Abstract
Ubiquitous high-mobility-group (HMGB) chromosomal proteins bind DNA in a non-sequence- specific fashion to promote chromatin function and gene regulation. Minor groove DNA binding of the HMG domain induces substantial DNA bending toward the major groove, and several interfacial residues contribute by DNA intercalation. The role of the intercalating residues in DNA binding, bending and specificity was systematically examined for a series of mutant Drosophila HMGB (HMG-D) proteins. The primary intercalating residue of HMG-D, Met13, is required both for high-affinity DNA binding and normal DNA bending. Leu9 and Tyr12 directly interact with Met13 and are required for HMG domain stability in addition to linear DNA binding and bending, which is an important function for these residues. In contrast, DNA binding and bending is retained in truncations of intercalating residues Val32 and Thr33 to alanine, but DNA bending is decreased for the glycine substitutions. Furthermore, substitution of the intercalating residues with those predicted to be involved in the specificity of the HMG domain transcription factors results in increased DNA affinity and decreased DNA bending without increased specificity. These studies reveal the importance of residues that buttress intercalating residues and suggest that features of the HMG domain other than a few base-specific hydrogen bonds distinguish the sequence-specific and non-sequence-specific HMG domain functions.
INTRODUCTION
The high-mobility-group (HMG) domain plays a critical role in the diverse and essential functions of eukaryotic transcription factors and non-histone chromosomal proteins. The 80-residue HMG domain defines the HMGB superfamily that is composed of two sub-families distinguished by their abundance, function and DNA specificity (Fig. 1A) (1–3). The HMG domain transcription factors, typified by the testis-determining factor, SRY (4–6) and the lymphoid enhancer factor-1 (LEF-1) (7), are of low abundance, restricted to relatively few cell types, usually contain a single HMG domain and function by binding site-specifically in promoter or enhancer regions of regulated genes (3). In contrast, the ubiquitous non-histone chromosomal proteins of the HMGB family, typified by HMGB1/2 (8), are characterized by their moderate DNA binding affinity with minimal sequence specificity (9,10), and recognition of pre-bent and modified DNA (11–16).
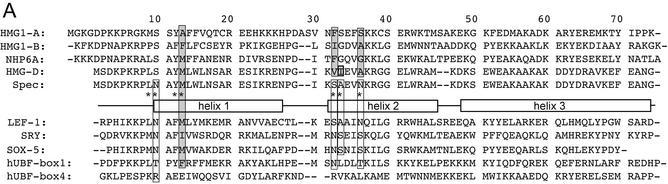
Figure 1.

HMG-box proteins. (A) Structural alignment of the sequences of HMGB1 boxes A (52) and B (53), NHP6A (42), HMG-D (54) and the ‘Spec’ protein used in this study, compared to the sequence-specific HMG proteins, LEF-1 (18), SRY (17) and Sox-5 (70) and human UBF HMG domains (71). The asterisks indicate residues that were mutated for this study. The sequences are aligned and numbered according the HMG-D structure with the alpha helices depicted by boxes (1,54). Residues shown from structural and modeling studies to intercalate the DNA are shaded in gray and the residues that differ between the sequence-specific and non-sequence-specific proteins are outlined in black. (B) Diagram showing the interaction of HMG-D-74 with linear DNA (21). The wild-type residues Met13, Ala36, Val32 and Thr33 partially intercalate the DNA at base-steps as indicated by arrows. (C) Diagram showing HMG-D-74 from the complex without the DNA in a view toward the DNA binding interface. Residues in gray contact the DNA and residues in black were examined in this study. (B) and (C) were constructed using Molscript (72) and Raster3D (73).
A unique mode of DNA bending is a dominant feature of the structures and functions of the sequence-specific and non-sequence-specific HMGB proteins (17–20). In fact, there can be exquisite sensitivity to precise DNA bending as has been observed for the testis-determining factor, SRY (5,6,21). SRY has a functional dependence on precise DNA bending, since the conservative sex-reversal mutant of SRY, M64I, is deficient in function and has a small but detectable change in DNA bending with no detectable change in DNA affinity (21,22). The chromosomal HMGB proteins function through interactions with DNA and other DNA binding proteins, whether direct or through induced alterations in DNA structure, that facilitate protein–protein interactions (reviewed in 12). For example, HMGB1/2 proteins facilitate chromatin function by interacting with nucleosomes (2,23), V(D)J recombination by interacting with RAG proteins (24) and regulation of transcription by interacting with many types of transcription factors (25–30). In order to facilitate multi-protein–DNA complexes known as enhancesomes, the HMG domain bends DNA bringing transcription factor binding sites into closer proximity (31,32). Biophysical studies, such as ligase-mediated circularization assays and fluorescence resonance energy transfer, have shown that the non-sequence-specific HMGB proteins including HMGB1 and individual HMGB domains (10,14,33,34), NHP6A (35,36), as well as HMG-D and HMG-Z (37–39) bend DNA by ∼60–110° per binding site.
Structural studies of several sequence-specific and non-sequence-specific HMG domains bound to linear DNA provided insights into DNA recognition and DNA bending (17–20,22). The 3-helix L-shaped HMG domain binds in a flattened, widened, bent and highly distorted minor groove, with burial of mixed hydrophobic and hydrophilic surfaces, ion pairs and hydrogen bonding contacts (Fig. 1B). Detailed analysis of crystallographic and NMR structures showing extreme DNA distortion and large DNA bend angles of up to 110° provided the structural basis for DNA bending for the HMG domain. DNA intercalation by multiple residues at two sites in the DNA is thought to be important for DNA bending in the non-sequence-specific proteins, whereas only one DNA intercalation site is observed in the sequence-specific HMG proteins (Fig. 1A) (reviewed in 40). Several of these intercalating residues also appear to be important specificity determinants because they differ consistently between the sequence-specific and non-sequence-specific HMG domain proteins (19,20,41,42).
In this study we systematically examine the specific contributions of interfacial intercalating residues of the HMG-D globular domain to DNA bending, binding and specificity using a mutagenesis and analysis approach. The integrity of the mutant HMG domain structures and protein stability were assessed using circular dichroism (CD) and thermal melting analyses. To distinguish whether the intercalating residues were necessary for DNA bending rather than just for stabilization of pre-bent-DNA binding, DNA affinity was measured using electrophoretic mobility shift assays (EMSAs) with two types of DNA fragments of similar sequence, a linear DNA sequence and a DNA fragment pre-bent by a disulfide crosslink (11,43). The effects of the mutations on DNA bending were determined by comparative ligase-mediated circularization studies designed to measure DNA bending propensity. An HMG-D specificity mutant having altered intercalating residues was designed and tested in the same way with additional studies conducted to evaluate DNA specificity. These results reveal novel features of the HMG domain relevant to DNA bending, dual DNA binding modes, HMG domain specificity and divergent HMG domain function.
MATERIALS AND METHODS
HMG-D-100 mutagenesis
DNA fragments encoding the HMG-D-100 gene with the L9A, M13A, M13V, M13I, M13L, M13F and M13T mutations were generated using standard PCR protocols with the pET13a plasmid containing the HMG-D-112 gene inserted between BamHI and NdeI sites (p112) as the template (44). Synthetic oligonucleotides (Operon Technologies, Inc.) were used as primers. One primer anneals to the first sixteen codons of the gene including the NdeI site (underlined), 5′-GAGATATACATATGTCTGATAAGCCAAAACGCCCACTCTCCGCCTACAGTCTGTGGCTC-3′, with the underlined codons substituted appropriately to produce the HMG-D-100 mutants at position 9 and 13. The second primer anneals near the C-terminus of the gene and introduces stop codons and a BamHI site (underlined), 5′-AAGGATCCCTATTACTTCTTGCTCTTCTT-3′. The desired PCR fragments were isolated from a 2% agarose gel by extraction (Qiagen), and digested with BamHI/NdeI (Invitrogen), as was the pET13a vector. The digested PCR products were ligated into the digested phosphatased (calf intestinal alkaline phosphatase; Promega) pET13a vector, and each ligation mixture was used to transform competent DH5α cells. Mutants Y12L, M13G, V32A, V32G, V32T, T33G, A36G and ‘Spec’ were made using either the Altered Sites II (Promega) or Quickchange (Stratagene) mutagenesis protocols with the p112 plasmid followed by PCR to truncate the construct to HMG-D-100 (38). The ‘Spec’ protein was made by first producing the S10N and V32N/T33S/A36N mutants separately using the Quickchange protocol with the following oligonucleotides and their complement strands: Spec-S10N-5′-GAGCCACAGCATGTAAGCGTTCAGTGGGCGTTTTGGC-3′ and Spec- V32N/T33S/A36S-5′-CACCACCGCGCTTGCTAACTTCG CTGTTTTTGATGCCGGGATTC-3′. PCR was used to combine the two mutants. All of the mutants were verified by DNA sequencing of the entire gene.
Protein expression and purification
The HMG-D-100 protein expression and purification protocol has been described previously (38). Briefly, the proteins were expressed in BL-21(DE3) cells transformed with the appropriate plasmid. After expression, the cell pellets were collected, and lysed by sonication and freeze/thaw procedures in a high salt lysis buffer A [50 mM HEPES (pH 7.9), 1.3 mM EDTA (pH 8.0), 670 mM NaCl, 0.8 mM PMSF, 1.3 mM benzamidine, 1.3 mM dithiothreitol (DTT)]. After dialysis of the lysate supernatant against buffer B [50 mM HEPES (pH 7.9), 1.3 mM EDTA (pH 8.0), 50 mM NaCl, 0.8 mM PMSF, 1.3 mM BZA, 1.3 mM DTT], the protein was separated by ammonium sulfate fractionation pelleting at 100% saturation and further dialyzed. Isocratic DEAE Sephacel chromatography (Amersham Biosciences) in buffer C [50 mM HEPES (pH 7.9), 1.0 mM EDTA, 50 mM NaCl] and SP-Sepharose column chromatography (Amersham Biosciences) with a 50 mM–1 M NaCl gradient in buffer C further purified the protein. To remove modifications and contaminants isocratic reverse phase-HPLC and SP-Sepharose chromatography were used. The protein was concentrated and exchanged into water using a stirred cell concentrator (Millipore). Samples were then analyzed by electrospray or MALDI mass spectrometry to confirm their masses, and UV spectroscopy to determine the protein concentration using an extinction coefficient of 19100 (Mcm)–1.
Circular dichroism
The CD spectra and thermal melting analyses were conducted with proteins at a concentration of between 6 and 12 µM in buffer containing 50 mM KCl, 10 mM Na/K phosphate, pH 7.0. CD spectra were recorded on a Jasco J-810 spectropolarimeter. Spectral scans were made at 20°C with a 1.5 nm bandwidth and a 0.1 nm step size, and data presented as molar ellipticity of HMG-D-100 protein. Thermal melting was carried out over a range of 0–80°C and refolding from 80 to 0°C at a rate of 1°C/min with a 1.5 nm bandwidth and a 0.2 nm step size. The ellipticity at 222 nm was recorded as a function of temperature for both the melting and refolding and individual spectra were recorded at 0, 20 and 80°C. There is a slight 2.5°C offset of the melting and refolding curves, but all of the proteins were similar in this respect. The melting temperature was determined to be the peak of the derivative of the melting spectrum as derived from curve fitting using an approach that also provided the van’t Hoff enthalpy (45).
DNA binding by EMSA
The linear duplex DNA fragment used for these studies has the sequence 5′-AGTTACTGAATTACGCTCAT-3′. The 20 bp disulfide crosslinked DNA fragment was kindly provided by Drs Scot Wolfe and Greg Verdine (11,43), xlnk: 5′-AGTTACTGXATTACGCTCAT-3′, 5′-ATGAGCGTXATTCAGTAACT-3′ where the Xs are cross-linked as described previously (33,34). LEF-1-DNA refers to the DNA fragment containing the LEF-1 binding site (underlined): 5′-TAGTTACTTCAAAGCGCTCAT-3′. Each DNA fragment was 32P-labeled at the 5′ end and purified as described previously (38). Of importance is extraction of the labeled DNA samples with phenol:chloroform:isoamyl alcohol (PCI) (25:24:1), and removal of the residual phenol using two ether extractions followed by ethanol precipitation. The DNA is resuspended in TE buffer (50 mM Tris–HCl pH 7.4, 1 mM EDTA) for storage at –20°C.
HMG-D-100 dilutions were made in 50 mM HEPES (pH 7.5), 0.1 M KCl, 1 mM EDTA, 100 µg/ml BSA and 50% glycerol to give a final concentration range of 0.0625 nM to 100 mM. Each 10 µl reaction mixture included 2 µg/ml BSA, 1 µl of 10× binding buffer [200 mM HEPES (pH 7.5), 500 mM KCl, 20 mM MgCl2], 1 µl of the appropriate protein dilution, and <1 nM radiolabeled DNA. The reactions were allowed to reach equilibrium during a 45 min incubation period at 22°C before being loaded onto a pre-run 6% native polyacrylamide gel (30:1, acrylamide: bis-acrylamide). After the samples had been electrophoresed at 125 V for 60 min in 0.33× TBE, the gel was dried and exposed to a phosphorimaging screen overnight. A Molecular Dynamics phosphorimaging system was used to digitize the gel images, and ImageQuant software (Molecular Dynamics, Inc.) was used to integrate band intensities corresponding to free and bound DNA at each protein concentration. The fraction of DNA bound was calculated and plotted for each protein concentration using Kaleidagraph (Synergy software). The resulting binding curves were fit to a single-site binding isotherm {Y = ([Pf] / Kd) / (1 + [Pf] / Kd)} using the same program. The free protein concentrations [Pf] and the fraction bound (Y) were calculated from the data whereas the equilibrium dissociation constant (Kd) was obtained as a result of the curve fit of the mean of all of the data points measured for each protein. The gels shown are typical gels, and show some smearing, which is common with non-sequence-specific DNA binding proteins that do not bind in one position on the DNA (38).
DNA bending—circularization assays
Circularization assays were conducted with an 11 nt duplex DNA fragment that has a cohesive two base 5′ overhang made of strands 5′-GCCATATTGAA-3′ and 5′-GCTTCAATATG-3′. Ligation products of a 10mer duplex, 5′-GCCTATTGAA-3′ and 5′-GCTTCAATAG-3′, were used as size controls. The oligonucleotides (Operon Technologies, Inc.) were purified, labeled and annealed as described previously (38). One microliter of a 10 µM stock of each mutant protein was added to 8 µl of 1 µM double-stranded DNA in ligase buffer (50 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 1 mM DTT) and allowed to incubate for ∼4 min. One unit of T4 DNA ligase (Invitrogen) was added and the reaction was allowed to proceed for 1 h at 22°C before DNA extraction and purification by PCI extraction, ether extraction and ethanol precipitation. The samples were redissolved in 10 µl of water and were digested for 20 min at 37°C with exonuclease III (Invitrogen) in (50 mM Tris–HCl, pH 8.0, 5 mM MgCl2, 1 mM DTT) to remove linear DNA fragments, followed by PCI extraction, ether extraction and ethanol precipitation. The samples were resuspended in water and loading buffer (50% glycerol, 1% xylene cyanol, 1% bromophenol blue), loaded onto a 40 cm 8% (30:1, acrylamide:bis-acrylamide; 1× TBE) native polyacrylamide gel, and electrophoresed for 4.0 h at 400 V. Gels were dried, exposed to a phosphorimaging plate, and analyzed using ImageQuant software (Molecular Dynamics, Inc.).
EMSA with purified DNA circles was conducted using individual circles that had been excised from preparative circularization gels, purified by PCI extraction, ether extraction and ethanol precipitation. Binding experiments were performed as described in the EMSA section.
Binding site selection
The DNA sequence of the 60 nt DNA fragment used in binding site selection contains an 18 bp stretch of randomized DNA sequence, 5′-GGGAGTCAAGCTTCCCACGAG(N)18GGTCACCTCGAATTCGCACCG-3′, flanked by fixed ends for PCR amplification with primers: 5′-GGGAGTCAAGCTTCCCACGAG-3′ (HindIII site underlined) and 5′-CGGTGCGAATTCGAGGTGACC-3′ (EcoRI site underlined). The initial labeled DNA duplex was prepared from the 60mer and the EcoRI primer using Klenow fragment and [32P]dATP. Binding site selection was accomplished by separating bound labeled DNA from unbound DNA using EMSA, followed by PCR amplification of the selected DNA using both primers. The binding reaction of the ‘Spec’ protein used conditions described in the EMSA section with a protein concentration of 100 nM initially and the addition of 100 ng of poly dIdC (Amersham Biosciences) as competitor DNA. The binding reaction was subjected to EMSA (5% native PAGE), and the DNA in the shifted band was extracted and purified using the Maxam and Gilbert crush and soak method followed by PCI extraction, ether extraction and ethanol precipitation. The purified DNA was PCR amplified using both primers and gel-purified for the next round of selection. After nine cycles of selection with a decrease in the amount of protein used to 20 nM, the final purified excised band was digested with EcoRI and HindIII, subcloned into EcoRI and HindIII digested and phosphatased BluescriptKS– plasmid (Stratagene), and the final product was used to transform DH5α cells. The sequences of 24 inserts were obtained and sequence and statistical analyses were carried out using the ‘Sequence Pattern Discovery’ server (46).
RESULTS AND DISCUSSION
The crystal structure of the HMG domain of HMG-D bound to DNA revealed a large hydrophobic interface area dominated by multiple partial DNA intercalations that appear to be important for the substantial DNA bending induced in unmodified linear DNA (Fig. 1B). In the duplex DNA fragment, GCGATATCGC, the largest bend angle at an individual base-step (T5-A6), 45°, is caused or stabilized by intercalation of Met13. The second most deformed base step (35° bend) in the binding site is at base-step G3-A4 that is intercalated by a pair of residues, Val32 and Thr33. Ala36 is closely associated with the base step, A4-T5, which shows the third largest bend angle, 15°, and may be involved in a partial intercalation that could help stabilize the overall DNA bend of the complex. To determine the role of these and other DNA contacting residues in DNA binding by the HMG domain, a series of amino acid substitutions was made at the individual positions and the proteins were examined for thermal stability and ability to bind and bend DNA (Fig. 1C). The HMG-D-100 protein, which is composed of a globular core domain and an unstructured C-terminal basic tail, was chosen as the framework for the mutagenesis, because it has the highest affinity for DNA of any of the HMG-D truncations studied to date, forming complexes that permitted reliable apparent affinity measurements to be made (38).
HMG domain stability depends on the ‘intercalation wedge’
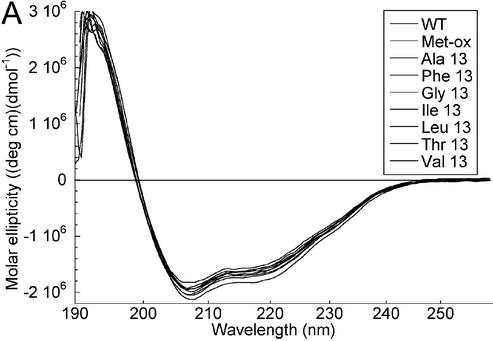
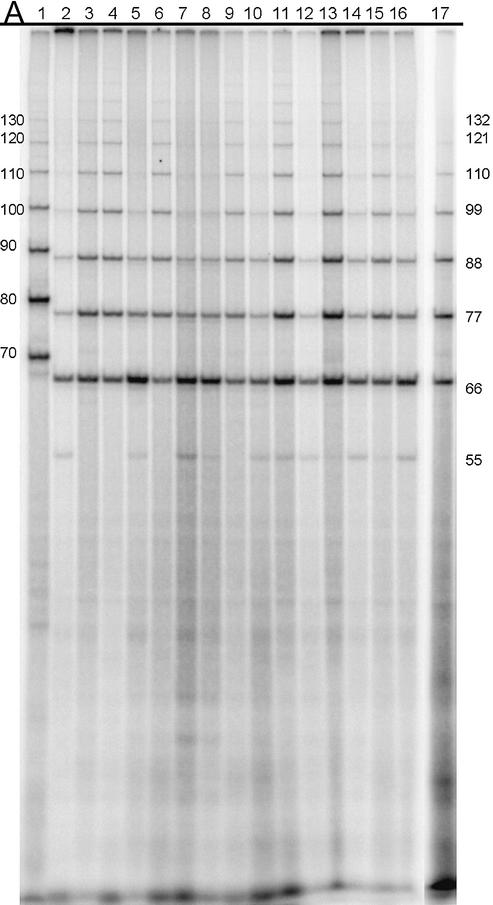
Mutant HMG-D-100 proteins with substitutions at position 13 have comparable secondary structure to the wild-type protein. Mutations that increased (Phe) or decreased (Leu, Ile, Val, Ala and Gly) the size of the intercalating residue, and a threonine substitution were produced using standard mutagenesis techniques and purified as described previously (Figs 1A and 2). The methionine sulfoxide (Met-ox)-containing protein was purified as described previously (44). CD studies give an average molar ellipticity of –20 000 (wavelength of 222 nm), indicating that HMG-D-100 contains ∼55% alpha helix with a small amount of random coil polypeptide (Fig. 3A) that is due to the unstructured basic tail (47). Position 13 occurs in helix 1 of HMG-D, and all of the mutants have CD spectra that are virtually indistinguishable from the wild-type protein, despite the fact that the threonine and glycine substitutions are not favorable alpha helix-forming residues (Fig. 3A). Therefore, the integrity of the HMG domain appears to be unaltered by any of the mutations at residue 13. A variety of residues observed at this primary intercalating position includes alanine in HMGB1-boxA, phenylalanine in HMGB1-boxA, isoleucine in SRY, and methionine in NHP6A, HMG-D and LEF-1 (1). Previous studies of the HMGB proteins showed that mutations produced at this position do not affect HMG domain structure, consistent with the HMG-D results (15,35,48,49).
Figure 2.
SDS–PAGE analysis of the mutant HMG-D proteins. Coomassie-stained 18% SDS–PAGE gels loaded with 1 µg of each protein. Lanes 1 and 11 are molecular weight markers (16 949, 14 404, 8160 are distinguishable), lanes 2–10 are the proteins WT, Met13ox, M13A, M13F, M13G, M13I, M13L, M13T, M13V, and lanes 12–19 are L9A, V32A, V32G, V32T, T33G, A36G, Spec, Y12L.
Figure 3.
Structure and stability of the HMG-D mutants. (A) CD scans overlaid for all of the M13 mutants. (B) CD scans overlaid for the L9A, A36G, V32A, V32G, V32T, V33G, Y12L and Spec mutants. (C) CD melting and renaturation curves for HMG-D wild-type protein (9.4 µM).
Thermal melting analyses by CD revealed the effect of the substitutions on the stability of the HMG domain. Thermal melting of the HMG domain is virtually reversible (Fig. 3C) under the conditions tested, and the melting temperature varies from 41.0°C for the wild-type protein to 35.0°C for the least stable Met13 mutant, M13T (Table 1). The wild-type and mutant proteins exhibit very slight premelting of the domain (<10%) at temperatures below the cooperative transition recorded in Table 1, with the exception of the M13G and M13F mutants that had significant premelting transitions below 20°C (data not shown). The M13F and M13G mutants also exhibited the lowest van’t Hoff enthalpy values of 27.6 and 30.8 kcal/mol compared to 39 kcal/mol for the wild-type protein. In general, the trend of melting temperatures, Met ∼ Leu > Phe > Ala > Ile > Val > Gly > Thr, follows the expected trend based on helix-forming propensities, Ala > Leu > Met > Phe > Ile > Val > Thr > Gly (50,51). Surprisingly, a very good helix-forming mutant, M13A, exhibits a lower than expected thermal stability, and the least stable mutant, M13T, is not the protein with the lowest expected helical propensity. Therefore, it is likely that alanine and threonine at position 13 destabilize the protein by a means other than destabilizing helix 1, perhaps through incorrect or missing interactions with neighboring residues (Fig. 1C).
Table 1. Thermal melting analysis of HMG-D and mutant proteins.
| Protein | Melting temperature (°C) | Difference from WT | ΔHvha (kcal/mol) | ΔΔHvha (kcal/mol) |
|---|---|---|---|---|
| Wild type | 41.0 | – | 39.0 | – |
| WT oxidized | 41.3 | –0.3 | 38.7 | 0.3 |
| M13A | 39.0 | 2.0 | 36.9 | 2.1 |
| M13F | 39.5 | 1.5 | 27.6 | 11.4b |
| M13G | 36.3 | 4.7 | 30.8 | 8.2b |
| M13I | 37.3 | 3.7 | 33.0 | 6.0 |
| M13L | 40.5 | 0.5 | 35.9 | 3.1 |
| M13T | 35.0 | 6.0 | 34.3 | 4.7 |
| M13V | 37.0 | 4.0 | 33.9 | 5.1 |
| Y12L | 30.3 | 10.7 | 31.8 | 7.2 |
| L9A | 37.8 | 3.2 | 38.7 | 0.3 |
| V32A | 44.7 | –3.7 | 40.7 | –1.7c |
| V32G | 42.2 | –1.2 | 37.2 | 1.8 |
| V32T | 41.8 | –0.8 | 37.6 | 1.4 |
| T33G | 41.8 | –0.8 | 36.9 | 2.1 |
| A36G | 38.3 | 2.7 | 37.0 | 2.0 |
| Spec | 40.8 | 0.2 | 37.9 | 1.1 |
aVan’t Hoff enthalpy determined by curve fitting according to John and Weeks (45).
bMelting curve showed greater premelting than other proteins (shoulder at ∼20°C in the derivative melting spectrum).
cNo premelting observed for this protein.
Close examination of the structural context of Met13 in the HMG domain reveals that the side chain has van der Waals interactions with Tyr12 and Leu9 in the free and bound proteins of several species of HMG domain (19,20,47,52,53). Tyr12 is adjacent to Met13 in helix 1 and is primarily solvent exposed in the absence of DNA. It has an important role itself in packing against Trp43 and helix 2 in the HMG domain core; in the DNA complex it interacts with a ribose and makes a hydrogen bond directly to the DNA, but was not necessarily expected to be important in free protein stability (19,20). Tyr12 was mutated to leucine, a large aliphatic residue, rather than alanine so that hydrophobic interactions with Trp43 could be maintained (Fig. 1C). The Y12L mutant forms a near wild-type protein as monitored by CD (Fig. 3B), but the stability is substantially lower than wild type, with a melting temperature of only 30.3°C (Table 1). Furthermore, Y12L is unable to bind DNA sufficiently well to determine its DNA affinity using EMSA, and so no further studies were conducted. The equivalent residue in NHP6A and HMGB1-box B has been mutated (20,42), and the deficiency in stability was measured for the HMGB1-box B F18A mutant, which melts at a temperature 19°C lower than the wild-type HMGB1-box B (49).
Leu9 is not in an alpha helix itself but is adjacent to the N-terminus of helix 1 in a region of irregular secondary structure (Fig. 1C). It makes van der Waals interactions with the Cβ atom of Met13 in the free protein (54). Leu9 was mutated to alanine, which would disrupt any contacts with Met13. The L9A mutant has equivalent secondary structure compared to the wild-type protein as measured by CD (Fig. 3B), but has a reduced melting temperature of 37.8°C. In HMGB1-box B, Pro15 is equivalent to Leu9, and mutation to alanine results in a much larger decrease in protein stability (9°C) compared to the L9A mutant of HMG-D, which decreases the melting temperature by only 3°C (49).
Among the known HMG domain sequences there is a correlation between the residues at positions 9 and 13. Particular non-polar residue pairs, Leu9–Met13 (HMG-D, NHP6A, LEF-1), Pro–Phe (HMGB1-box B) and Met–Ala (HMGB1-box A) appear in the majority of the non-sequence-specific HMG-proteins, and in addition, Met–Met, Met–Ile (SRY), and a few others occur in the sequence-specific proteins (1). These residues interact in structural studies of HMGB1-boxA, HMGB1-boxB and NHP6A, and together with residue 12, have been called the ‘intercalation wedge’, because they appear to be important for the specific bent-DNA structure observed in structural studies (17,20,55). Interest ingly, recent structural analyses of one of the human upstream binding factor (UBF) HMG domains, domain 5, reveals that the equivalent residues are polar and charged, potentially able to form ion pairs that could also stabilize the HMG domain (PDB entry 1L8Z; W. Yang, Y. Xu, J. Wu, W. Zeng and Y. Shi, unpublished results). Therefore, we suggest that the interaction of residues 9, 12 and 13 is an important component of HMG domain stability in addition to the role that this group may play in DNA binding and bending.
Contribution of the primary ‘intercalation wedge’ to DNA bending and DNA binding
By analogy with SRY and HMGB1, Leu9 and Tyr12 have been proposed to contribute to Met13 intercalation through buttressing the DNA binding interaction. To determine how these residues contribute to DNA binding and whether DNA bending and binding are correlated in HMG-D, DNA binding affinities of the HMG-D-100-mutants to a linear and a pre-bent DNA substrate were measured. EMSAs were used to separate bound from free DNA (Figs 4A and 5A), and Kd values were calculated based on between four and eight replicates (Figs 4B and 5B).
Figure 4.
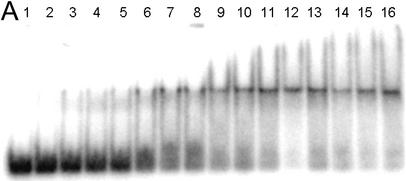
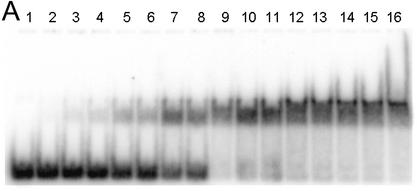
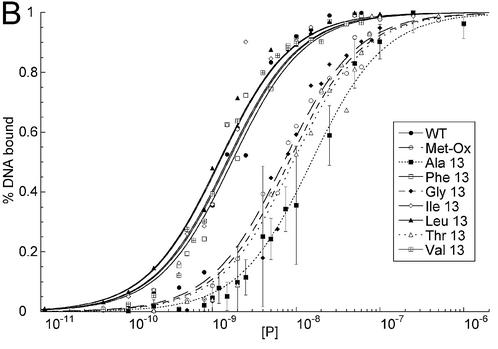
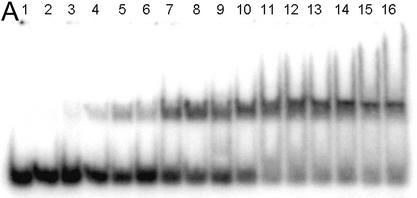
EMSA of HMG-D mutants with linear DNA. (A) Image of a typical EMSA gel used to obtain data for binding curve analysis. Lane 1 contains no protein; lanes 2–16 contain V32A protein at the following final concentrations: 0.4, 0.8, 1.0, 2.0, 5.0, 8.0, 10.0, 20.0, 40.0, 60.0, 80.0, 100.0, 250.0, 500.0 nM and 1.0 µM, respectively. (B) Binding curve overlays for the M13 series of mutants with error bars superimposed for M13V. (C) Binding curve overlays for the L9, A36, V32 and T33 series of mutants with error bars superimposed for L9A.
Figure 5.
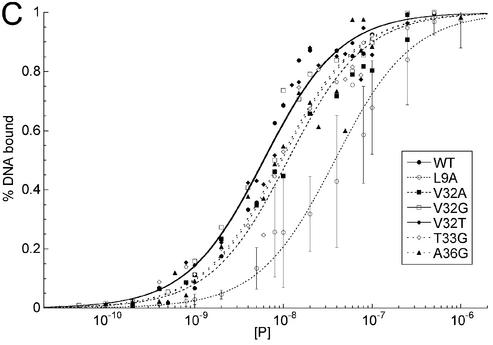
EMSA of HMG-D mutants with disulfide crosslinked ‘pre-bent’ DNA. (A) Image of a typical EMSA gel used to obtain data for binding curve analysis. Lane 1 contains no protein; lanes 2–16 contain M13L protein at the following final concentrations: 0.01, 0.05, 0.1, 0.2, 0.4, 0.8, 1.0, 2.0, 5.0, 10.0, 15.0, 25.0, 50.0, 100.0 and 250.0 nM, respectively. (B) Binding curve overlays for the M13 series of mutants with error bars superimposed for M13A. (C) Binding curve overlays for the A36, V32 and T33 series of mutants with error bars superimposed for L9A.
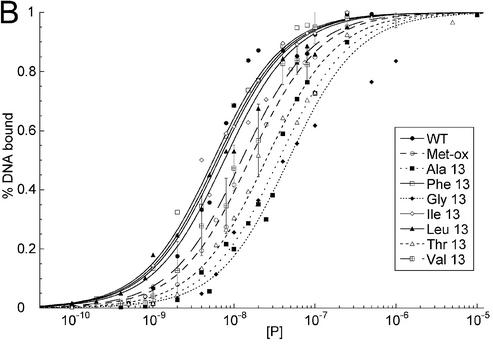
A linear DNA fragment of 20 nt was used to determine whether particular residues are required to bend and bind linear DNA. The trend in mutant affinity is wild type (WT) ∼ Phe ∼ Ile ∼ Leu > Val > OX >> Thr > Ala > Gly, where M13G affinity is 7.7-fold weaker than WT HMG-D. The mutants, in order of overall affinity, cluster into a group containing aliphatic side chains and Phe13 that have affinity most similar to the wild type (<2.5-fold differences). The remaining mutants, containing either beta-branched hydrophilic or small residues, bind with decreasing affinity depending on size. The isostructural mutants, M13V and M13T, have affinities that differ from each other by 2-fold, suggesting that the threonine hydroxyl has only a slight effect on DNA binding.
The DNA binding affinities of the same series of mutants was determined for a pre-bent DNA fragment of nearly identical sequence containing a disulfide crosslink (11) providing information about the importance of particular residues for pre-bent DNA binding. The DNA is underwound by 35° and has a smooth bend toward the major groove of 30°, and was previously used in HMG-D binding and NMR structural studies (11,47). The mutants followed a similar trend in affinity for the pre-bent DNA, Leu ∼ Ile > Val ∼ Phe ∼ WT > OX > Gly ∼ Thr > Ala, but they cover a range of Kd differences from 0.7- to 8.9-fold (Fig. 5B and Table 2). Interestingly, the conservative mutants, Phe, Leu, Ile and Val, actually bind better to the pre-bent DNA than the wild-type protein with a methionine at position 13. The Met-ox13 mutant bound relatively worse to the pre-bent compared to the linear DNA, and the most hydrophilic and the smallest sized mutants bound the least well of all. Each of the Kd values reported in Table 2 has a <10-fold difference in DNA affinity compared to the wild-type protein. This result was surprising, because it suggests that abolishing intercalation itself through residue 13 is not completely detrimental to DNA binding. In studies with other HMG domain proteins, it has been shown that loss of electrostatic interactions due to a single basic residue has a much greater effect on HMG domain DNA binding affinity than observed here for loss of DNA intercalation (56).
Table 2. DNA affinity of HMG-D mutants for pre-bent and linear DNA fragments.
| Protein | Kd xlnk DNA (nM) (n) | Kdmut/KdWT | Kd linear DNA (nM) (n) | Kdmut/KdWT | Kd LEF-1 DNA (nM) (n) | Kdmut/KdWT |
|---|---|---|---|---|---|---|
| WT | 1.71 ± 0.18 (4) | 6.08 ± 0.67 (4) | 1.79 ± 0.17 (7) | |||
| Ox 13 | 7.13 ± 0.49 (4) | 4.2 | 15.5 ± 0.90 (4) | 2.5 | ||
| M13A | 15.2 ± 0.92 (7) | 8.9 | 35.5 ± 2.64 (5) | 5.8 | ||
| M13F | 1.54 ± 0.14 (7) | 0.9 | 5.57 ± 0.34 (5) | 0.9 | ||
| M13G | 8.20 ± 0.84 (6) | 4.8 | 46.9 ± 5.72 (5) | 7.7 | ||
| M13I | 1.22 ± 0.14 (4) | 0.7 | 6.51 ± 0.48 (5) | 1.1 | ||
| M13L | 1.19 ± 0.11 (5) | 0.7 | 7.61 ± 0.59 (8) | 1.3 | ||
| M13T | 9.21 ± 0.85 (6) | 5.4 | 24.8 ± 1.55 (3) | 4.1 | ||
| M13V | 1.48 ± 0.10 (6) | 0.9 | 12.4 ± 0.93 (6) | 2.0 | ||
| L9A | 2.53 ± 0.25 (4) | 1.5 | 38.3 ± 3.83 (5) | 6.3 | ||
| V32A | 1.05 ± 0.07 (4) | 0.6 | 11.3 ± 1.06 (6) | 1.9 | ||
| V32G | 1.89 ± 0.18 (7) | 1.1 | 6.03 ± 0.52 (5) | 1.0 | ||
| V32T | 0.79 ± 0.10 (5) | 0.5 | 5.95 ± 0.47 (6) | 1.0 | ||
| T33G | 3.03 ± 0.37 (4) | 1.8 | 9.64 ± 0.86 (8) | 1.6 | ||
| A36G | 2.09 ± 0.19 (7) | 1.2 | 9.17 ± 0.10 (7) | 1.5 | ||
| Y12L | ND | ND | ||||
| Spec | 0.697 ± 0.065 (4) | 0.11 | 0.350 ± 0.029 (4) | 0.19 |
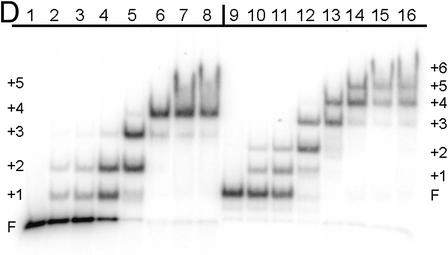
To determine the DNA bending propensity of the mutants, a ligase-mediated circularization assay that is quite sensitive to slight differences in DNA bending was used (37,38). DNA circles are formed from multimers of an 11 nt oligomer with two-base cohesive ends in the presence of DNA ligase and 1:1 ratio of mutant HMG-D-100 protein to DNA duplexes. The relative affinity for the DNA is not a consideration of the experiment, as the concentration of the protein and the DNA are at least 10-fold higher than the weakest measured Kd. The circular DNA fragments were separated by native PAGE (Fig. 6A), and the percent of each DNA circle formed as a function of circle size was used as a measure of the degree of DNA bending propensity. The plot in Figure 6B clearly shows that there are two patterns of DNA circle sizes observed, those that cluster near the wild type and produce between 71 and 88% 55 and 66 bp circles, and those that form relatively fewer 66 bp circles, in the range of 36–48%. Examination of HMG-D binding to 66 and 77 bp circles was conducted by EMSA to determine how many distinct complexes could form (Fig. 6D). The series of bands indicates that the number of proteins bound to the different sized circles are at least four for the 66 bp circle and five for the 77 bp circle. Based on this, an estimate of the DNA bend angle is at least 72°, but probably closer to 90°, values that are consistent with observations from FRET analysis and the HMG-D–DNA crystal structure (19,39). The discontinuity of the pattern of bound circles, the slight jump between bands 2 and 3, was observed previously for HMG-D and HMGB1, but the explanation is unclear (14). The mutations that result in clustering near the wild-type DNA bending propensity are the aliphatic and aromatic substitutions, and those that bend less well are the hydrophilic and small side chain groups. The bending propensity trend correlates with the affinity of the mutants for both linear and pre-bent DNA, but has a bimodal distribution similar to the pre-bent DNA binding results (Fig. 5B).
Figure 6.
DNA bending of HMG-D mutants assayed by ligase-mediated circularization assay. (A) Electrophoresis gel image of the circles formed by the 11mer DNA in the presence of HMG-D-100 proteins; lanes 2–17, WT, Met13ox, M13A, M13F, M13G, M13I, M13L, M13T, M13V, L9A, V32A, V32G, V32T, T33G, A36G and Spec. A control lane of the 10mer circularized in the presence of the WT HMG-D-100 is in lane 1. (B) Circularization curves for the M13 series of mutants. (C) Circularization curves for L9, A36, V32 and T33 series of mutants and Spec. (D) EMSA of HMG-D bound to the 66 bp DNA circles (lanes 1–8) and 77 bp circles (lanes 9–16). Protein concentrations (WT) for each group are 0, 0.05, 0.1, 0.5, 1.0, 5.0, 25.0 and 100.0 nM, respectively.
To examine the role of the buttressing residue Leu9, DNA binding and bending experiments were conducted (Figs 4C and 5C). The L9A mutant binds linear DNA with an affinity comparable to that of M13A (Fig. 4C and Table 2). In contrast, L9A binds to crosslinked DNA only slightly less well than the wild type, which suggest that Leu9 does not have an important role in binding to pre-bent DNA, but is required for binding to linear DNA (Fig. 5C and Table 2). We also found that the L9A mutant was among the most deficient mutant proteins in the DNA bending assay (Fig. 6A and C). An explanation for these results is that intercalation by Met13 is important for binding to linear DNA and pre-bent DNA, but the potentially stabilizing effect of Leu9 on Met13 is only required for linear DNA binding, which has a higher energetic cost of DNA bending than partially pre-bent DNA. It has not been possible in the past to separate the effects of intercalation by Met13 from the stabilizing effects of Leu9, except by this subtle difference in binding to linear versus pre-bent DNA. A most dramatic example of the importance of the interplay between these two residues is seen in the SRY sex-reversal mutant M64I (17,21,57). In the M64I mutant the Met of the Met–Ile pair is mutated to isoleucine, which results in a shift in the position and deeper intercalation of the isoleucine with an associated decrease in DNA bending of ∼13° (22).
The role of the intercalation wedge in DNA bending and binding of HMG-D is similar to its function in other HMG domain proteins (15,16,20,35,49). This systematic study of residues at position 13 confirms the notion that hydrophilic residues are not favorable intercalators. For the HMG domains that are known to bind and bend DNA, non-polar residues are absolutely conserved at this site, as are the buttressing residues, equivalent to Leu9 and Tyr12. Furthermore, we show that wild-type Met13 intercalation and DNA bending depends on interactions with the Leu9 position. In contrast, several of the UBF protein HMG domains contain hydrophilic and charged residues at the ‘intercalating wedge’ positions, which based on our analysis argues against the normal DNA binding function for these particular domains (Fig. 1A) (1,58). There is increasing evidence that the HMG-1/2 HMG domain has protein–protein interaction surfaces in addition to its DNA binding surface (24,25,29,30,59). Therefore, it would not be surprising to find HMG domains that function primarily as protein interaction modules, as may exist in the multi-HMG domain proteins, such as UBF (60), which interact with DNA itself, and numerous other proteins (reviewed in 61).
A major contribution of secondary intercalating residues to DNA bending and binding
In addition to intercalation by Met13, the structure of HMG-D-bound to DNA shows partial DNA intercalation by a pair of residues, Val32 and Thr33, and the possibility of an additional DNA intercalation by Ala36 at the adjacent base step (19). To investigate the importance of these residues, mutations to glycine were made at all three positions, and two additional mutations, V32A and V32T, were made to examine the importance of Val32 intercalation in more detail. CD spectra and melting analyses (Fig. 2B and Table 1) on each of these mutants confirmed the stability of the HMG domain. The spectra were virtually indistinguishable from the wild-type protein, and the melting temperatures fell into three distinct ranges. The V32A mutant was significantly more stable than the wild type with a melting temperature of 44.7°C, the A36G mutant was less stable than the wild type with a melting temperature of 38.3°C, and V32T, V32G and V33G mutants had comparable stability to the wild-type protein. Residues Val32 and Thr33 are found near the N-terminus of helix 2, and Ala36 is in the middle of helix 2. The difference in melting temperature follows the expected trend based on the alpha helix-forming propensity of the amino acid substitutions for A36G and V32A, but not for the others (50,51). This observation is consistent with the position of Ala36 in the middle of the helix already having a good helix former, and residues Val32 and Thr33 that are already rather poor helix formers residing at the end of the helix. There are no other residues within the van der Waals interaction distance of Val32, which suggests that the stability of helix 2 is important in the stability of the overall protein fold. HMG-D has a destabilizing residue at this position, which suggests that there is a balance between its particular role in DNA bending and protein stability.
These mutants had nearly the same trends in binding affinity for the pre-bent and linear DNA fragments, and had overall smaller effects on DNA affinity compared to the substitutions of the primary intercalating site in the protein, Met13 (Table 2). Therefore, the intercalating positions 32, 33 and 36, at the edge of the protein–DNA interface, are not as sensitive to mutation as Met13, which is in the center of the binding site (Figs 4C, 5C and Table 2). The T33G mutation resulted in nearly 2-fold weaker binding to both linear and pre-bent DNA. In contrast, V32G binds to both DNA fragments similarly to the wild type. V32A binds well to pre-bent DNA, but it binds with almost 2-fold lower affinity to linear DNA than the wild-type protein. Interestingly, the V32T protein has significantly increased affinity for the pre-bent DNA, which suggests that it may form additional stabilizing contacts with the DNA perhaps through hydrogen-bonding interactions. The difference between residues 32 and 33 in DNA binding suggests a more important role for Thr33 than had been appreciated from structural studies and comparison to the HMGB1-boxA protein, for which intercalation by residue 32 dominates DNA binding in this region of the protein.
The DNA bending analysis in Figure 6C clearly shows again that there are two modes of DNA bending as seen in the distribution of circle sizes, the wild-type circle sizes, and a group of mutants that form relatively fewer 66 bp circles. The residues that cluster in the wild-type DNA bending group are the mutants V32A, V32T and A36G, and the bending deficient proteins are the glycine mutants of Val32 and Thr33. The results of this bending study for Val32 are consistent with DNA bending studies conducted with other HMG domain proteins (15,20,35,62). However, our results and conclusions regarding the HMG-D V32A mutant differ slightly from those obtained by Payet et al. (16), who found in a similar, but not identical assay system, that V32A was not the same as the wild type in bending; this difference could be due to the different DNA sequences used in the assay, which resulted in larger circles sizes of 75 bp in contrast to the 55 bp circles that we observed here. Because we found that T33G and V32G failed to produce the wild-type pattern of DNA circles in the bending assay, intercalation or van der Waals DNA interactions must be required for wild-type DNA bending for both of these residues. It is interesting to note that although DNA bending was compromised, DNA-binding affinity was only minimally (<2-fold) affected in these two mutants, which indicates that the additional energetic cost of bending the DNA may only just be compensated by favorable DNA interactions. Finally, Ala36 appeared to intercalate, just barely, in the HMG-D-74–DNA complex crystal structure; mutation to glycine had no effect on DNA bending in the bending propensity assay. The slight effects on linear and pre-bent DNA binding, indicate instead that the beta carbon of Ala36 is most likely involved in DNA interactions by increasing van der Waal’s contacts between the protein and the DNA.
The involvement of this secondary intercalation wedge in DNA binding and bending by HMG-D differs from the behavior of the comparable regions in the well studied NHP6A and HMGB1-box A proteins, which both have a phenylalanine at position 32 followed by a glycine, alanine or serine. The specificity of HMG domain proteins for cisplatin-modified DNA and four-way junction DNA appears to be related to the size of the residue at position 32, because HMGB1 boxA and NHP6A also have a greater preference for cisplatinated DNA than HMGB1 boxB and HMG-D, which have smaller residues at position 32 (15,35,48,49,63). The importance of both Val32 and Thr33 in DNA bending reveals one mechanism by which the HMG-D domain achieves such high bending propensity when compared to other HMG proteins. The HMG-D HMG domain appears to bend DNA better than other HMG domains, as judged by its unique ability to circularize 55 bp DNA, and from solution and crystallographic studies (19,39). We suggest that Thr33 intercalation is responsible for this additional bending, and note that HMG-D is the only well studied HMG protein with an intercalating residue at both positions 32 and 33; other HMG proteins with a small intercalating residue at position 32 usually have a non-intercalating glycine or alanine at position 33.
Relationship of intercalating residues to DNA specificity
Amino acid sequence differences between the sequence-specific HMG domain proteins, such as LEF-1 and SRY and the non-sequence-specific HMG domain proteins, such as HMG-D and the HMGB proteins occur both in structural regions of the proteins and in the residues directly involved in creating the protein–DNA interface (18,22,64). The residues that are consistently different between these two groups at the DNA interface, 10, 32, 33 and 36, we previously called specificity determinants (19,40) based on their observed interactions in the crystal structure of the HMG-D–DNA complex and NMR structures of the LEF-1–DNA, SRY–DNA and NHP6A–DNA complexes (Fig. 1B) (18–20,22). Two of these residues, 32 and 33, intercalate the DNA in the non-sequence-specific proteins (Fig. 1B), but are hydrophilic residues capable of forming direct hydrogen bonds with the DNA, as has been seen for both SRY and LEF-1 with DNA (1,18,22). We have shown that the side chain of Ala36 is relevant in DNA binding, probably for purposes of surface complementarity, but not necessarily for DNA bending. In the sequence-specific proteins this residue is a hydrogen-bonding residue such as asparagine or serine. Finally, residue 10 is nearly always serine in the non-sequence-specific proteins and asparagine in the sequence-specific HMG proteins. Therefore, these four substitutions of LEF-1 were introduced into HMG-D, S10N/V32S/T33A/A36N, generating a hybrid HMG protein that is named ‘Spec’.
The Spec protein folds with equivalent secondary structure to the wild-type HMG-D as shown by CD (Fig. 3B), and it has comparable stability, with a melting temperature of 40.8°C, compared to the native HMG-D that has a melting temperature of 41.0°C (Table 2). In order to determine whether Spec has any sequence specificity, a binding site selection study was conducted. Nine rounds of selection using PCR to amplify products that were gel-purified from EMSA (65) with the Spec protein revealing increased DNA affinity for the pool of PCR products by qualitative inspection of the amount of shifted DNA at lower concentrations of protein (data not shown). Somewhat surprisingly, a particular DNA binding site was not selected as seen by the lack of a sequence pattern in the sequences of the subcloned selected inserts. There were no significant trinucleotide sequences selected, but a similar dinucleotide distribution with a slight trend of more GC-rich sequences was observed when compared to that seen previously with native HMG-D (Table 3) (9).
Table 3. Binding site selection of the HMG-D mutant Spec.
| Dinucleotide | Frequencya | Dinucleotide complement | Frequency | Frequency of pair |
|---|---|---|---|---|
| GC | 10.6 | CG | 6.9 | 17.5 |
| GG | 10.8 | CC | 5.9 | 16.7 |
| CA | 8.6 | TG | 6.1 | 14.7 |
| AC | 7.6 | GT | 6.9 | 14.5 |
| AG | 7.1 | CT | 4.9 | 12 |
| TA | 5.9 | AT | 4.4 | 10.3 |
| GA | 5.9 | TC | 3.4 | 9.3 |
| TT | 3.2 | AA | 1.5 | 4.7 |
aThese values represent the average over all nucleotide pairs within the randomized region of the oligonucleotide, where the average dinucleotide frequency should be 6.25.
DNA binding experiments were conducted to determine whether Spec had any specificity for the LEF-1 binding site, since the engineered ‘specificity determinants’ were from LEF-1. Competitive EMSA experiments (data not shown) and affinity measurements of the Spec protein with the linear DNA oligonucleotide (non-specific site) compared to a similar length oligonucleotide LEF-1 binding site show very little, if any, preferential binding to the LEF-1 binding site (Fig. 7). Interestingly, the Spec protein binds with ∼10-fold greater affinity (0.69 nM) than the wild-type HMG-D (6.08 nM) to the non-specific DNA, and ∼5-fold better (0.35 nM) than the wild-type HMG-D (1.79 nM) to the specific LEF-binding site DNA. The wild-type HMG-D also has a higher affinity for the LEF-1 binding site of ∼3-fold. Despite higher affinity for DNA of the Spec protein, it bends DNA less well than the wild-type HMG-D protein in the DNA circularization assay (Fig. 6A and C).
Figure 7.
Binding of HMG-D-100 and ‘Spec’ protein to the LEF-1 binding site and linear non-specific DNA. (A) Image of a typical EMSA gel used to obtain data for binding curve analysis. Lane 1 contains no protein; lanes 2–16 contain ‘Spec’ protein at the following final concentrations: 0.01, 0.02, 0.04, 0.08, 0.1, 0.2, 0.4, 0.8, 1.0, 4.0, 8.0, 10.0, 40.0, 100.0 and 250.0 nM, respectively. (B) Binding curve overlays for the four binding experiments with error bars superimposed on the Spec–linear DNA binding curve.
These mutagenesis and selection experiments were conducted with the expectation of having produced a protein with greater DNA binding specificity for the LEF-1 target site DNA than native HMG-D. The observation of higher affinity without a greater degree of specificity strongly suggest that specificity in DNA recognition of this family of proteins is more complicated than mere direct readout. It appears that the Spec protein may have increased binding affinity because of new hydrogen bonding potential at the DNA interface. Clearly this is not sufficient for true specificity, and the results of early specificity experiments of Crane-Robinson and coworkers are illuminating in this regard (64). A chimeric HMGB1–LEF-1 protein with the N-terminal 11 residues and last 25 C-terminal residues of the LEF-1 HMG domain maintained much of the specificity of the LEF-1 parent protein. The swapped region contained residue 10 that was examined in this study, but also contained the structural region of the HMG domain known as the ‘minor wing’. The wing encompasses the secondary hydrophobic core of the HMG domain as described in early NMR studies (54), and has lower stability in the sequence-specific proteins than the non-sequence-specific proteins (66,67). This region appears to fold upon DNA binding, which may provide a large contribution to DNA specificity through the mechanism of indirect readout (67,68).
CONCLUSIONS
This study shows that DNA binding and bending depend on individual intercalating residues as well as global features of the HMG domain, such as shape complementarity and protein stability. Although, several of the HMG domain intercalating residues are implicated as specificity determinants, they are not, by themselves, sufficient to provide specificity in the context of the HMG-D protein fold. Therefore, specificity of the HMG domain is most likely to be due to additional global factors such as the ability of the sequence-specific HMG domain to achieve better shape complementarity with the DNA surface (64,67).
DNA intercalation is not absolutely required for DNA bending or binding by the HMG domain, but it is important for wild-type DNA binding and bending. There is a clear and direct correlation between DNA binding affinity and bending propensity for the primary intercalating residue Met13, suggesting that in HMG-D, intercalation is important for high affinity DNA binding. A buttressing residue, Leu9, is not required for binding to pre-bent DNA, which is consistent with it having a stabilizing effect on Met13 as part of the intercalation wedge. For the secondary intercalating wedge the correlation between DNA binding and bending is not as clear. The contribution of the non-polar intercalation wedges to protein stability as well as DNA binding and bending is a potentially general role for these residues. The equivalent residues of divergent HMG domains, such as those in UBF, have a polar ‘intercalation wedge’ that could stabilize the HMG domain through electrostatic interactions, and may provide a surface for protein interaction rather than DNA binding.
HMG domain proteins mediate the formation of higher order enhancesomes together with a variety of transcription factors (31,32). To accommodate such different DNA sequences and contexts requires flexibility in DNA recognition. The bimodal distribution of bending propensity among the mutants suggests that there may be at least two modes of DNA binding by HMG domain proteins, as has been seen in the structures of HMG-1A and HMG-D bound to DNA (19,40,69). A particular mode may be preferred by different HMG domains, which have different degrees of DNA bending potential depending on DNA sequence and protein interaction context. The HMG domain is a deceptively simple structure and we are only beginning to understand the complexity of its DNA binding. What remains is to understand interdomain relationships in DNA binding by multi-HMG domain proteins as well as to uncover how HMG domains facilitate transcription factor function.
Acknowledgments
ACKNOWLEDGEMENTS
We appreciate the helpful suggestions and contributions of members of the Churchill Laboratory, including Xun Shi and Tony Armstrong for technical assistance, and Dr Linda Dow for protein biochemistry. We thank Drs Greg Verdine and Scot Wolfe for the gift of the disulfide crosslinked DNA, Dr Robert Hodges for helpful discussions, and we acknowledge the University of Colorado Health Sciences Center Biophysics Core Facility for use of the CD instrument. We are grateful for support from the NIH (Shannon Award and RO1-GM59456), American Cancer Society, the American Heart Association for a Grant-In-Aid to M.E.A.C., the NIH Molecular Biophysics pre-doctoral training grant and a University of Illinois Graduate College Travel Grant to F.V.M.
REFERENCES
- 1.Baxevanis A.D. and Landsman,D. (1995) The HMG-1 box protein family: classification and functional relationships. Nucleic Acids Res., 23, 1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustin M. and Reeves,R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol., 54, 35–100. [DOI] [PubMed] [Google Scholar]
- 3.Grosschedl R., Giese,K. and Pagel,J. (1994) HMG domain proteins—architectural elements in the assembly of nucleoprotein structures. Trends Genet., 10, 94–100. [DOI] [PubMed] [Google Scholar]
- 4.Sinclair A.H., Berta,P., Palmer,M.S., Hawkins,J.R., Griffiths,B.L., Smith,M.J., Foster,J.W., Frischauf,A.M., Lovell-Badge,R. and Goodfellow,P.N. (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature, 346, 240–244. [DOI] [PubMed] [Google Scholar]
- 5.Goodfellow P.N. and Lovell-Badge,R. (1993) SRY and sex determination in mammals. Annu. Rev. Genet., 27, 71–92. [DOI] [PubMed] [Google Scholar]
- 6.Haqq C.M., King,C.Y., Ukiyama,E., Falsafi,S., Haqq,T.N., Donahoe,P.K. and Weiss,M.A. (1994) Molecular basis of mammalian sexual determination: Activation of Müllerian inhibiting substance gene expression by SRY. Science, 266, 1494–1500. [DOI] [PubMed] [Google Scholar]
- 7.Giese K., Amsterdam,A. and Grosschedl,R. (1991) DNA-binding properties of the HMG-domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev., 5, 2567–2578. [DOI] [PubMed] [Google Scholar]
- 8.Bianchi M.E., Falciola,L., Ferrari,S. and Lilley,D.M.J. (1992) The DNA binding site of HMG1 protein is composed of 2 similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J., 11, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill M.E.A., Jones,D.N.M., Glaser,T., Hefner,H., Searles,M.A. and Travers,A.A. (1995) HMG-D is an architecture-specific protein that preferentially binds to DNA containing the dinucleotide TG. EMBO J., 14, 1264–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paull T.T., Haykinson,M.J. and Johnson,R.C. (1993) The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev., 7, 1521–1534. [DOI] [PubMed] [Google Scholar]
- 11.Wolfe S.A., Ferentz,A.E., Grantcharova,V., Churchill,M.E.A. and Verdine,G.L. (1995) Modifying the helical structure of DNA by design: recruitment of an architecture-specific protein to an enforced DNA bend. Chem. Biol., 2, 213–221. [DOI] [PubMed] [Google Scholar]
- 12.Thomas J.O. and Travers,A.A. (2001) HMG1 and 2 and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci., 26, 167–174. [DOI] [PubMed] [Google Scholar]
- 13.Webb M. and Thomas,J.O. (1999) Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J. Mol. Biol., 294, 373–387. [DOI] [PubMed] [Google Scholar]
- 14.Webb M., Payet,D., Lee,K.B., Travers,A.A. and Thomas,J.O. (2001) Structural requirements for cooperative binding of HMG1 to DNA minicircles. J. Mol. Biol., 309, 79–88. [DOI] [PubMed] [Google Scholar]
- 15.He Q., Ohndorf,U.M. and Lippard,S.J. (2000) Intercalating residues determine the mode of HMG1 domains A and B binding to cisplatin-modified DNA. Biochemistry, 39, 14426–14435. [DOI] [PubMed] [Google Scholar]
- 16.Payet D., Hillisch,A., Lowe,N., Diekmann,S. and Travers,A. (1999) The recognition of distorted DNA structures by HMG-D: a footprinting and molecular modeling study. J. Mol. Biol., 294, 79–91. [DOI] [PubMed] [Google Scholar]
- 17.Werner M.H., Huth,J.R., Gronenborn,A.M. and Clore,G.M. (1995) Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell, 81, 705–714. [DOI] [PubMed] [Google Scholar]
- 18.Love J.J., Li,X., Case,D.A., Giese,K., Grosschedl,R. and Wright,P.E. (1995) Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature, 376, 791–795. [DOI] [PubMed] [Google Scholar]
- 19.Murphy F.V. IV, Sweet,R.M. and Churchill,M.E.A. (1999) The structure of a chromosomal high mobility group protein–DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J., 18, 6610–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masse J.E., Wong,B., Yen,Y.M., Allain,F.H., Johnson,R.C. and Feigon,J. (2002) The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J. Mol. Biol., 323, 263–284. [DOI] [PubMed] [Google Scholar]
- 21.Pontiggia A., Rimini,R., Harley,V.R., Goodfellow,P.N., Lovell-Badge,R. and Bianchi,M.E. (1994) Sex-reversing mutations affect the architecture-specificity of SRY–DNA complexes. EMBO J., 13, 6115–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy E.C., Zhurkin,V.B., Louis,J.M., Cornilescu,G. and Clore,G.M. (2001) Structural basis for SRY-dependent 46-X,Y sex reversal: modulation of DNA bending by a naturally occurring point mutation. J. Mol. Biol., 312, 481–499. [DOI] [PubMed] [Google Scholar]
- 23.Nightingale K., Dimitrov,S., Reeves,R. and Wolffe,A. (1996) Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J., 15, 548–561. [PMC free article] [PubMed] [Google Scholar]
- 24.van Gent D.C., Hiom,K., Paull,T.T. and Gellert,M. (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J., 16, 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boonyaratanakornkit V., Melvin,V., Prendergast,P., Altmann,M., Ronfani,L., Bianchi,M.E., Taraseviciene,L., Nordeen,S.K., Allegretto,E.A. and Edwards,D.P. (1998) High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol., 18, 4471–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge H. and Roeder,R.G. (1994) The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem., 268, 17136–17140. [PubMed] [Google Scholar]
- 27.Melvin V.S. and Edwards,D.P. (1999) Coregulatory proteins in steroid hormone receptor action: the role of chromatin high mobility group proteins HMG-1 and -2. Steroids, 64, 576–586. [DOI] [PubMed] [Google Scholar]
- 28.Zappavigna V., Falciola,L., Citterich,M., Mavilio,F. and Bianchi,M. (1996) HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J., 15, 4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 29.Zwilling S., Konig,H. and Wirth,T. (1995) High-mobility-group protein-2 functionally interacts with the POU domains of Octamer transcription factors. EMBO J., 14, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayaraman L., Moorthy,N.C., Murthy,K.G.K., Manley,J.L., Bustin,M. and Prives,C. (1998) High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev., 12, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellwood K.B., Yen,Y.M., Johnson,R.C. and Carey,M. (2000) Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol., 20, 4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsouras K., Wong,B., Arayata,C., Johnson,R.C. and Carey,M. (2002) The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol., 22, 4390–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stros M. (1998) DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem., 273, 10355–10361. [PubMed] [Google Scholar]
- 34.Jamieson E.R., Jacobson,M.P., Barnes,C.M., Chow,C.S. and Lippard,S.J. (1999) Structural and kinetic studies of a cisplatin-modified DNA icosamer binding to HMG1 domain B. J. Biol. Chem., 274, 12346–12354. [DOI] [PubMed] [Google Scholar]
- 35.Yen Y.M., Wong,B. and Johnson,R.C. (1998) Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities: role of the unique N-terminus and putative intercalating methionine. J. Biol. Chem., 273, 4424–4435. [DOI] [PubMed] [Google Scholar]
- 36.Tang L., Li,J., Katz,D.S. and Feng,J.A. (2000) Determining the DNA bending angle induced by non-specific high mobility group-1 (HMG-1) proteins: a novel method. Biochemistry, 39, 3052–3060. [DOI] [PubMed] [Google Scholar]
- 37.Payet D. and Travers,A.A. (1997) The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J. Mol. Biol., 266, 66–75. [DOI] [PubMed] [Google Scholar]
- 38.Churchill M.E.A., Changela,A., Dow,L.K. and Krieg,A.J. (1999) Interactions of high mobility group box proteins with DNA and chromatin. Methods Enzymol., 304, 99–133. [DOI] [PubMed] [Google Scholar]
- 39.Lorenz M., Hillisch,A., Payet,D., Buttinelli,M., Travers,A. and Diekmann,S. (1999) DNA bending induced by high mobility group proteins studied by fluorescence resonance energy transfer. Biochemistry, 38, 12150–12158. [DOI] [PubMed] [Google Scholar]
- 40.Murphy F.V.I. and Churchill,M.E.A. (2000) Nonsequence-specific DNA recognition: a structural perspective. Struct. Fold. Design, 8, R83–R89. [DOI] [PubMed] [Google Scholar]
- 41.Balaeff A., Churchill,M.E.A. and Schulten,K. (1998) Structure prediction of a complex between the chromosomal protein HMG-D and DNA. Proteins, 30, 113–135. [DOI] [PubMed] [Google Scholar]
- 42.Allain F.H.-T., Yen,Y.-M., Masse,J.E., Schultze,P., Dieckmann,T., Johnson,R.C. and Feigon,J. (1999) Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J., 18, 2563–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferentz A.E., Keating,T.A. and Verdine,G.L. (1993) Synthesis and characterization of disulfide cross-linked oligonucleotides. J. Am. Chem. Soc., 115, 9006–9014. [Google Scholar]
- 44.Dow L., Changela,A., Hefner,H. and Churchill,M. (1997) Oxidation of a critical methionine modulates DNA binding of the Drosophila melanogaster high mobility group protein, HMG-D. FEBS Lett., 414, 514–520. [DOI] [PubMed] [Google Scholar]
- 45.John D.M. and Weeks,K.M. (2000) van’t Hoff enthalpies without baselines. Protein Sci., 9, 1416–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rigoutsos I. and Floratos,A. (1998) Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics, 14, 55–67. [DOI] [PubMed] [Google Scholar]
- 47.Dow L.K., Jones,D.N., Wolfe,S.A., Verdine,G.L. and Churchill,M.E. (2000) Structural studies of the high mobility group globular domain and basic tail of HMG-D bound to disulfide cross-linked DNA. Biochemistry, 39, 9725–9736. [DOI] [PubMed] [Google Scholar]
- 48.Stros M. and Muselikova,E. (2000) A role of basic residues and the putative intercalating phenylalanine of the HMG-1 box B in DNA supercoiling and binding to four-way DNA junctions. J. Biol. Chem., 275, 35699–35707. [DOI] [PubMed] [Google Scholar]
- 49.Taudte S., Xin,H. and Kallenbach,N.R. (2000) Alanine mutagenesis of high-mobility-group-protein-1 box B (HMG1-B). Biochem. J., 347, 807–814. [PMC free article] [PubMed] [Google Scholar]
- 50.O’Neil K.T. and DeGrado,W.F. (1990) A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science, 250, 646–651. [DOI] [PubMed] [Google Scholar]
- 51.Padmanabhan S., Marqusee,S., Ridgeway,T., Laue,T.M. and Baldwin,R.L. (1990) Relative helix-forming tendencies of nonpolar amino acids. Nature, 344, 268–270. [DOI] [PubMed] [Google Scholar]
- 52.Hardman C.H., Broadhurst,R.W., Raine,A.R.C., Grasser,K.D., Thomas,J.O. and Laue,E.D. (1995) Structure of the A-domain of HMG1 and its interactions with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry, 34, 16596–16607. [DOI] [PubMed] [Google Scholar]
- 53.Weir H.M., Kraulis,P.J., Hill,C.S., Raine,A., Laue,E.D. and Thomas,J.O. (1993) Structure of the HMG box motif in the B-domain of HMG1. EMBO J., 12, 1311–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones D.N.M., Searles,A., Shaw,G.L., Churchill,M.E.A., Ner,S.S., Keeler,J., Travers,A.A. and Neuhaus,D. (1994) The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster. Structure, 2, 609–627. [DOI] [PubMed] [Google Scholar]
- 55.Weiss M.A., Ukiyama,E. and King,C.Y. (1997) The SRY cantilever motif discriminates between sequence- and structure-specific DNA recognition: alanine mutagenesis of an HMG box. J. Biomol. Struct. Dyn., 15, 177–184. [DOI] [PubMed] [Google Scholar]
- 56.Stros M. (2001) Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry, 40, 4769–4779. [DOI] [PubMed] [Google Scholar]
- 57.King C.-Y. and Weiss,M.A. (1993) The SRY high-mobility-group box recognizes DNA by partial intercalation in the minor groove: a topological mechanism of sequence specificity. Proc. Natl Acad. Sci. USA, 90, 11990–11994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landsman D. and Bustin,M. (1993) A signature for the HMG-1 box DNA-binding proteins. Bioessays, 15, 539–546. [DOI] [PubMed] [Google Scholar]
- 59.Sutrias-Grau M., Bianchi,M.E. and Bernues,J. (1999) High mobility group protein 1 interacts specifically with the core domain of human TATA box-binding protein and interferes with transcription factor IIB within the pre-initiation complex. J. Biol. Chem., 274, 1628–1634. [DOI] [PubMed] [Google Scholar]
- 60.Jantzen H.M., Admon,A., Bell,S.P. and Tjian,R. (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature, 244, 830–836. [DOI] [PubMed] [Google Scholar]
- 61.Moss T. and Stefanofsky,V.Y. (2002) At the center of eukaryotic life. Cell, 109, 545–548. [DOI] [PubMed] [Google Scholar]
- 62.Teo S.H., Grasser,K.D. and Thomas,J.O. (1995) Differences in the DNA-binding properties of the HMG-box domains of HMG1 and the sex determining factor SRY. Eur. J. Biochem., 230, 943–950. [DOI] [PubMed] [Google Scholar]
- 63.Wong B., Masse,J.E., Yen,Y.M., Giannikoupolous,P., Feigon,J. and Johnson,R.C. (2002) Binding to cisplatin-modified DNA by the Saccharomyces cerevisiae HMGB protein NHP6A. Biochemistry, 41, 5404–5414. [DOI] [PubMed] [Google Scholar]
- 64.Read C.M., Cary,P.D., Preston,N.S., Lnenicek-Allen,M. and Crane-Robinson,C. (1994) The DNA sequence-specificity of HMG boxes lies in the minor wing of the structure. EMBO J., 13, 5639–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bianchi A., Stansel,R.M., Fairall,L., Griffith,J.D., Rhodes,D. and de Lange,T. (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J., 18, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Houte L.P.A., Chuprina,V.P., van der Wetering,M., Boelens,R., Kaptein,R. and Clevers,H. (1995) Solution structure of the sequence-specific HMG-box of the lymphocyte transcriptional activator Sox-4. J. Biol. Chem., 270, 30516–30524. [DOI] [PubMed] [Google Scholar]
- 67.Weiss M.A. (2001) Floppy SOX: mutual induced fit in HMG (high-mobility group) box-DNA recognition. Mol. Endocrinol., 15, 353–362. [DOI] [PubMed] [Google Scholar]
- 68.Travers A.A. (1989) DNA conformation and protein-binding. Annu. Rev. Biochem., 58, 427–452. [DOI] [PubMed] [Google Scholar]
- 69.Ohndorf U.-M., Rould,M.A., He,Q., Pabo,C.O. and Lippard,S.J. (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature, 399, 708–712. [DOI] [PubMed] [Google Scholar]
- 70.Cary P.D., Read,C.M., Davis,B., Driscoll,P.C. and Crane-Robinson,C. (2001) Solution structure and backbone dynamics of the DNA-binding domain of mouse Sox-5. Protein Sci., 10, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Y., Yang,W., Wu,J. and Shi,Y. (2002) Solution structure of the first HMG box domain in human upstream binding factor. Biochemistry, 41, 5415–5420. [DOI] [PubMed] [Google Scholar]
- 72.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 73.Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]