Abstract
Regulation of gene expression involves sequence elements in nucleic acids. In promoters, multiple sequence elements cooperate as functional modules, which in combination determine overall promoter activity. We previously developed computational tools based on this hierarchical structure for in silico promoter analysis. Here we address the functional organization of post-transcriptional control elements, using the HIV-1 genome as a model. Numerous mutagenesis studies demonstrate that expression of HIV structural proteins is restricted by inhibitory sequences (INS) in HIV mRNAs in the absence of the HIV-1 Rev protein. However, previous attempts to detect conserved sequence patterns of HIV-1 INS have failed. We defined four distinct sequence patterns for inhibitory motifs (weight matrices), which identified 22 out of the 25 known INS as well as several new candidate INS regions contained in numerous HIV-1 strains. The conservation of INS motifs within the HIV genome was not due to overall sequence conservation. The functionality of two candidate INS regions was analyzed with a new assay that measures the effect of non-coding mRNA sequences on production of red fluorescent reporter protein. Both new INS regions showed inhibitory activity in sense but not in antisense orientation. Inhibitory activity increased by combining both INS regions in the same mRNA. Inhibitory activity of known and new INS regions was overcome by co-expression of the HIV-1 Rev protein.
INTRODUCTION
Eukaryotic gene expression is a complex mechanism that can be regulated on the transcriptional, post-transcriptional, translational and post-translational levels. It is well known from the analysis of promoters and enhancers that several distinct elements [e.g. distinct transcription factor (TF)-binding sites] cooperate to achieve a common goal or function (e.g. regulation of transcription initiation) by specific interaction with proteins (1–3). Similarly, cooperative interactions of elements in nucleic acids are also involved in restricting expression of cellular genes on the post-transcriptional level. Such inhibitory sequences (INS) are active within mRNAs and thus show a preference for the coding strand of the DNA.
Well known examples for such INS are found in cellular mRNAs like c-fos, c-myc and granulocyte macrophage colony-stimulating factor (GM-CSF) in which multiple iterations of the AUUUA pentamer sequence, mainly within the 3′-untranslated region (3′-UTR), are responsible for the observed inhibitory effects (4–6). Binding of cellular RNA-binding proteins, like AU-A, HuR and HuA, counteracts the inhibitory effect of these elements (7–10).
INS are also contained in the mRNAs of various viruses, including hepatitis B virus (HBV), human papilloma virus type 1 (HPV1), bovine papilloma virus (Bovine HP1), and retroviruses such as human immunodeficiency virus type 1 (HIV-1), simian retrovirus (SRV1) and Mason-Pfizer monkey virus (MPMV). These viruses use various rescue mechanisms to overcome the inherent inhibitory effects on their transcripts, involving cellular proteins and in some cases also viral factors, as described below (11–16). We dissected HIV-1 INS into several components, INS region, INS element and INS motif, definitions of which are given in Materials and Methods.
The importance of INS activity in HIV replication is well established (17), although mechanistic details of cellular and viral INS functions remain obscure. INS restrict the expression of HIV structural proteins, which are encoded by unspliced and singly spliced mRNAs. The inhibitory activity of INS is overcome by the viral regulatory factor Rev, which is encoded by multiply spliced mRNAs species. Rev binds to an RNA element in the env gene called the Rev response element (RRE) and mediates nuclear export and efficient expression of its target RNA. Inhibition of gene expression by INS has been proposed to involve increased splicing efficiency, prevention of nuclear export of unspliced transcripts and degradation of INS-containing mRNAs or a combination thereof (18–21). Proteins shown to bind HIV-1 INS include poly A-binding protein (22) and hnRNPA1 (23), but the roles of these proteins in overall inhibition of gene expression are not clear.
In an effort to characterize the sequence hallmarks of HIV-1 INS, the effects of mutagenesis of various sections of the HIV genome on gene expression were studied. INS-containing regions were identified by loss of repressive activity and loss of Rev dependence after mutation and were defined in the HIV gag, pol and env genes (24–27) (gag, INS1 and INS2; pol, INS3 and CRS; env, RRE; see Fig. 1). A total of 25 INS elements (each 20–40 nt long) were determined as the smallest functional units of the INS regions. The inhibitory effect of HIV-1 INS elements could be transferred to heterologous mRNAs (27–30). Each of these short elements was able to inhibit viral gene expression individually. However, the maximal inhibitory effect of the INS regions (i.e. INS1, INS2, INS3, CRS) (25,27) required the cooperative interaction of multiple INS elements, reflecting the organization in the HIV-1 genome. Presence of additional but poorly characterized INS elements within the pol and env genes was also suggested (27).
Figure 1.
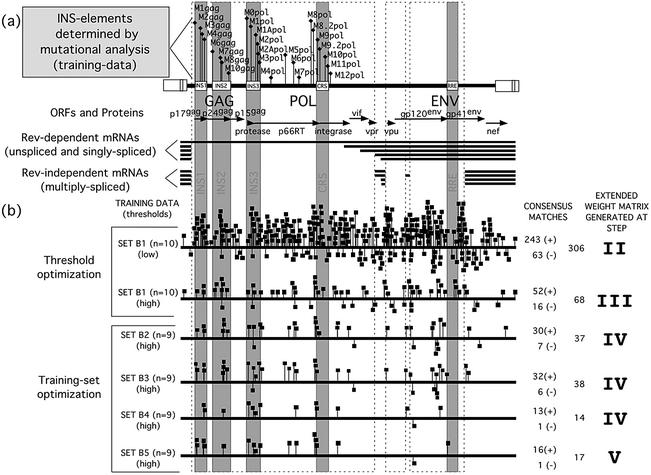
HIV-1 INS elements, INS regions, genomic organization and gene expression and weight matrix generation scheme. (a) The corresponding INS element names are depicted above the HIV genome. Reading frames [p17gag, p24gag, p15gag, protease, reverse-transcriptase (p66RT), integrase, gp120env, gp41env and several accessory proteins] and the corresponding INS regions (INS1, INS2, INS3 and CRS) containing experimentally identified and verified INS elements are indicated above the two HIV-1 transcript classes. (b) An example of the matrix generation scheme, which is described in detail in Results.
Despite the unquestionable function of these elements, all efforts to define HIV-1 INS on the sequence level have failed so far, which may in part be due to the fact that there may be more than one distinct INS sequence pattern. The development and application of a new strategy for the analysis of sequences containing several different functional sites enabled us for the first time to identify and separate four subsets of sequences, each containing one conserved INS motif shared by several INS elements. The resulting descriptions were not only able to detect the well characterized HIV-1 INS elements but also identified new candidate INS elements within the pol and env genes of HIV-1. We subsequently verified the functionality and cooperativity of two candidate INS regions by quantitative analysis of their inhibitory effect on the expression of fluorescent reporter proteins in transfection experiments. The counteraction of this inhibitory activity by Rev was also demonstrated.
MATERIALS AND METHODS
Terminology
Since, in the literature, similar terms have been used so far to describe stretches of nucleic acids that differ in size and composition, this paper uses the following terminology for INS.
Functional descriptions. INS element: the smallest functionally defined unit that shows INS functionality. An INS element may contain more than one INS motif. INS region: regions of the HIV-1 genome with inhibitory activity, that contain more than a single INS element.
Bioinformatics description. INS motif: short sequence consensus defined by a weight matrix in this study that is associated with INS elements and INS regions. An INS motif is analogous to a TF-binding site with regard to length and sequence conservation.
Sequence training sets
The positive training set for the generation of the matrices consisted of sequences from the HIV-1 full-length proviral sequence of HXB2R. Only those regions of the HXB2R ‘wild-type’ sequence were used which corresponded to the 25 areas analyzed by mutagenesis (indicated by gray shading in Fig. 1) (27).
Default parameters were used for the initial analysis in all programs. We used the complete sequence of the HIV-1 full-length proviral sequence of HXB2R (accession number K03455) as the primary test sequence for the pre-evaluation and optimization of the matrices. The detailed secondary analysis and evaluation was performed with the larger positive test set consisting of 26 HIV-1 full-length proviral sequences (Hiv3202a12-U34603, Hiv3202a21-U34604, Hivant70-L20587, Hivbcsg3c-L02317, Hivcam1-D10112, Hivd31-U43096, Hiveli-K03454, Hivhan-U43141, Hivhxb2r-K03455, Hivibng-L39106, Hivjrcsf-M38429, Hivjrfl-M74978-M75007-U63632, Hivlai-K02013, Hivlw123-U12055, Hivmal-K03456, Hivmanc-U23487, Hivmn-M17449, Hivmvp5180-L20571, Hivndk-M27323, Hivny5cg-M3843, Hivoyi-M26727, Hivrf-M17451, Hivth475a-L31963, Hivu455-M62320, Hivweau160-U21135, Hivz2z6-M22639) from GenBank.
As a negative control we used the sequence of the Rev-independent virus carrying mutations within the INS1, INS2, INS3, CRS and some additional mutations within the pol gene sequence between INS2 and INS3 and between INS3 and CRS (Fig. 1) (27).
The AUUUA matrix was generated from the previously described and experimentally verified AUUUA inhibitory elements from the chicken c-fos proto-oncogene (M37000), the Xenopus laevis c-myc (M14455), the human (V01512) and the rat c-fos (X06769) cellular oncogenes, and from the GM-CSF genes from human (M10663) and mouse (X03019) (6,31–34).
The program CoreSearch was used with default parameters, starting with 7-tuples [for a detailed explanation of the parameters, see Wolferstetter et al. (35)].
The elements were taken from the 3′-UTR sequences of the chicken c-fos proto-oncogene (three elements), the Xenopus laevis c-myc mRNA (five elements), the human cellular oncogene c-fos (one element) and the rat c-fos oncogene (one element), and from the GM-CSF mRNAs of human (seven elements) and mouse (10 elements).
This matrix was created from sequences of 25 nt in length, with the AUUUA penta-nucleotide in the center and 10 nt of flanking sequences on either side.
Tools for computational analysis
Sequence alignments were performed with the tools of the GCG-sequence analysis software package of the Genetics Computer Group, Inc. (GCG; Madison, WI) (36). From our own developments we used the program CoreSearch and ConsInd (37). CoreSearch delivers a description of a conserved sequence element in the form of a weight matrix from a set of seven or more sequences. The matrix can then be used for the analysis of large data sets (i.e. GenBank) with the ConsInspector program (37–40). ConsInd utilizes a refined anchored alignment to generate similar matrices.
CoreSearch requires sequences of at least 100–120 nt in length. Therefore, we selected the sequences (INS elements) covered by the mutagenic oligonucleotide and added ∼30 nt of the surrounding wild-type sequence at each end, which resulted in an overall length ranging from 102 to 148 nt. We will refer to these areas by name, which consists of the letter M (for mutant), followed by a number [indicating the relative position within the open reading frame (ORF)] and the name of the reading frame it is located in. As shown in Figure 1, the INS elements were numbered successively from the 5′ end to the 3′ end of each ORF starting at 1 in the gag-ORF and at 0 in the pol-ORF.
The program MatInd (41) generated the matrix description derived from the HIV-1 INS elements (INS motifs), which were used by the MatInspector program (41) to identify and score matches to the matrix in multiple sequence data sets. As input data, the program uses sequences contained in GenBank as well as user-defined data sets.
The SeqEd sequence editor of the GCG software package was used to superimpose the results obtained from the INS motif analysis onto the genomic organization of HIV-1. The computational analysis was performed primarily on DEC workstations and additional analyses were performed on Apple-Macintosh computers.
Cell lines and transfections
HeLa cells are a human cervical adenocarcinoma cell line with epithelial morphology and HLtat cells are HeLa cells stably transfected with pL3tat HIV-1 tat expression plasmid (42,43). Cells were kept under standard cell culture conditions using Dulbecco’s modified Eagle medium with 10% FCS and 2 mM Glutamax I (Life Technologies, Karlsruhe, Germany). For microscopy and imaging purposes, cells were cultured in medium without phenol red as described (44). Cells were transfected with plasmid DNA using the calcium phosphate co-precipitation technique as described (45) (CellPhect kit; Pharmacia, Erlangen, Germany). Transfection mixtures typically contained ∼1 µg of DNA of the plasmid harboring the sequence region of interest (adjusted to ensure equimolar amounts of INS-containing plasmids in each experiment), 100 ng of pFRED143, 100 ng of pCsRevsg143 and sufficient carrier plasmid (Bluescript-derivative pBSPL) to obtain 17 µg of total DNA. Transfection efficiencies were determined by FACS analysis of green fluorescent protein (GFP)-expressing cells and ranged between 10 and 40% transfected cells. The same transfection protocol was used for the Rev-rescue experiments, except that the Fugene6 (F. Hoffmann-La Roche AG, Basel, Switzerland) system was used according to the manufacturers protocol. Cell fluorescence was analyzed by flow cytometry and/or epifluorescence microscopy 24 h after transfection.
Constructs for transfection assays
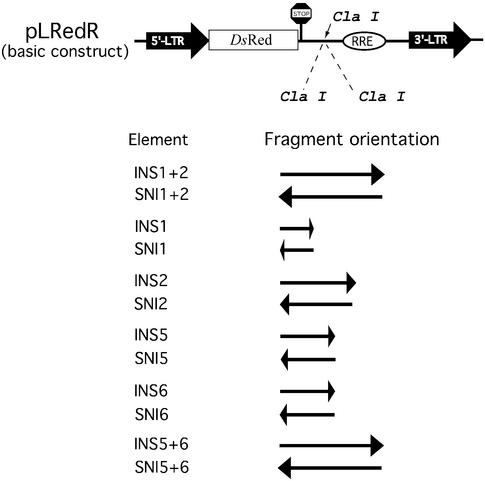
INS reporter plasmids are depicted in Figure 5. Plasmid pLRedR was constructed by replacing the gag-coding region of p37R (27) with the ORF for the red fluorescent protein (RFP) of Discosoma sp. (46), amplified from pDsRed1-N1 (BD Biosciences, Clontech, Heidelberg, Germany). All DsRed-expressing constructs contain two translational STOP codons immediately downstream of the DsRed ORF to ensure efficient termination of translation. A ClaI restriction site between the DsRed ORF and the RRE was used to insert various INS regions from the HIV-1 genome (27). Plasmid pLRed(p17/p24INS)R contains most of the p37 gag (p17gag+p24gag; including INS1+2) sequence without the ATG start codon (bases 379–1424 of HXB2 genome) taken from p37R (27), whereas pLRed(p17INS)R and pLRed(p24INS)R contain most of the DNA sequence for p17gag (bases 379–729, including INS1) or p24gag (bases 730–1424, including INS2), respectively.
Figure 5.
Reporter plasmids for functional INS analysis. The construct pLRedR contains the HIV 5′-LTR as promoter/enhancer, an ORF encoding a red fluorescent reporter protein (RFP; DsRed1), multiple additional translational STOP codons for termination of the DsRed ORF, the RRE and the 3′-LTR of HIV for termination of transcription. Different INS regions (Fig. 4) were inserted in sense and antisense orientation into the unique ClaI restriction site of the basic reporter plasmid pLRedR.
Two predicted INS region candidates (INS5 and INS6) were amplified by PCR from plasmid HXB2-(frameshift BamHI) either separately or as a combination of both (47), using specific 5′ and 3′ primers that carried a unique ClaI restriction site. Cloning of the amplified INS regions into the ClaI-linearized pLRedR vector resulted in various INS reporter plasmids carrying single or double insertions of the amplified INS sequences, either in sense or in antisense orientation (Fig. 5). Constructs were selected and verified by restriction analysis and DNA sequencing (Sequiserve: Dr Willi Metzger, Vaterstetten, Germany). The GFP expression construct pFRED143 was used for transfection control (48,49). Plasmid pCsRevsg143 (50) was used for expression of Rev-GFP and was used as transfection control in Rev-rescue experiments.
FACS analysis
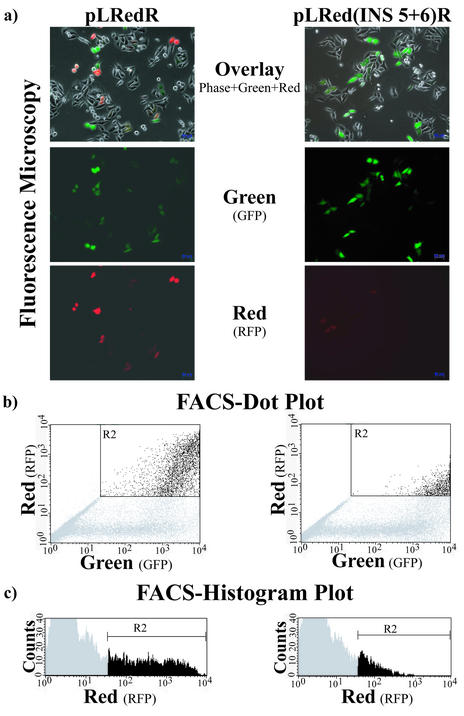
For flow cytometry, cells were trypsinized, washed, resuspended in PBS and kept on ice. Flow cytometry was performed using a BD-FACSCalibur cell sorter (BD-Systems, Heidelberg, Germany) and FACScan software CellQuest (BD Immunocytometry Systems, San Jose, CA). On average, between 20 000 and 50 000 cells were analyzed using the FL-1 (530/30) to detect green fluorescence (transfection control) and FL3 (650LP) for red fluorescence (reporter protein). The population of dual-fluorescent (red and green) cells was identified.
To assess inhibitory activities of INS regions, FACS analysis was carried out with cells cotransfected with INS reporter plasmids and pFRED143. Median intensities of green and red fluorescence were calculated from histogram-data. The ratio of green to red fluorescence was calculated as a measure for the relative reporter protein expression. Statistical analysis was carried out by the Mann–Whitney test for non-parametric independent two-group comparisons (PrismGraph, GraphPad Software, San Diego, CA). Two-tailed P-values were calculated to assess the significance of the differences between sense and antisense results of all constructs.
To assess Rev-dependent rescue of INS activity, parallel cell cultures were transfected with the INS reporter plasmids and either pCsRevsg143 for expression of Rev-GFP (Rev-positive) or pFRED143 for expression of unfused GFP (Rev-negative). The percentage of dual-fluorescent cells (red and green) in the transfected cell population (green) was determined. This data was used to calculate Rev-mediated induction of reporter protein expression.
Digital fluorescence microscopy
Epifluorescence microscopy was performed using a computer-controlled inverted research microscope (Zeiss Axiovert 200M; Carl Zeiss, Oberkochem, Germany) with AxioVision 3.1 software (Carl Zeiss Vision GmbH, Hallbergmoos, Germany), an AxioCam HRm CCD camera (Carl Zeiss) and a 10×/0.30 Plan Neofluar Objective (Carl Zeiss). Images for green and red fluorescence were taken with the appropriate filters: EGFP (Ex, BP 475/40, FT 500; Em, BP 530/50) and DsRed (Ex, BP 545/25, FT 570; Em, BP 605/70). Images were processed and arranged for presentation with standard graphics software (Adobe Illustrator; Adobe Systems, San Jose, CA).
RESULTS
The goal of our study was to develop a methodology for the recognition of multiple distinct motifs contributing to a common regulatory function from a single set of sequences containing the motifs in undefined subsets. As an example, we chose the INS regions of HIV-1 (Fig. 1) and combined computational and functional analyses to characterize and verify the sequence features of INS activity.
Detailed experimental mutational analysis of INS elements within the HXB2R sequence provided us with the data necessary for a computational approach aimed at the identification of conserved sequence features of INS elements. To this end, we took advantage of several software tools previously developed by us for the detection of conserved core sequences (cores) within sequences of low overall similarity (35,37,41).
Strategy for development of matrix descriptions
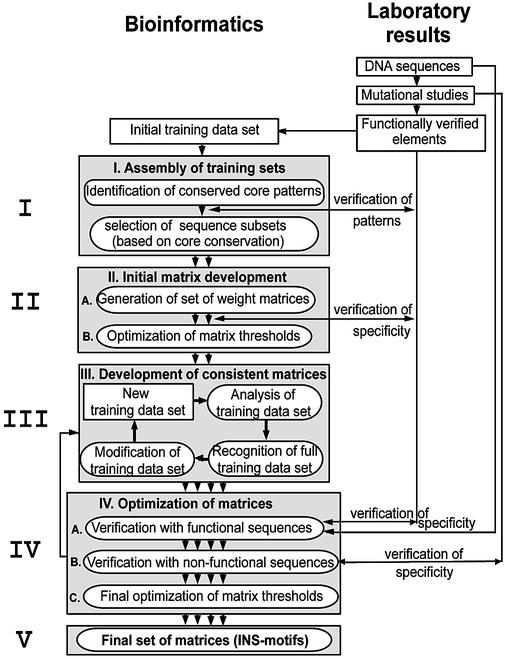
Step I: assembly of initial training sets. The first step in this analysis (see Fig. 2 for an overview of the whole strategy) was to identify conserved core sequences in all or at least in a subset of the sequences by using the CoreSearch program. Subsequently, we discriminated between core sequences with verified functions and potentially non-functional ones, taking advantage of published data from mutation analysis (27). This allowed separation of training set(s) of sequences, each of which contained only one conserved core sequence. This approach provided an excellent positive training set of functional sequences and an equally well defined negative training set of non-functional sequences.
Figure 2.
Strategy for the generation of INS weight matrices (INS motifs). This figure shows a schematic representation of the general strategy for the identification of the HIV-1 INS motifs (from top to bottom). The approach started in the laboratory with the detailed mutational analysis, which is indicated in the right half of the figure while the multiple steps of the computational, bioinformatics approach are represented on the left. Arrows between both sites indicate crosscheck analyses between experimental data and the computational results. Circular iterations were necessary between steps II and III and within step III (see text for details).
Step II: initial matrix development. Since more than one conserved core sequence was found, individual matrices were developed for each conserved core sequence by appropriate programs (ConsInd and MatInd). Both programs are able to generate matrices directly from a training set of sequences.
Step III: development of consistent matrices. A good matrix should be able to detect all sequences used for the generation of the matrix (training data set) as a minimal requirement. However, this can almost always be achieved by lowering the thresholds for matrix similarity. Therefore, our goal was to develop matrices capable of recognizing their complete training set with the same threshold settings that will be used for the analysis of new sequence data (test data). A matrix fulfilling this condition will be called a consistent matrix from here on.
First, the training set of sequences was analyzed with the default matrix thresholds used by the programs MatInspector and ConsInspector. If individual sequences were not recognized, either the thresholds were reduced or the sequences were removed from the training set and a new matrix was compiled from the reduced training set. This finally resulted in matrices that recognized all sequences within their training set.
These matrices were then analyzed with the test sequence (HIV-1 HXB2 genomic sequence, ∼10 kb). The matrices and thresholds were adjusted to retain the maximum number of matches to the training set while minimizing the total amount of false positive matches in the test sequence. At this point all matches outside known INS elements were regarded as false positives.
Finally, the training set was reanalyzed with the adjusted thresholds. If sequences were not recognized, a reduced training set was defined and another cycle of analysis was started [with (step I)].
Matrices that passed through the whole cycle without changes in their training set were consistent and used in further steps. Matrices retaining less than five training sequences (in the case of MatInd matrices) or seven sequences (in the case of ConsInd matrices) were discarded.
Step IV: optimization of matrices. The consistent matrices were finally verified with the initial positive and negative training sets. Thresholds were optimized to yield the minimum amount of false positive matches while retaining the true positive matches. Consistency of the matrices was rechecked after threshold optimization.
Application of strategy to HIV-1 INS elements
The first step (step I) of the initial analysis was started with a positive training set of 25 functionally verified INS elements (M1gag–M10gag and M0pol–M12pol) of the HIV-1 HXB2R genomic sequence (initial training data set, Fig. 1). These regions covered the INS1, INS2, INS3, CRS (INS regions) as well as the region between INS3 and CRS, as indicated in Figure 1. CoreSearch analysis revealed six different conserved core sequences AAACA, TTATA, ATAGA, TAGAT, AAAAG or ATAAA. Only the core sequences AAAAG and ATAAA were present in enough training sequences to allow initial definition of weight matrices. Based on the two different cores the input sequences were split into two separate sequence sets (SET A and SET B), which were used in the following steps.
Development of weight matrices from the training sets
The initial matrix development (step II) was carried out with the programs MatInd and MatInspector. The MatInd program generated two matrices (defined length of 15 nt) from these data sets. The resulting matrix A consisted of 21 sequences sharing the AAAAG core sequence and can be summarized by the IUPAC string AANAAAAAGNAMNN. Matrix B, summarized by the IUPAC string RNNAAATAAAANAYA, was generated from 11 sequences sharing the ATAAA core sequence.
For the development of consistent matrices (step III) we used the complete sequence of the HIV-1 HXB2R full-length provirus as the test sequence. Several consecutively refined sequence data sets were required to reach the consistency criteria described above. The best training data set for matrix A consisted of six sequences from five different INS elements [M3gag, M4gag (2×), M10gag, M7pol and M8.2pol] in gag and pol. The resulting consistent matrix was able to identify 32 matches within the HIV-1 HXB2R test sequence, using the MatInspector default threshold settings (core similarity 0.8, matrix similarity 0.85) of which 31 were located on the coding strand (also known as sense or plus strand). Optimization for higher specificity was continued, using matrix similarity as the filter until the number of false positive matches (i.e. matches in regions with no INS function) was minimized without loss of any of the experimentally verified true positive matches. This was achieved at a matrix similarity of 0.90. Under these conditions we found 15 matches to matrix A located exclusively on the coding strand of the HIV-1 HXB2R sequence, including all known and several new candidates for INS elements. This finding coincided with the expected prevalence of INS elements on the coding-DNA strand.
Matrix B was less specific and recognized only five of the 11 sequences used for the generation of the matrix and it was therefore inconsistent with the training data. MatInd cannot select sequences during matrix generation. Therefore, matrix quality depends greatly on the manual selection of sequences in the training set. In contrast to MatInd, the ConsInd program is able to reject sequences from the training data set. ConsInd requires longer sequences than MatInd, which were available for our sequence sets. Therefore, we repeated steps I–III with the ConsInd program.
Refined matrix analysis of training data sets
Steps II, III and IV. To generate more specific weight matrices we used the initial training data sets A and B and left sequence selection to the ConsInd program. Therefore, no manual selection was required. The resulting matrix is always consistent with the training data and can be used directly for further analyses.
SET-B (ATAAA core) resulted in a first stable weight matrix termed INS-B (INS motif), which was evaluated using the ConsInspector program on the HIV-1 HXB2R test sequence. The effects of each step of the consistency analysis on the specificity of the developing matrix are shown in Figure 1. During INS-B matrix development more than 300 matches were initially found in the HXB2R test sequence using the default settings of the ConsInspector program. Gradual optimization of the parameters and subsequent refinements of the sequence training sets reduced the total number of matches dramatically from over 300 to 17 until the final optimized matrix INS-B was reached (SET B5 in Fig. 1). This also included optimization of the parameters on the experimentally verified positive and negative training sets (step IV).
Weight matrix INS-B recognized all training sequences M3gag, M8gag, M0pol, M2pol, M2Apol, M6pol and M8-2pol (fulfilling the criterion for consistency). Additional matches corresponding to M1gag and M1pol were identified. Another six matches represented candidates for new INS elements, since they are located in regions that might include potential functional INS (indicated as dashed-line boxes in Figs 1b and 3).
Figure 3.
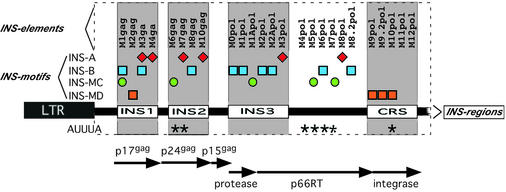
Distinct internal composition of INS regions in the GAG/POL-ORF of HXB2R. To the right of the weight matrix name, INS motif matches are indicated as symbols as described in Figure 4. The corresponding INS element names are depicted above and the INS regions indicated below. Reading frames [p17gag, p24gag, p15gag, protease, reverse-transcriptase (p66RT), integrase] and the corresponding INS regions (INS1, INS2, INS3 and CRS) containing experimentally identified and verified INS elements are indicated.
Matrix INS-A (INS motif, AAAAG core) was developed from the 21 sequences of SET A in exactly the same manner as described for INS-B. The final sequence set of the optimized matrix INS-A included seven sequences, one each from M3gag, M7gag, M10gag, M3pol and M8pol, and two from M4gag. A total of 17 matches could be identified in the HIV-1 HXB2R test sequence with the final parameters. Therefore, INS-A identified 10 additional matches in the test sequence besides its own training set. The matrices INS-A and INS-B generally recognize separate elements (Fig. 3). Even in the single case in which INS-A and INS-B are contained in the same element (M3gag), they recognize two separate and distinguishable core sequences.
Re-evaluation of sequences not detected by INS-A and INS-B
Generation of the INS-MC matrix motif. Experimental data indicated that sequences beyond the 14 INS elements recognized by the two matrices INS-A and INS-B (for details see Fig. 3) show strong inhibitory activities within the HIV-1 HXB2R sequence. Therefore, the remaining sequences that contribute to the INS effect were analyzed further to identify additional matrices.
A consistent matrix termed INS-MC was developed by MatInd analysis from a sequence training data set containing five sequences (M1gag, M6gag, M1Apol, M5pol and M7pol) that were not included in the other two matrices. In addition to the training data set, two further matches could be identified in the HXB2R test sequence by the MatInspector program, starting at nucleotide positions 6038 and 7234, respectively. Both matches are located within gp120 of the env-coding region (Fig. 4). The combined results of the analysis in the gag/pol region of the HIV-1 HXB2R test sequence with the matrix INS-MC, INS-A and INS-B are shown in Figure 3. Using these three matrices, 18 of the 26 sequences (70%) that are believed to contribute to INS-dependent down-regulation were recognized.
Figure 4.
Comparative mapping of INS regions and motifs in 26 full-length HIV-1 proviral sequences. In this graphical representation of the alignment of 26 HIV-1 full-length proviral genome sequences, the top line illustrates the organization of the HIV-1 genome (ORFs, line 1). Line 2 shows the localization of the INS regions (rectangles), with the known regions marked in orange and the predicted regions uncolored. Line 3 shows the positions of the INS elements (orange boxes) in the HIV-1 HXB2 genome. INS elements were identified by loss of Rev-dependent expression after mutation (27). The lines below represent the following individual HIV-1 sequences (listed from top to bottom, with the corresponding clades indicated in parentheses): hxb2r (B), lw123 (B), 3202a12 (B), 3202a21 (B), mn (B), cam1 (B), th475a (B), bcsg3 (B), lai (B), weau160 (B), han (B), d31 (B), jrcsf (B), jrfl (B), manc (B), oyi (B), ny5cg (B), rf (B), ndk (D), z2z6 (D), eli (D), u455 (A), mal (ADI), ibng (AG), mvp5180 (O) and ant70 (O). Different geometrical shapes and colors indicate matches to the INS matrices (INS motifs). INS motifs for INS-A are marked as red diamonds, for INS-B as filled skyblue squares, for INS-MC as green circles, and for INS-MD as orange boxes. Since INS elements and INS regions are functional on the mRNA level only the sense strand (plus strand) is shown. Open boxes below the aligned sequences indicate the newly predicted INS regions (INS4, INS5, INS6, INS7, INS8, INS9, INS10). A complete and more detailed version of this Figure is available on our homepage (www.gsf.de/biodv/).
Generation of a matrix recognizing the CRS-INS (motif INS-MD). INS-A, INS-B and INS-MC were unable to recognize the INS elements located within the CRS and the M2gag element within INS2. Therefore, we used the remaining INS sequences, corresponding to these four elements (M2gag, M9.2pol, M10pol and M11pol), to develop an additional INS weight matrix. This resulted in a consistent matrix termed INS-MD. With the optimized settings of 0.71 (core similarity) and 0.87 (matrix similarity) six matches could be identified in the HIV-1 HXB2R test sequence. Four were contained in well characterized INS elements in gag (INS-M2gag) and pol (CRS) and two additional matches were identified at nucleotide positions 4424 (integrase region of the pol-ORF) and at 7422 within gp41env (env-ORF). All these matches are located in regions with demonstrated INS activities, which make them good candidates for functional matches.
The four sites defined here by the matrices INS-A, INS-B, INS-MC and INS-MD recognized 22 out of the 25 (>88%) INS elements for which experimental data are available and yielded 18 additional matches predominantly in areas known or expected to contain further INS elements.
Generation of the AUUUA matrix. HIV-1 mRNAs also contain several AUUUA penta-nucleotide elements shown to function as inhibitory elements in several cellular mRNAs. Therefore, we investigated occurrence of these AUUUA sequences in HIV-1 mRNAs, although no experimental data regarding their role in inhibiting HIV-1 expression is available.
From 32 experimentally verified AUUUA sequences derived from the 3′-UTR of six different mRNAs, 27 were suitable for the generation of the consistent AUUUA matrix that can be summarized by the IUPAC string WWWNTTATNTATTTATTTATTTANN (6,51,52). In contrast, no matches were detected with the same sequence set used as negative control during experimental analysis (6,31–34).
The analysis of the HXB2R test sequence revealed that, in contrast to the INS matrices, more AUUUA matrix matches were found on the non-coding antisense strand (15 matches) than on the coding sense strand (nine matches). All nine sense strand matches coincided with eight of the other INS elements of HIV-1 HXB2R [M6gag, M7gag, M4pol, M5pol, M6Pol, M7pol (two matches), M8pol and M10pol] (Fig. 4).
Conservation analysis of INS motifs in 26 HIV-1 full-length proviral sequences. If the INS motifs were predicted correctly, they should also be conserved in other HIV-1 genomes. A multiple alignment of 26 HIV-1 full-length proviral sequences was carried out using the tools included in the GCG sequence analysis software package of the University of Wisconsin Genetics Computer Group (36). This multiple sequence alignment was necessary to counteract the heterogeneous 5′ ends of the sequences, which prevent direct comparison of the match positions of the matrices in individual sequences. The results from all the INS motif analyses were then superimposed onto the genomic organization of HIV-1 (Fig. 4). Matrix analyses of the 26 full-length proviral sequences were performed using the optimized parameters for each matrix and resulted in the identification of 1219 matches in the full-length proviral sequence. Again, it was striking that 1148 (∼94%) of these matches corresponded to matches on the sense strand and only 71 (∼6%) to matches on the antisense strand, which we considered to be false positive matches, since this strand does not encode viral proteins. For better differentiation, matrix matches are indicated with different shapes and colors in Figure 4 as described in the figure legend.
In the majority of HIV genomic sequences, we observed that the plus-strand matches to the INS motifs are located in similar genomic regions in different HIV-1 isolates from different clades (as indicated in Fig. 4). In about 43 locations, INS motifs matching the same matrix were conserved in at least 10 or more HIV genomic sequences. Nineteen of these were confined to the well characterized INS regions (INS1, INS2, INS3, CRS and RRE). In a few cases, matches conserved in only a few HIV genomic sequences could be detected. Some of them seemed to be in close proximity to functionally conserved INS regions, which they might be complementing, while the others were scattered single matches. Matches to all four INS motifs were observed in all the clades analyzed and no apparent clade-specific differences in the INS motif match distribution was observed, except for the lack of INS motif matches within the gag region of the most distant O-clade.
In addition to known INS regions, several new candidate INS regions were identified. The composition of two regions indicated as INS5 and INS6 appeared most similar to known functional INS regions (Fig. 4). Therefore, these regions were selected for experimental verification.
Verification of INS function of in silico predicted candidate INS regions INS5 and INS6. The two INS regions, INS5 and INS6, were cloned into plasmid pLRedR containing a gene for the RFP and the RRE (Fig. 5). Candidate INS regions were inserted downstream of multiple translational STOP codons terminating the reading frame of the RFP to ensure that the INS region candidates were not contained within the translated region (see Materials and Methods; Fig. 5). Constructs were transfected into HLtat cells together with a construct for expression of the GFP as transfection control. Cells were evaluated for red and green fluorescence by FACS analysis, resulting in the distribution of cell populations shown in Figure 6. The ratio of green to red fluorescence was calculated (detailed in Materials and Methods). For each INS-bearing construct, this ratio was normalized to the value of pLRedR (no INS region, Fig. 5), which was set to 1. Constructs bearing the INS region candidates in sense orientation were compared with identical constructs bearing the INS region candidates in antisense orientation.
Figure 6.
Evaluation of the influence of INS regions on expression of red fluorescent reporter protein by flow cytometry. HLtat cell cultures were transfected with different RFP reporter plasmids (see Fig. 5) and a GFP-expression plasmid as transfection control. The fluorescence of cells [shown for single cells in (a)] was evaluated by flow cytometry (b). The y-axis represents red fluorescence, reflecting expression of the reporter protein from the gene with the INS region, whereas the x-axis displays green fluorescence of the transfection control protein. Fewer cells in the upper right quadrant indicate a greater down-modulating effect of the INS region in the construct. Quantitative evaluation was carried out based on the histogram data shown in (c). Counts indicate cell numbers (y-axis) and the x-axis represents fluorescence intensity. The median of red fluorescence intensity was determined from the range labeled R2 (black bars). The gray areas were excluded as background (level observed with non-transfected cells).
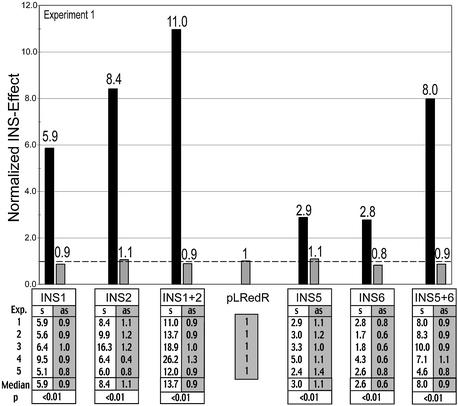
The graph in Figure 7 shows a comparison of the inhibitory effects of the original, functionally determined INS regions INS1 and INS2 in gag, which have previously been shown to work only in sense orientation (25,27), and the newly predicted candidate INS regions INS5 and INS6 in pol. The graph illustrates the results of a representative experiment from the series of independent experiments shown in the table (Fig. 7). The inhibitory activities of INS1, INS2 as well as the combined INS1 and INS2 (INS1+INS2), were confirmed in this assay (25,27,28). All inhibitory effects were statistically highly significant using a non-parametric test (P < 0.01). In addition, this assay allowed for the first time a direct comparison of individual activities of INS1 and INS2. The inhibitory effect of the combined INS1 and INS2 (INS1+2) was more pronounced than that of the individual elements (Fig. 7).
Figure 7.
Inhibitory effects of known and predicted INS regions. To analyze inhibitory activity of known and predicted INS regions of the HIV-1 genome, cells were transfected in parallel with various reporter plasmids containing INS regions or with the pLRedR control plasmid, which lacks INS (plasmids shown in Fig. 5). Flow cytometry was used to determine the ratio of green to red fluorescence (detailed in Materials and Methods). For each INS-bearing construct, this ratio was normalized to the value of pLRedR, which was set to 1. Inhibitory activity was evaluated for the known INS regions, INS1 and INS2, individually and in combination (INS1+2) (left side) and similarly for the newly predicted INS region candidates (INS5 and INS6) and their combination (right side). All INS regions were analyzed in sense (black bars, s) and antisense orientation (gray bars, as). FACS data were obtained from five independent experiments, all of which are shown in the table below the graph. The statistical significance of the difference between sense and antisense was determined as detailed in Material and Methods (Mann–Whitney non-parametric test) and is given below the table. The graph shows the values of experiment 1.
Both candidate INS regions (INS5 and INS6) also showed inhibitory activity when tested individually. A synergistic effect was observed for the combined INS5+6 INS region, corresponding to the arrangement of these regions in the HIV-1 genome. As shown for the other INS regions, the inhibitory effects of INS5 and INS6 and of the combined INS5+6 region were observed only in sense, not in antisense orientation, further confirming their functionality as INS regions.
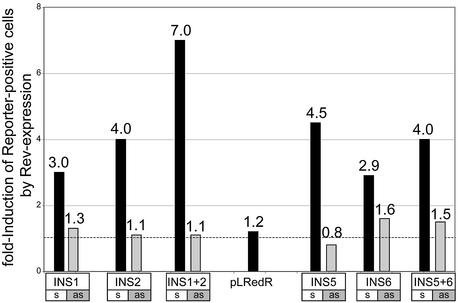
Co-expression of the HIV-1 Rev protein with the INS-containing constructs increased the number of reporter protein positive cells for all constructs that contained the INS regions in sense orientation (Fig. 8). The relative number of reporter protein positive cells bearing the antisense INS constructs remained almost unaffected by the co-expression of Rev, as would be expected (Fig. 8). This demonstrates that the inhibitory function of the candidate INS is overcome by the HIV-1 Rev protein, as shown previously for known INS regions (27).
Figure 8.
Rev rescue of INS inhibitory activities. To analyze whether the inhibitory activity of known and predicted INS regions of the HIV-1 genome can be overcome by HIV-1 Rev protein, reporter plasmids with the indicated INS regions were cotransfected in parallel with either pCsRevsg143 for expression in the presence of Rev-GFP or pFRED143 for expression in the absence of Rev. Reporter plasmid pLRedR, which lacks INS (Fig. 5), was analyzed as a control. Cells showing green and red fluorescence were counted and the Rev-induced increase in the number of dual-fluorescent cells determined (detailed in Materials and Methods). The effect of Rev on all INS regions was analyzed in sense (black bars, s) and antisense orientations (gray bars, as).
DISCUSSION
Previous attempts to generate a sequence consensus for INS elements failed, because they were based on the assumption that all INS elements conform to a single motif. Here we demonstrate that the INS elements as well as INS regions are actually composed of at least four distinct sequence patterns (INS motifs). The strategy described here allowed an a priori separation of the sequences of INS elements into four subsets and subsequent definition of the four subset-specific INS motifs (matrices INS-A, INS-B, INS-MC and INS-MD).
The basic strategy followed in this study is not limited to analysis of HIV-1 INS elements and can be used for the development of motif descriptions from other sets of nucleotide sequences with common functions (like transport or protein binding). The consistency criterion we introduced in this study prevents generation of spurious matrices, as these would be eliminated by consecutive reduction of the training set. Consistency requires no more than an initial training set of sequences and at least one test sequence (which might be any sequence of sufficient length) to provide an estimation of the number of additional matches. Availability of well characterized test sequences as in this study is an advantage, but not a prerequisite. The results also demonstrate that the sequence conservation of RNA sequence elements can be described by the methodology developed for DNA elements (e.g. TF-binding sites).
An important feature of our approach was the incorporation of data characterizing the effect of defined mutations on INS function. The inclusion of this data significantly improved the specificity of the final matrices describing INS motifs. Without this functional data, we would have had to carry out the consistency analysis for many more matrices (most of which would have been eliminated during this process) and final optimization would have been impossible.
The matrix definition by CoreSearch used in this study typically requires 7–20 sequences to define a matrix with high confidence (seven independent sequences, more if there is overall similarity between some of the sequences). CoreSearch relies on the uneven distribution of mismatches in real binding sites in order to select the best candidates excluding all sequences with no recognizable core (35) (because core length is used as a parameter).
However, matrix scores are not directly correlated with biological activity. Empirically or statistically determined thresholds allow discrimination between candidates likely to be functional and spurious matches. However, there is no way to predict relative strengths of the biological effects solely from in silico ratings.
Threshold settings can be very important for this kind of approach. Rather than relying on some predefined standards we varied thresholds systematically and used constraints to determine the optimal values. Matrices were required to recognize the whole training set at optimized thresholds as well as produce the best ratio of matches in known INS regions versus additional matches in the control HIV-1 genome. Since these constraints work against each other, the danger of over-fitting is greatly reduced.
Analyses of HIV sequences from different clades revealed that most of the INS motifs are phylogenetically conserved and cluster to form INS regions (Fig. 4). Two of these previously unknown clusters were defined as independent candidate INS regions and the INS function of these regions was subsequently verified. These experiments also confirmed that the new candidates showed features expected of INS regions: they function only in sense orientation and do not require the context of a reading frame, indicating that they act at the RNA level in a translation-independent manner (25,27,29). The HIV-1 sense strand is more A-rich than the antisense strand. However, the strand bias is not likely to be a mere consequence of the different A content of both strands as shuffling of the sequence within 10 bp windows (preserving even the AT profile over the whole sequence) resulted in a clear decrease of strand bias of the INS matrix matches while the strand distribution of the AUUUA matrix remained unaffected (data not shown). The physical overlap of INS elements and reading frames within the HIV genome is similar to the sequence structure of retroviral long terminal repeats (LTRs) (53). Here, regions responsible for transcriptional initiation (enhancer and promoter) overlap physically with the region governing transcriptional termination (polyadenylation) but they are functionally independent. We also show a dose-dependent effect (combination exerts a stronger inhibition) for candidate INS regions, INS5 and INS6 as example, which confirms features of other INS regions (e.g. INS1+2) (this study; 25,27,29). This finding is in line with the observed clustering of INS motifs within the INS regions.
The sites detected by our matrices within the INS elements also bear several hallmarks characteristic of other regulatory sequence elements and regions (e.g. TF-binding sites in promoters): (i) they are conserved among different HIV-1 genomes at similar locations (hallmark criterion 1); (ii) they occur in clusters (hallmark criterion 2); (iii) their conservation is independent of the conservation of the genomic nucleotide sequences they are embedded in (data not shown; hallmark criterion 3).
Our analysis also provided some insight into the internal structure of the HIV-1 INS regions that show clear preferences among the four defined INS motifs (see Fig. 3). The fact that the CRS is composed quite differently (three matches to INS-MD) might indicate that the CRS-INS functions differently than the INS1, INS2, INS3 and the RRE and that the order and combination of individual INS motifs is important as well.
Although the function of the INS elements has been studied in great detail, not much is known about the factors involved in the inhibitory effect. It appears that INS function is mediated by cellular factors since it does not require HIV infection and is even functional in non-primate cells (54). So far, only two cellular INS-binding factors have been identified (22,55,56). However, the presence of distinct, cooperative motifs that comprise the HIV-1 INS regions remind us of cooperating TF-binding sites in promoters, where multiple distinct binding sites and factor interactions account for the function (1). The organization and synergistic activity of the INS regions is reminiscent of promoter modules (1), although it is unclear whether INS motifs are also organized with similar stringency with respect to order and distance.
We demonstrate in this study that it is now possible to identify and define several distinct elements from a set of (mixed) sequences following the strategy outlined here, provided that experimental evidence is available. In our example, new candidate INS regions (INS5 and INS6) of the HIV genome were predicted and verified in functional assays. The strategy developed here is not dependent on biological features specific to HIV-1. The key features required for applicability of this strategy are relatively short sequence stretches, which are moderately conserved on the sequence level. This is typically the case in DNA as well as in RNA when sequence-specific nucleic acid binding proteins are involved. In all such cases our approach is generally applicable for defining the sequence characteristics of functionally conserved nucleotide sequences of viral or cellular origin, and extends the range of available pattern detection methods to the RNA world.
Acknowledgments
ACKNOWLEDGEMENTS
We thank George Pavlakis for plasmids p37R and pFred143. This work was supported by DFG grant ‘Informatic methods for the analysis and interpretation of large amounts of genomic data’ (Grant 2370/1-1) and SFB 464.
REFERENCES
- 1.Werner T. (1999) Models for prediction and recognition of eukaryotic promoters. Mamm. Genome, 10, 168–175. [DOI] [PubMed] [Google Scholar]
- 2.Werner T. (2000) Computer-assisted analysis of transcription control regions. Matinspector and other programs. In Misener,S. and Krawetz,S.A. (eds), Bioinformatics Methods and Protocols. Humana Press, Totowa, NJ, Vol. 132, pp. 337–349. [DOI] [PubMed]
- 3.Werner T. (2001) Analyzing Regulatory Regions in Genomes. Wiley-VCH, New York, Heidelberg.
- 4.Shyu A.B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- 5.Peng S.S., Chen,C.Y. and Shyu,A.B. (1996) Functional characterization of a non-AUUUA AU-rich element from the c- jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol. Cell. Biol., 16, 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaw G. and Kamen,R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- 7.Myer V.E., Fan,X.C. and Steitz,J.A. (1997) Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J., 16, 2130–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz D.A., Theodorakis,N.G., Cleveland,D.W., Lindsten,T. and Thompson,C.B. (1994) AU-A, an RNA-binding activity distinct from hnRNP A1, is selective for AUUUA repeats and shuttles between the nucleus and the cytoplasm. Nucleic Acids Res., 22, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan X.C. and Steitz,J.A. (1998) HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl Acad. Sci. USA, 95, 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan X.C. and Steitz,J.A. (1998) Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J., 17, 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furth P.A. and Baker,C.C. (1991) An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol., 65, 5806–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grüter P., Tabernero,C., von Kobbe,C., Schmitt,C., Saavedra,C., Bachi,A., Wilm,M., Felber,B.K. and Izaurralde,E. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell, 1, 649–659. [DOI] [PubMed] [Google Scholar]
- 13.Huang J. and Liang,T.J. (1993) A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol. Cell. Biol., 13, 7476–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saavedra C., Felber,B. and Izaurralde,E. (1997) The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol., 7, 619–628. [DOI] [PubMed] [Google Scholar]
- 15.Tang H., Gaietta,G.M., Fischer,W.H., Ellisman,M.H. and Wong-Staal,F. (1997) A cellular cofactor for the constitutive transport element of type D retrovirus. Science, 276, 1412–1415. [DOI] [PubMed] [Google Scholar]
- 16.Tang H., Xu,Y. and Wong-Staal,F. (1997) Identification and purification of cellular proteins that specifically interact with the RNA constitutive transport elements from retrovirus D. Virology, 228, 333–339. [DOI] [PubMed] [Google Scholar]
- 17.Freed E.O. and Martin,M.A. (2001) HIVs and their replication. Chapter 59. In Knipe,D.M., Howley,P.M., Griffin,D.E., Lamb,R.A., Martin,M.A., Roizman,B. and Strauss,S.E. (eds), Field’s Virology, 4th Edn. Lippicott Williams and Wilkins, Philadelphia, PA, Vol. Specific Virus Families, pp. 1971–2042.
- 18.Wodrich H. and Krausslich,H.G. (2001) Nucleocytoplasmic RNA transport in retroviral replication. Results Probl. Cell Differ., 34, 197–217. [DOI] [PubMed] [Google Scholar]
- 19.Reddy T.R., Tang,H., Xu,W. and Wong-Staal,F. (2000) Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene, 19, 3570–3575. [DOI] [PubMed] [Google Scholar]
- 20.Boris-Lawrie K., Roberts,T.M. and Hull,S. (2001) Retroviral RNA elements integrate components of post-transcriptional gene expression. Life Sci., 69, 2697–2709. [DOI] [PubMed] [Google Scholar]
- 21.Kong W., Tian,C., Liu,B. and Yu,X.F. (2002) Stable expression of primary human immunodeficiency virus type 1 structural gene products by use of a noncytopathic sindbis virus vector. J. Virol., 76, 11434–11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afonina E., Neumann,M. and Pavlakis,G.N. (1997) Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem., 272, 2307–2311. [DOI] [PubMed] [Google Scholar]
- 23.Najera I., Krieg,M. and Karn,J. (1999) Synergistic stimulation of HIV-1 Rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J. Mol. Biol., 285, 1951–1964. [DOI] [PubMed] [Google Scholar]
- 24.Nasioulas G., Zolotukhin,A.S., Tabernero,C., Solomin,L., Cunningham,C.P., Pavlakis,G.N. and Felber,B.K. (1994) Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J. Virol., 68, 2986–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz S., Campbell,M., Nasioulas,G., Harrison,J., Felber,B.K. and Pavlakis,G.N. (1992) Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol., 66, 7176–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosen C.A., Terwilliger,E., Dayton,A., Sodroski,J.G. and Haseltine,W.A. (1988) Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc. Natl Acad. Sci. USA, 85, 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider R., Campbell,M., Nasioulas,G., Felber,B.K. and Pavlakis,G.N. (1997) Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol., 71, 4892–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S., Felber,B.K. and Pavlakis,G.N. (1992) Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol., 66, 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz S., Felber,B.K. and Pavlakis,G.N. (1992) Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol. Cell. Biol., 12, 207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasioulas G., Hughes,S.H., Felber,B.K. and Whitcomb,J.M. (1995) Production of avian leukosis virus particles in mammalian cells can be mediated by the interaction of the human immunodeficiency virus protein Rev and the Rev-responsive element. Proc. Natl Acad. Sci. USA, 92, 11940–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson T. and Treisman,R. (1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature, 336, 396–399. [DOI] [PubMed] [Google Scholar]
- 32.Raymond V., Atwater,J.A. and Verma,I.M. (1989) Removal of an mRNA destabilizing element correlates with the increased oncogenicity of proto-oncogene fos. Oncogene Res., 5, 1–12. [PubMed] [Google Scholar]
- 33.Jones T.R. and Cole,M.D. (1987) Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol. Cell. Biol., 7, 4513–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole M.D. and Mango,S.E. (1990) Cis-acting determinants of c-myc mRNA stability. Enzyme, 44, 167–180. [DOI] [PubMed] [Google Scholar]
- 35.Wolfertstetter F., Frech,K., Herrmann,G. and Werner,T. (1996) Identification of functional elements in unaligned nucleic acid sequences by a novel tuple search algorithm. Comput. Appl. Biosci., 12, 71–80. [DOI] [PubMed] [Google Scholar]
- 36.Devereux J., Haeberli,P. and Smithies,O. (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res., 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frech K., Herrmann,G. and Werner,T. (1993) Computer-assisted prediction, classification and delimitation of protein binding sites in nucleic acids. Nucleic Acids Res., 21, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quandt K., Grote,K. and Werner,T. (1996) GenomeInspector: a new approach to detect correlation patterns of elements on genomic sequences. Comput. Appl. Biosci., 12, 405–413. [DOI] [PubMed] [Google Scholar]
- 39.Frech K., Dietze,P. and Werner,T. (1997) ConsInspector 3.0: new library and enhanced functionality. Comput. Appl. Biosci., 13, 109–110. [DOI] [PubMed] [Google Scholar]
- 40.Frech K., Quandt,K. and Werner,T. (1997) Finding protein-binding sites in DNA sequences: the next generation. Trends Biochem. Sci., 22, 103–104. [DOI] [PubMed] [Google Scholar]
- 41.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felber B.K., Drysdale,C.M. and Pavlakis,G.N. (1990) Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J. Virol., 64, 3734–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz S., Felber,B.K., Benko,D.M., Fenyo,E.M. and Pavlakis,G.N. (1990) Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol., 64, 2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumann M., Afonina,E., Ceccherini-Silberstein,F., Schlicht,S., Erfle,V., Pavlakis,G.N. and Brack-Werner,R. (2001) Nucleocytoplasmic transport in human astrocytes: decreased nuclear uptake of the HIV Rev shuttle protein. J. Cell Sci., 114, 1717–1729. [DOI] [PubMed] [Google Scholar]
- 45.Graham F.J. and Van der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–460. [DOI] [PubMed] [Google Scholar]
- 46.Matz M.V., Fradkov,A.F., Labas,Y.A., Savitsky,A.P., Zaraisky,A.G., Markelov,M.L. and Lukyanov,S.A. (1999) Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol., 17, 969–973. [DOI] [PubMed] [Google Scholar]
- 47.Hadzopoulou-Cladaras M., Felber,B.K., Cladaras,C., Athanassopoulos,A., Tse,A. and Pavlakis,G.N. (1989) The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol., 63, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stauber R.H., Horie,K., Carney,P., Hudson,E.A., Tarasova,N.I., Gaitanaris,G.A. and Pavlakis,G.N. (1998) Development and applications of enhanced green fluorescent protein mutants. Biotechniques, 24, 462–466, 468–471. [DOI] [PubMed] [Google Scholar]
- 49.Stauber R.H. and Pavlakis,G.N. (1998) Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology, 252, 126–136. [DOI] [PubMed] [Google Scholar]
- 50.Ludwig E., Silberstein,F.C., van Empel,J., Erfle,V., Neumann,M. and Brack-Werner,R. (1999) Diminished rev-mediated stimulation of human immunodeficiency virus type 1 protein synthesis is a hallmark of human astrocytes. J. Virol., 73, 8279–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C.Y., Xu,N. and Shyu,A.B. (1995) mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol., 15, 5777–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoecklin G., Hahn,S. and Moroni,C. (1994) Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J. Biol. Chem., 269, 28591–28597. [PubMed] [Google Scholar]
- 53.Werner T. and Brack-Werner,R. (1998) Lentivirus long terminal repeats (LTRs): functions and common features. In Saksena,N.K. (ed.), Human Immunodeficiency Viruses: Biology, Immunology and Molecular Biology. Medical Systems S.p.A, Genoa, Italy, pp. 57–84.
- 54.Pavlakis G.N. (1997) The molecular biology of Human Immunodeficiency Virus Type 1. In DeVita,V.T., Hellman,S., Rosenberg,S.A., Essex,M.E., Curran,J.W., Fauci,A.S. and Freeman,J.S. (eds), AIDS: Biology, Diagnosis, Treatment and Prevention, 4th Edn. Lippincott-Raven Publishers, Philadelphia, PA, pp. 45–74.
- 55.Black A.C., Luo,J., Watanabe,C., Chun,S., Bakker,A., Fraser,J.K., Morgan,J.P. and Rosenblatt,J.D. (1995) Polypyrimidine tract-binding protein and heterogeneous nuclear ribonucleoprotein A1 bind to human T-cell leukemia virus type 2 RNA regulatory elements. J. Virol., 69, 6852–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Black A.C., Luo,J., Chun,S., Bakker,A., Fraser,J.K. and Rosenblatt,J.D. (1996) Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes, 12, 275–285. [DOI] [PubMed] [Google Scholar]