (Fig. 1A). Both mRNA synthesis and genomic replication are performed within the nucleus of an infected cell by a ribonucleoprotein (RNP) complex composed of nucleoproteins (NP) and a virus-encoded heterotrimeric RNA-dependent RNA polymerase (RdRp), which contains proteins PB1, PB2 and PA (1).
Figure 1.
(A) Strategies for mRNA synthesis and genomic replication of the viral RNA genome of influenza A virus. The newly synthesized cRNA associates with NP and RdRp to form a cRNP complex, which then produces vRNAs. The newly synthesized vRNAs then bind NP and RdRp to form a vRNP complex, which gives rise to viral mRNA. The mRNA strands have the canonical 5′ cap and 3′ poly(A) tail to allow nuclear export and translation by the host translation machinery. In contrast, the cRNAs do not have such modifications, but are full-length copies of the vRNA molecules. (B) Terminal sequences of vRNA from the influenza A virus. Boxed sequences are converged in all of the influenza A virus variants. Numbering of the 3′ strand is followed by a prime notation (′). The sequence shown is that of vRNA segment 8 of influenza A/PR/8/34. (C) Secondary structure of the 31 nt vRNA promoter, which was determined previously (left), and the 31 nt cRNA promoter (middle) and 25 nt cRNA promoter (right), which were studied in this paper. Watson–Crick base pairs and wobble base pairs are distinguished by bars and circles, respectively. The terminal 5 bp of the 31 nt cRNA promoter have been changed to two G-C pairs in the 25 nt cRNA promoter model; these are boxed.
For initiation of RNA synthesis, the influenza A virus RdRp binds specifically to promoter structures formed by the 5′ and 3′ termini of each genomic RNA segment. Despite the high levels of sequence variability in influenza viruses, the two opposite arms of the vRNA promoters are highly conserved for all genomic segments among most influenza A virus variants (Fig. 1B). Regulatory signals for replication, transcription initiation and termination, polyadenylation and packaging are believed to reside within these terminal sequences (2–4).
The termini of the corresponding cRNA molecules could also form an irregular duplex analogous to that of the vRNA. However, the sequences of the cRNA termini differ from those of vRNA and, therefore, the two promoters form distinct structures. Because genomic replication starts with the synthesis of complementary RNP (cRNP) and a dramatic shift from cRNA to vRNA synthesis occurs, it is implied that a shift of template and recognition of the cRNA promoter by the newly synthesized polymerase and NP occur. The observed asymmetric pool sizes for cRNA versus vRNA molecules in infected cells (∼1:10, respectively) (5,6) also imply that the viral RdRp differentially recognize them. In fact, it is widely accepted that inherent asymmetry between the vRNA and cRNA promoter structures results in the differential recognition by influenza RdRp (5,6).
The internal loop among all residues in RNA promoters is known as an important site not only for RdRp binding to both RNA promoters (7) but also for the differential recognition by RdRp (4,8). Even though only vRNPs are packaged into virions, cRNP can be packaged into a virion by changing only the A*(UU) internal loop motif to the (AA)*U motif observed in the vRNP (4). The endonuclease activity of the viral polymerase, which is activated only by the vRNA promoter, also requires the presence of the (AA)*U sequence (8). By changing the A*(UU) of the cRNA to (AA)*U, the endonuclease activity induced by the cRNA promoter is significantly increased in vitro (8). These imply that the internal loop can be one of the most important factors for the viral RdRp to recognize two promoters differently.
We previously reported the three-dimensional (3D) structure and dynamic properties of the vRNA promoter based on NMR studies (9,10). These studies revealed that nucleotides at or near the asymmetric internal loop have a flexible conformation and identified a novel (AA)*U motif and a C-A mismatch that form within this promoter as well as the characteristic bending property at the fourth position from the 3′ terminus. Even though a structural basis for the requirement for specific interaction with the RdRp is revealed, the structure of the cRNA promoter is still an essential requirement for understanding the structural and dynamic basis for differential recognition of two RNA promoters by the RdRp.
In this study, we present the solution structure of the RNA oligonucleotide model of the conserved internal loop sequence of the influenza A virus cRNA promoter (PDB accession no. 1M82). Our results revealed structural and dynamic properties shared by the internal loops of the vRNA and cRNA promoters that could be important for their interaction with the RdRp. The comparison of cRNA and vRNA structures can provide insight into how differential binding of RdRp to the two promoters could occur.
MATERIALS AND METHODS
RNA sample preparation
Unlabeled and uniformly 13C, 15N-labeled samples of the 25 nt cRNA promoter were prepared by in vitro transcription as described previously (11,12). The unlabeled 31 nt cRNA promoter was prepared by cleaving a substrate RNA (41 nt) with a trans-cleaving hammerhead ribozyme designed for the 31 nt cRNA sequence (9). NMR samples contained 20 mM sodium phosphate buffer (pH 6.0) with 0.1 mM EDTA. The water NOESY spectrum was collected on a 1.0 mM unlabeled sample in 90% H2O/10% D2O. All other experiments were performed on a 1.0 mM unlabeled sample in 100% D2O or a 1.0 mM 13C, 15N-labeled sample in 100% D2O.
NMR spectroscopy
NMR spectra were recorded on Bruker AMX 500 MHz, Bruker DRX 600 or 800 MHz, or Varian Inova 600 MHz spectrometers, processed using FELIX (Biosym/MSI) and analyzed using SPARKY 3.95 (University of California, San Francisco). All spectra were recorded at 300 K unless otherwise specified. Two-dimensional NOESY spectra were recorded in water at 280 K with 150 ms mixing time. The NOESY build-up spectra were recorded with three different mixing times (80, 120 and 300 ms). The NOESY spectra recorded with the 80 and 200 ms mixing times were also recorded at 290 and 310 K. 31P-1H HetCOR, DQF-COSY, TOCSY (τm = 60 ms), 3D 1H-13C NOESY-HSQC (τm = 200 ms), 3D HCCH-COSY-TOCSY (13) and 3D HCCH-COSY spectra were also acquired and analyzed.
The 13C T1ρ measurements of aromatic carbons (C6/8) of the cRNA promoter were performed at 600 MHz on the Varian Inova spectrometer using the pulse sequence provided through RNA pack. 1H-13C correlation intensities decayed exponentially as a function of the 13C spin lock mixing time, and the T1ρ values for the aromatic carbon resonances were calculated by a non-linear least squares fit of the exponential decay. Ten different spin-lock times between 5 and 60 ms were used; the spectrum at the 10 ms spin-lock was repeated four times for error analysis.
Spectral analysis and structure calculations
Distance constraints obtained in the D2O NOESY experiments at 80, 120 and 200 ms mixing times and 3D NOSEY-HSQC experiments at 200 ms mixing time were grouped into four classes based on their intensities: strong (0–3 Å), medium (0–4 Å), weak (0–5 Å) and very weak (0–6 Å). Peaks present only at the longest mixing time (200 ms) were given even looser upper bounds (0–7 Å). The pyrimidine H5-H6 cross-peak in the 80 ms spectra provided a reference for establishing the ‘strong’ category, while the H1′-H3′ cross-peaks were used as a reference for medium intensity peaks. NOE cross-peaks involving exchangeable protons were classified as very weak, except between the uracil imino and adenine H2, and the guanosine imino and cytosine amino in Watson–Crick base pairs.
Hydrogen bonding and planarity restraints were imposed for the seven Watson–Crick base pairs in the stems and the G5-U21 and C6-G20 base pairs in the internal loop on the basis of the imino spectra and the observed NOEs and chemical shifts. The δ dihedral angles were derived from analysis of the 3JH1′–H2′ in DQF-COSY spectra. All χ values were constrained to –158 ± 15°, based on the medium to weak intra residue H1′-H6/8 NOE that ruled out the syn conformation, except for G14 (syn) and U18 (unconstrained). Other dihedral angle restraints (for α, β, ε and ζ) were obtained from 31P-1H HetCOR. 3JP–H5′ 3JP–H5′ and 3JH3′(i – 1)–P(i) couplings were measured to obtain information about the β and ε conformations and the α and ζ angles were restrained on the basis of 31P chemical shifts. None of the backbone dihedral angles were constrained in the internal loop, although the β and ε angles were loosely constrained based on 1H-31P HetCOR data.
Structure calculations were performed with CNS (14). 100 starting structures were generated with distance geometry using full structure embedding. The structures were then subjected to a torsion angle dynamics (15) protocol of 40 ps at 20 000 K, followed by 40 ps of cooling to 1000 K and 10 ps of cooling to 300 K. The distance force constant was 50 kcal mol–1 Å–2, and the dihedral angle constant was scaled to 400 kcal mol–1 Å–2 when cooling.
RESULTS
Design of cRNA promoter sequence for NMR studies
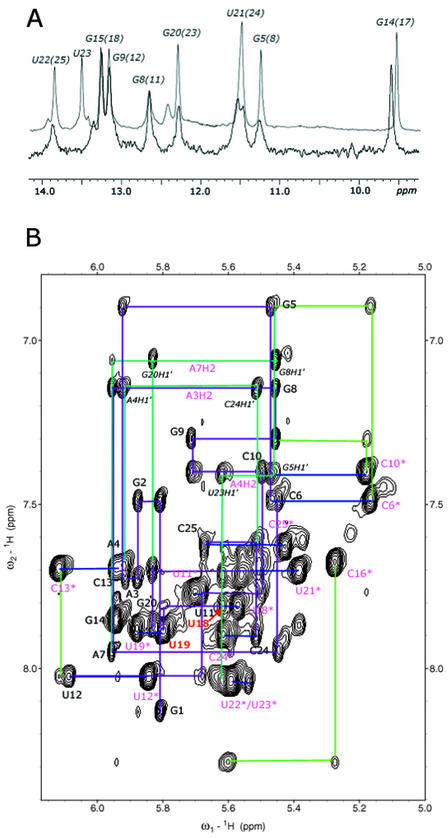
The sequences and secondary structures of the cRNA and vRNA promoters are shown in Figure 1C. Two G-U pairs in the terminal stem of the vRNA promoter are replaced by A-C mismatches in the cRNA promoter. As a consequence, the terminal region of the cRNA promoter is extremely unstable, and none of the five putative base pairs could be observed in the water NOESY spectra, even at low temperature (Fig. 2A). The single-strand nature of the terminal region makes it impossible to determine the entire structure of the 31 nt cRNA promoter. Therefore, we were forced to study a shorter fragment (25 nt cRNA) composed of residues 6–13 and 18–26 in the 31 nt cRNA promoter.
Figure 2.
(A) Superimposed imino region spectrum of the 31 nt cRNA promoter and 25 nt cRNA promoter. The spectrum was recorded at 4°C in 20 mM sodium phosphate buffer (pH 6.0) with 0.1 mM EDTA using a Bruker DRX 600 MHz spectrometer. (B) Fingerprint (aromatic-H1′) region in a NOESY spectrum of the 25 nt cRNA promoter; sequential assignments are indicated in violet. The spectrum was recorded at 300 K in 20 mM sodium phosphate buffer (pH 6.0) with 0.1 mM EDTA using a Bruker DRX 800 MHz spectrometer. The mixing time was 200 ms.
The 25 nt cRNA promoter showed the same base pairing as the 31 nt cRNA promoter except for the terminal region, as revealed by the analysis of NOESY spectra collected in water. Figure 2A shows that the imino proton spectrum of the 31 nt cRNA promoter is almost the same as that of the 25 nt cRNA promoter. Also, we observed the same chemical shifts of the residues in the internal loop and the proximal stem in the D2O NOESY spectrum (data not shown). This implies that the deletion of the terminal region did not induce structural changes within the conserved internal loop of the cRNA promoter that we aimed to study.
We are therefore confident that the sequence used in the present study represents a valid model of the internal loop of the influenza A virus cRNA promoter.
Resonance assignments and structure determination
All imino proton resonances of the 25 nt cRNA promoter except G1, U18 and U19 were assigned in water NOESY spectra using well established methods (16) and all amino protons from Watson–Crick paired bases were also identified. The G1 imino proton was absent, as is usually observed for terminal base pairs, but the two U imino protons expected within the internal loop were also invisible. Characteristically strong NOE between G5 and U21 imino protons and the behavior of the G5 amino proton resonances support the formation of the putative G5-U21 wobble base pair.
All of the base protons, H1′, H2′ and most of H3′, H4′ and H5′/H5″ resonances were assigned in D2O experiments (for more information, see Materials and Methods). The A3 H2 and A4 H2 were identified by strong NOEs to the U23 and U22 imino protons in the water NOESY spectra, respectively. The A7 H2 was identified by NOEs to the G8 H1′ and G20 H1′ and confirmed in the 1H-13C HSQC spectrum. Aromatic-H1′ and H2′ sequential assignments through all nucleotides were made from 2D and 3D NOESY spectra (Fig. 2B) (16). H1′-H2′ cross peaks of U18 and U19 were observed in the DQF-COSY spectra. U18 has a large 3JH1′–H2′ (∼10 Hz) and therefore the ribose sugar adopts primarily the C2′-endo pucker. U19 has an intermediate value of 3JH1′–H2′ and its ribose sugar is interconverting rapidly between the C3′-endo and C2′-endo conformations. We were able to identify and assign several phosphorous resonances in 31P Het-COR spectra. A7, U19 and G20 showed 31P chemical shifts separated from other residues.
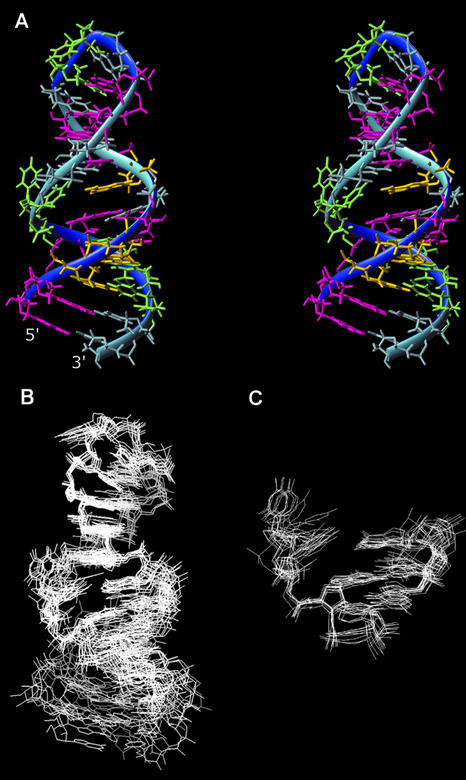
NOE distance and torsion angle constraints for structure calculation were collected as described in Materials and Methods and used in the program CNS to calculate the RNA structure. A set of 31 final structures was derived from 100 starting structures. These were based on 592 NOE-derived distance restraints, 140 torsion angle restraints, 54 planarity restraints and 48 base pair restraints. Average pairwise RMSD of all nucleotides is 2.0 Å, but the average pairwise RMSDs for the terminal stem and the proximal stem is 0.9 and 0.7 Å, respectively (Table 1). Thirty-one final structures were analyzed by MOLMOL (Institut für Molekularbiologie und Biophysik, Switzerland).
Table 1. Experimental and structural statistics.
| Experimental constraints | |
| Number of NOE distance restraints | 592 |
| Intra-residue | 247 |
| Inter-residue | 345 |
| Mean number per residue | 23 |
| Dihedral restraints (α, β, γ, δ, ε, ζ and χ) | 140 |
| Base pair restraints including hydrogen bonding | 48 |
| Base planarity restraints | 54 |
| Total number of restraints | 834 |
| Structure analysis | |
| RMSD for all heavy atoms from average structure (Å) | |
| Terminal stem (residues 1–4, 22–25) | 0.9 ± 0.2 |
| Proximal stem (residues 8–10, 15–17) | 0.7 ± 0.2 |
| Internal loop (residues 5–7, 18–21) | 1.4 ± 0.4 |
| UUCG tetraloop (residues 11–14) | 0.7 ± 0.2 |
| All nucleotides | 2.0 ± 0.5 |
| Average NOE violations (Å) | 0 (>0.5 Å) |
| Average dihedral angle violations (°) | 0 (>5°) |
| Mean deviation from covalent geometry | |
| Bond lengths (Å) | 0.002 |
| Angles (°) | 0.6 |
| Improper (°) | 0.3 |
Description of the overall structure
The stereoview of the lowest energy member of the family of 31 converged structures is shown in Figure 3A. The structure consists of the terminal double-helical stem, the internal loop, the proximal stem and the UUCG tetraloop. The structure of the UUCG tetraloop is very similar to the previously observed structure (17) and will not be discussed further. The G5-U21 wobble pair and C6-G20 normal Watson–Crick pair in the internal loop region, as well as all expected Watson–Crick base pairs in both stem regions, were confirmed from the pattern of NOE interactions and chemical shifts observed in the NOESY spectra recorded in water. In addition, NOEs observed in D2O NOESY spectra showed that both stems have the expected A-form geometry (Fig. 2B).
Figure 3.
(A) Stereo view of the lowest energy member of the family of 31 converged structures. Adenines are colored in yellow, guanines in pink, uridines in green and cytidines in light blue. (B) Superimposed overall structures (from residue 5 to 21) of the 10 lowest energy members of the structure ensemble. (C) Superimposed internal loop structures of the 10 lowest energy members of the structure ensemble.
Widened major groove of the internal loop
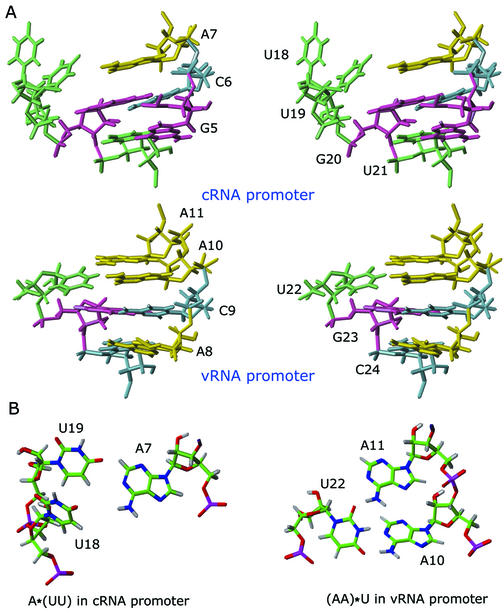
We previously reported that the internal loop of the vRNA promoter consists of a novel (A10 A11)*U22 motif and a C24-A8 mismatch (9); the counterparts in the cRNA promoter are an A7*(U18 U19) sequence and a G5-U21 wobble pair. The G5-U21 is well stacked above the C6-G20 pair (Fig. 4A). Within the internal loop region, A7, U18, U19 and G20 have the sequential NOEs characteristic of right-handed double helices. The cross-peaks between A7 H2 and G20 H1′ and the upfield chemical shift of the A7 H2 resonance (7.06 p.p.m.) revealed that the A7 base stacks on top of the C6-G20 base pair and that the A7 base is involved in inter-strand stacking with G20. Consistent with this observation, the A7 as well as U18 and U19 of the cRNA promoter are shifted towards the minor groove (Fig. 4A).
Figure 4.
(A) Major groove stereoview of the internal loop of the cRNA promoter (top) and the vRNA promoter (PDB accession no. 1fo7). (B) Top views of the A*(UU) from the cRNA promoter (left) and (AA)*U from the vRNA promoter structure (right).
Because the U18 and U19 bases are tilted upward and displaced into the minor groove, the helix axis is slightly redirected from the terminal stem toward the proximal stem; this qualitative observation was confirmed by a quantitative analysis with CURVES (18). However, the distortion is not large compared to other RNAs with asymmetric internal loops, such as the HIV TAR sequence (19). Since there are only two unpaired bases on the 3′-side of the loop and the U18 base is not well stacked on the neighboring bases, the physical separation between the base pairs adjoining the two stem regions on that strand is reduced (19). Moreover, the C2′-endo sugar conformation of U18 and the interchanging sugar conformation of U19 enable the backbone to proceed with a remarkably small overall distortion compared to A-form RNA, at the cost of an increased displacement of the bases into the minor groove. This base displacement results in a much wider and deeper major groove compared to normal A-form RNA.
U19 exhibits significantly weaker internucleotide NOE compared to other nucleotides in the internal loop (Fig. 2B), suggesting that U18 might be extra-helical. The U18 base was almost perpendicular to the neighboring base pairs in 29 out of 31 structures (Fig. 4A). The remaining two structures had the U18 base stacked into the helix, while the U19 base was moved into the minor groove even more significantly (data not shown).
There have been studies on the motifs formed by two uridines and one adenine in RNA (20–22). These studies showed that the bases constituting this motif are apart from one another in the sequence or that a distant uridine is hydrogen bonded to one A-U base pair. However, there have been no reports of a motif composed of one adenine and two consecutive uridines on the opposite strand, as we have studied here. In our structures, the A7 base does not form hydrogen bonds with U18 or U19, even though it is co-planar with U19 (Fig. 4B). Consistent with this, we failed to observe any resonance for the U18 or U19 imino proton resonances (Fig. 2A). The novel (AA)*U motif of the vRNA promoter studied previously also failed to show any specific hydrogen bonding between the bases. Altogether, these results suggest that neither the cRNA nor the vRNA internal loop is a rigid conformation in the absence of the protein (19,23,24).
Dynamic structure of the internal loop residues
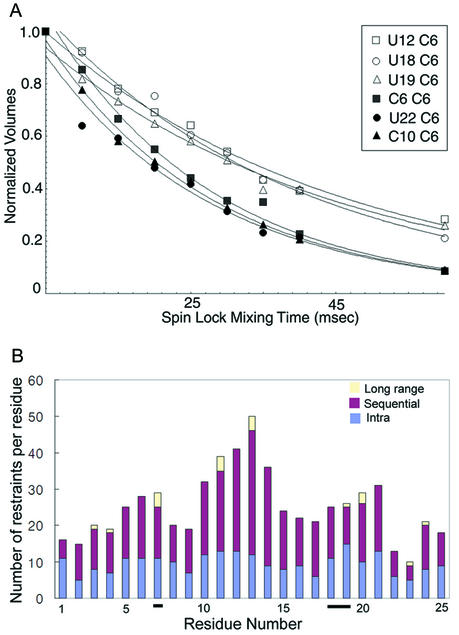
In order to investigate more quantitatively the dynamic characters of the residues in the internal loop, we measured rotating frame spin-lattice relaxation times (T1ρ) (Fig. 5A). For the pyrimidine C6 carbon atoms in the helical region of the cRNA promoter (residues C10, U22, U23 and U24), T1ρ values were nearly identical within the experimental error, varying from 22.6 to 23.4 ms. However, T1ρ values for U12 C6 (40.8 ms) in the UUCG loop and U19 C6 (40.9 ms) in the internal loop are much longer than those for residues in the helical regions. These results confirm that the bases of U19 and U12 display significant motion on the pico- to nanosecond timescale (25). The T1ρ value of U18 is also slightly longer than those for residues in the helical region (34.7 ms), indicating the presence of internal motion for this residue as well.
Figure 5.
(A) Nucleotides within the influenza A virus cRNA promoter experience internal motion on different timescales. Normalized intensity of H6(8)/C6(8) cross-peaks versus 13C spin-lock mixing time in T1ρ experiments for C6, C10, U12, U18, U19 and U22. The non-linear least squares fit of the data for each residue is shown with a solid line. (B) The plot of the number of NOE restraints per residue over the whole RNA. Sequential and long range NOE were counted both sides of two residues. A7, U18 and U19 are underlined. Total numbers of restraints of those residues are not significantly smaller than other residues, while the number of sequential NOEs of U19 is relatively small and most of the restraints of internal loop residues were given loosely.
Considering instead purine residues, the T1ρ value for the C8 carbon of A7 (25.6 ms) was the smallest among all resolved purines (G2, A3, A4, A7, G15 and G20). However, the difference between A7 and the other purines was not large (e.g. one of the differences was 3.83 ms), suggesting that A7 may experience motion on a slower timescale (micro- to millisecond).
T1ρ values for a particular carbon type (C6 or C8) are expected to be identical for all nucleotides that are rigid with respect to the molecular framework. Therefore, variations in T1ρ values for different residues within the internal loop strongly suggest that these residues have distinguishable dynamic characters, with differing motional timescales for each residue. Previous studies showed that motion on the micro- to millisecond timescale decreases T1ρ, while disorder on the pico- to nanosecond timescale increases T1ρ (25,26). In this respect, these results suggest that the observation of relatively large local RMSDs for the internal loop, as shown in Table 1, is not due to insufficient NMR information (Fig. 5B), but rather reflects genuine motional properties of this region of the structure (26).
DISCUSSION
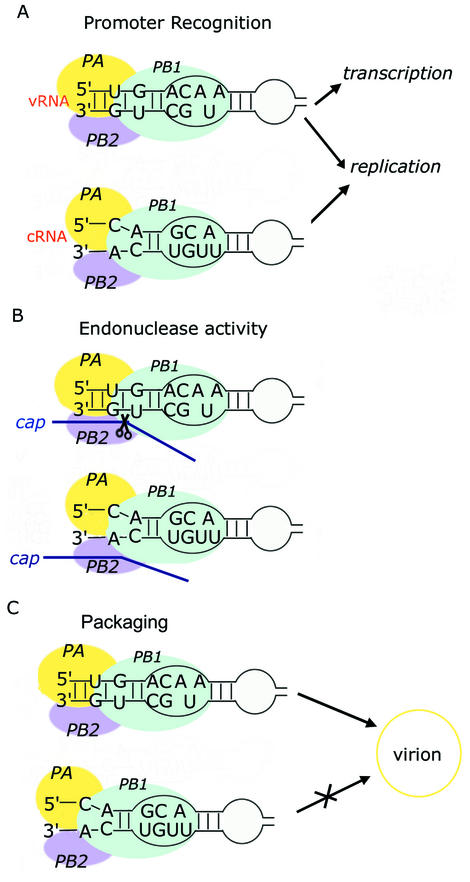
Promoter binding of the RdRp is a prerequisite for positioning the catalytic core of the enzyme at the polymerization start site during initiation of RNA synthesis. Cross-linking studies using influenza A virus RdRp that had been prepared from recombinant vaccinia virus showed that PB1 and PB2 can bind to both the vRNA and cRNA promoters (27). The binding regions within PB1 that are necessary for polymerase activity have also been identified (28). Residues located at the N-terminus of the protein interact with both RNA promoters, but PB1 also contains amino acid residues that selectively bind either vRNA or cRNA. The central region of PB1 is necessary for binding to the cRNA promoter, while the C-terminal region is crucial for binding to the vRNA promoter. The RNA residues within the two promoters that are necessary for binding RdRp were also identified. For the 31 nt vRNA promoter, A8-A11 and/or C20-G23 and U28-U31 are crucial for RdRp binding. For the 31 nt cRNA promoter, C9-A10 and C3-A5 are required for binding RdRp (27,29). Most of these residues belong to the internal loop in both RNA promoters (Fig. 6A).
Figure 6.
The differential recognition of the vRNA and cRNA promoters by influenza RdRp. (A) The RdRp can bind to both the vRNA and cRNA promoters. (B) The endonuclease activity of the RdRp activated by the vRNA promoter requires the presence of (AA)*U in the internal loop and the stable terminal stem. (C) The packaging signal of vRNP is the (AA)*U in the internal loop of vRNA promoter.
Our studies of the cRNA and vRNA promoters show that both internal loops have highly dynamic structures, and the major grooves of both RNAs are locally widened as described above (9). Remarkably, this distortion results in similar spatial cavities on the 3′ side of the internal loop, even though the symmetry of the loop is inverted (the extra nucleotide is on opposite strands in the vRNA and cRNA promoters) (Fig. 4A). The dynamic nature and widened major groove of the two internal loops are frequently observed in other protein-binding RNAs (19,30). From these data, it can be expected that the protein interacts with the widened major groove side of the internal loop. The importance of the asymmetric internal loop structures is confirmed by biochemical data showing that the 5′ arm of either RNA does not bind to the viral polymerase when paired with a perfectly complementary 3′ strand (27). This observation suggests that the organized structure and consequent flexibility of the internal loop is more important for protein binding than the sequence of the loop itself. In this respect, the A8-C24 mismatch of the 31 nt vRNA promoter and G5-U21 wobble pair of the 25 nt cRNA promoter may play a similar role to maintain the highly dynamic but pre-organized internal loop structure. Because the terminal region of the cRNA promoter is extremely unstable, the G5-U21 pair could be particularly important for the maintenance of the internal loop structure.
In each step of the infectious replication process, the newly synthesized viral polymerase must distinguish between the two RNA promoters, because vRNA synthesis from cRNP and mRNA synthesis from vRNP occur simultaneously and, therefore, both RNAs are present at the same time (Fig. 1) (31). Other viral and possibly cellular factors, such as the NP, participate in the replication process and may be important in providing this functional distinction between the two promoters (32). However, a recent study showed that the NP is not required for the initiation of de novo replication (33), suggesting that the different biochemical processes performed by the RdRp on the two different promoters should be determined by distinct interactions between the RNA promoters and the viral polymerase in their complex.
Previous biochemical studies about the packaging signal (4) and endonuclease activity (8) provide evidence that the 5′ bulge [5′-(AA)*U-3′] in the internal loop of the vRNA promoter and also the 3′ bulge [5′-A*(UU)-3′] in the internal loop of the cRNA promoter are important for differential recognition (Fig. 6B and C). As described above, the detailed conformations of the two internal loops are different from each other, even though they share global structural properties such as the locally widened major groove (Fig. 4A). In particular, the U18 and U19 bases of the cRNA promoter are not stacked into the helix but are displaced into the minor groove while all of the bases in (AA)*U of the vRNA promoter are stacked within the helix.
Studies on other RNA–protein complexes revealed that complex formation requires conformational changes in the RNA and often in the protein as well (23,24). Looped-out bases in both the Rev-RRE (Rev responsive element) complex and TAR RNA are critical for protein binding because they are stabilized by stacking and/or hydrogen bonding interactions with protein side chains triggering the conformational transitions (34). For the influenza A virus polymerase, biochemical studies have provided evidence for conformational changes in PB1 that occur upon interaction with either vRNA or cRNA (28). Therefore, flexibility within the internal loop, in particular within the two uridines of the internal loop of the cRNA promoter, may favor a conformational change that is probably required for protein binding in the cRNP complex.
There have been three distinct promoter models for the influenza virus RNA: the panhandle (9,35), RNA-fork (36,37) and corkscrew (5,38). The corkscrew model has been suggested especially for the promoter–polymerase open complex. Also, these models have been applied to the case of the cRNA promoter (4,7). The promoter regions that correspond to our internal loop are common for all three models; the differences in these models are found at the very end of the promoter structure. In the panhandle model, the 5′ and 3′ ends are base paired with each other, while they are not in the RNA-fork model. In the corkscrew model, hairpin loop structures are present in both the 5′ and 3′ termini. In our studies, the terminal region of the cRNA is quite unstable and cannot form a rigid structure, while that of the vRNA promoter has a stable stem (9). In company with the internal loop, this unstructured terminal region could be involved in differentiating the cRNA from the vRNA promoter. Regarding the initiation of viral replication, the endonuclease activity of RdRp normally required for initiation of transcription is not necessary because the replication is not cap-dependent. It has been reported that full activation of the endonuclease activity is dependent on the presence of both 5′ and 3′ termini of the vRNA promoter, while either termini alone does not effectively trigger the activity (39). Also, studies on other virus RNA replications have shown that the single-strand nature of the 3′ end is essential for the initiation of de novo synthesis (40,41). These studies suggest that the differences in the terminal region of both RNA promoters contribute to the effective viral replication by regulating RdRp activities, such as the endonuclease activity (Fig. 6B).
Taken together, the structural data presented here and previously published biochemical data allow us to formulate several working hypotheses on how the same RdRp regulates viral replication by interacting with distinct but similar promoter structures. The internal loops of both RNA promoters provide common protein binding sites, and the common displacement into the minor groove and locally widened major groove are likely to be the important common features that allow recognition of RdRp to occur. However, the structural and dynamic differences within the vRNA and cRNA promoters may induce two distinct RNP conformations. These structural differences of the initial interaction are likely to be amplified in the final complex and may thus be responsible for the divergent biological functions associated with the cRNA and vRNA promoter complexes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr Paul Cole for help with labeled sample preparation. This work was supported by the National Creative Research Initiative from the Ministry of Science and Technology, the Republic of Korea. C.-J.P., S.-H.B. and M.-K.L. were supported partially by the BK21 project.
PDB accession no. 1M82
REFERENCES
- 1.Lamb R.A. and Choppin,P.W. (1983) The gene structure and replication of influenza virus. Annu. Rev. Biochem., 52, 467–506. [DOI] [PubMed] [Google Scholar]
- 2.Fodor E., Pritlove,D.C. and Brownlee,G.G. (1994) The influenza virus panhandle is involved in the initiation of transcription. J. Virol., 68, 4092–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon L.L., Pritlove,D.C., Sharps,J. and Brownlee,G.G. (1998) The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J. Virol., 72, 8214–8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tchatalbachev S., Flick,R. and Hobom,G. (2001) The packaging signal of influenza viral RNA molecules. RNA, 7, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flick R., Neumann,G., Hoffmann,E., Neumeier,E. and Hobom,G. (1996) Promoter elements in the influenza vRNA terminal structure. RNA, 2, 1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 6.Mukaigawa J., Hatada,E., Fukuda,R. and Shimizu,K. (1991) Involvement of the influenza A virus PB2 protein in the regulation of viral gene expression. J. Gen. Virol., 72, 2661–2670. [DOI] [PubMed] [Google Scholar]
- 7.Pritlove D.C., Fodor,E., Seong,B.L. and Brownlee,G.G. (1995) In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J. Gen. Virol., 76, 2205–2213. [DOI] [PubMed] [Google Scholar]
- 8.Leahy M.B., Zecchin,G. and Brownlee,G.G. (2002) Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J. Virol., 76, 2019–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae S.H., Cheong,H.K., Lee,J.H., Cheong,C., Kainosho,M. and Choi,B.S. (2001) Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proc. Natl Acad. Sci. USA, 98, 10602–10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong H.K., Cheong,C., Lee,Y.S., Seong,B.L. and Choi,B.S. (1999) Structure of influenza virus panhandle RNA studied by NMR spectroscopy and molecular modeling. Nucleic Acids Res., 27, 1392–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao C., Zheng,M. and Rüdisser,S. (1999) A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA, 5, 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W., Kakalis,L.T., Jiang,L., Jiang,F., Ye,X. and Majumdar,A. (1998) 3D HCCH-COSY-TOCSY experiment for the assignment of ribose and amino acid side chains in 13C labeled RNA and protein. J. Biomol. NMR, 12, 559–564. [DOI] [PubMed] [Google Scholar]
- 14.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve, R,W, Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 15.Stein E.G., Rice,L.M. and Brünger,A.T. (1997) Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson., 124, 154–164. [DOI] [PubMed] [Google Scholar]
- 16.Varani G., Aboul-ela,F. and Allain F.H. (1996) NMR investigation of RNA structure. Prog. NMR Spectrosc., 29, 51–127. [Google Scholar]
- 17.Allain F.H. and Varani,G. (1995) Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol., 250, 333–353. [DOI] [PubMed] [Google Scholar]
- 18.Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irrregular nucleic acids. J. Biomol. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 19.Aboul-ela F., Karn,J. and Varani,G. (1996) Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res., 24, 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 21.Carter A.P, Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Wimberly,B.T. and Ramakrishnan,V. (2000) Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 22.Deng J., Xiong,Y., Sudarsanarkumar,C., Shi,K. and Sundaralingam,M. (2001) Crystal structures of two forms of a 14mer RNA/DNA chimer duplex with double UU bulges: a novel intramolecular U*(A·U) base triple. RNA, 7, 1425–1431 [PMC free article] [PubMed] [Google Scholar]
- 23.Frankel A.D. (1999) If the loop fits…. Nature Struct. Biol., 12, 1081–1083. [DOI] [PubMed] [Google Scholar]
- 24.Williamson J.R. (2000) Induced fit in RNA-protein recognition. Nature Struct. Biol., 10, 834–837. [DOI] [PubMed] [Google Scholar]
- 25.Dayie K.T., Wagner,G. and Lefpvre,J.F. (1996) Theory and practice of nuclear spin relaxation in proteins. Annu. Rev. Phys. Chem., 47, 243–282. [DOI] [PubMed] [Google Scholar]
- 26.Legault P., Hoogstraten,C.G., Metlitzky,E. and Pardi,A. (1998) Order, dynamics and metal-binding in the lead-dependent ribozyme. J. Mol. Biol., 284, 325–335. [DOI] [PubMed] [Google Scholar]
- 27.Tiley L.S., Hagen,M., Mathews,J.T. and Krystal,M. (1994) Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5-end of the viral RNAs. J. Virol., 68, 5108–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez S. and Ortin,J. (1999) Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. EMBO J., 18, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fodor E., Seong,B.L. and Brownlee,G.G. (1993) Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residue 9 to residue 12 of the promoter. J. Gen. Virol., 74, 1327–1333. [DOI] [PubMed] [Google Scholar]
- 30.Gosser Y., Hermann,T., Majumdar,A., Hu,W., Frederick,R., Jiang,F., Xu,W. and Patel,D.J. (2001) Peptide-triggered conformational switch in HIV-1 RRE RNA complexes. Nature Struct. Biol., 8, 146–150. [DOI] [PubMed] [Google Scholar]
- 31.Lee K.H. and Seong,B.L. (1998) The position 4 nucleotide at the 3′ end of the influenza virus neuraminidase vRNA is involved in temporal regulation of transcription and replication of neuraminidase RNAs and affects the repertoire of influenza virus surface antigens. J. Gen. Virol., 79, 1923–1934. [DOI] [PubMed] [Google Scholar]
- 32.Klumpp K. Ruigrok,R.W. and Baudin,F. (1997) Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J., 16, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M.T., Bishop,K., Medcalf,L., Elton,D., Digard,P. and Tiley,L. (2002) Definition of the minimal viral components required for the initiation of unprimed RNA synthesis by influenza virus RNA polymerase. Nucleic Acids Res., 30, 429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leulliout N. and Varani,G. (2001) Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry, 40, 7947–7956. [DOI] [PubMed] [Google Scholar]
- 35.Baudin F., Bach,C., Cusack,S. and Ruigrok,R.W. (1994) Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J., 13, 3158–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fodor E., Pritlove,D.C. and Brownlee,G.G. (1995) Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol., 69, 4012–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.J., Fodor,E., Brownlee,G.G. and Seong,B.L. (1997) Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol., 78, 353–357. [DOI] [PubMed] [Google Scholar]
- 38.Flick R. and Hobom,G. (1999) Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol., 80, 2565–2572. [DOI] [PubMed] [Google Scholar]
- 39.Hagen M., Chung,T.D., Butcher,J.A. and Krystal,M. (1994) Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol., 68, 1509–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dreher T.W. (1999) Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol., 37, 151–174. [DOI] [PubMed] [Google Scholar]
- 41.Chen D. and Patton,J.T. (1998) Rotavirus RNA replication requires a single-stranded 3′ end for efficient minus-strand synthesis. J. Virol., 72, 7387–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]