Abstract
Distamycin binds the minor groove of duplex DNA at AT-rich regions and has been a valuable probe of protein interactions with double-stranded DNA. We find that distamycin can also inhibit protein interactions with G-quadruplex (G4) DNA, a stable four-stranded structure in which the repeating unit is a G-quartet. Using NMR, we show that distamycin binds specifically to G4 DNA, stacking on the terminal G-quartets and contacting the flanking bases. These results demonstrate the utility of distamycin as a probe of G4 DNA–protein interactions and show that there are (at least) two distinct modes of protein–G4 DNA recognition which can be distinguished by sensitivity to distamycin.
INTRODUCTION
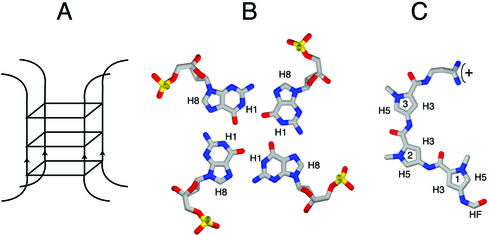
G-quadruplex or ‘G4’ DNA is a four-stranded structure stabilized by G-quartets. In a G-quartet, four guanines interact via Hoogsteen bonding to form a planar ring (Fig. 1) (1). Single-stranded DNA (2–6) or RNA (7) containing runs of consecutive guanine residues readily and spontaneously self-associate in vitro to form G-quadruplex structures, as diagrammed in Figure 1A. These structures may contain one or more individual nucleic acid strands, in parallel or antiparallel configuration. G4 DNA is very stable once formed, deriving its chemical stability from hydrogen bonding within each quartet, stacking of the hydrophobic quartets upon one another and coordination of a monovalent cation in the central channel (reviewed in 8). Because the structure of G4 DNA is different from that of duplex DNA, it is impervious to attack by enzymes which target single-stranded or duplex DNA, suggesting that specialized enzymes may maintain G-rich regions of the genome or eliminate G4 structures that might otherwise interfere with transcription, replication, translation or recombination. Consistent with this, a number of proteins have been shown to bind, cleave or unwind G4 DNA with considerable specificity (see, for example, 9–22).
Figure 1.
Structure of G4 DNA, a G-quartet and distamycin. (A) Schematic representation of a parallel tetrameric G-quadruplex. In this scheme three G-quartet planes stack; in the actual structures each subsequent plane is offset, creating a twist of the four equivalent grooves. (B) Top view of a G-quartet, showing four guanosine residues forming a planar array stabilized by Hoogsteen base pairing. (C) Distamycin.
Three regions of the genome are G-rich and have considerable potential to form G4 DNA: the telomeres, rDNA and mammalian immunoglobulin heavy chain switch regions. In addition, some genes encode G-rich mRNAs capable of forming G4 RNA, which appears to confer specific function in translation (23–27). Recent structural analysis has shown that the mammalian telomeric repeat TTAGGG spontaneously forms parallel G4 DNA in physiological salts (13). Formation of G4 DNA could regulate telomere maintenance by telomerase, by rendering the telomeric tail unavailable as a primer for telomere maintenance. This has fueled the notion that compounds that bind and stabilize G4 DNA might be of therapeutic value.
Highly conserved polypeptide domains have been implicated in high affinity G4 DNA binding. The abundant nucleolar protein nucleolin, which binds very tightly (Kd = 1 nM) to G4 DNA (15), contains four RBDs (RNA binding domains; also called RNA recognition motifs, or RRMs), a domain comprised of characteristic β-sheet structures thought to provide a platform for protein–nucleic acid contacts, and nine C-terminal Arg-Gly-Gly (RGG) repeats (28,29). By deletion analysis, G4 DNA binding has been mapped to two distinct domains of nucleolin, one comprised of the RBDs (Kd = 0.5 nM) and the other of the C-terminal RGGs (Kd = 3.3 nM) (15). Two candidate mammalian telomere binding proteins, hnRNP A1 (30,31) and hnRNP D (19), also bind to G4 DNA and contain RBD and RGG domains. In addition, FMRP, a negative regulator of translation encoded by the FMR1 gene, contains a C-terminal RGG domain which binds to G4 RNA (Kd = 10 nM) (24,31). Collectively, the RGG domains of nucleolin and FMRP are found to associate with four-strand parallel, two-strand hairpin and single-strand fold-back G4 DNA and G4 RNA (15,24,25).
There is also at least one additional conserved domain that recognizes G4 DNA that occurs in the RecQ helicase family. This is a small family of conserved enzymes with homology to Escherichia coli RecQ. All helicases in the RecQ family tested thus far, including E.coli RecQ, Saccharomyces cerevisiae Sgs1p and human BLM and WRN, unwind G4 DNA as well as or better than double-stranded (ds)DNA (13,14,17,21,22,32). None of the RecQ helicases contain any RBD or RGG domains, but these proteins bind G4 DNA with high affinity (5 nM). The RecQ G4 DNA recognition domain thus constitutes a third conserved region for protein–G4 DNA interaction.
Despite the variety of proteins that interact with G4 DNA with high affinity, the structural motifs in G4 DNA that proteins recognize have not been defined. It is not even known if different proteins recognize the same or distinct determinants. In order to define the modes of protein–G4 DNA interaction, we have sought to identify small molecules that inhibit protein binding to G4 DNA and could be used as convenient probes of activity.
Distamycin is a small molecule (Fig. 1C) which binds with high affinity to the minor groove of B-form DNA, recognizing AT-rich tracts (33,34), and has been a valuable tool for studying protein–dsDNA interactions. Here we demonstrate that distamycin inhibits G4 DNA binding by the RGG domain of nucleolin, but does not interfere with G4 DNA binding by the region of nucleolin that contains the four RBD domains, or by BLM helicase. This shows that there are (at least) two distinct modes of protein–G4 DNA recognition. By NMR analysis we show that the distamycin rings stack efficiently on the terminal G-planes of G4 DNA, intercalating between these G-planes and the bases that flank the G-quartet structures. This contrasts with the interaction of distamycin with duplex DNA, which involves an extensive hydrogen bonding network in the minor groove and no intercalation. Taken together, these results identify distamycin as a ligand that is a useful probe for protein–G4 DNA interactions and suggest a mechanism which RGG proteins may generally employ to bind to G-quadruplex structures.
MATERIALS AND METHODS
Protein purification and G4 DNA binding and unwinding reactions
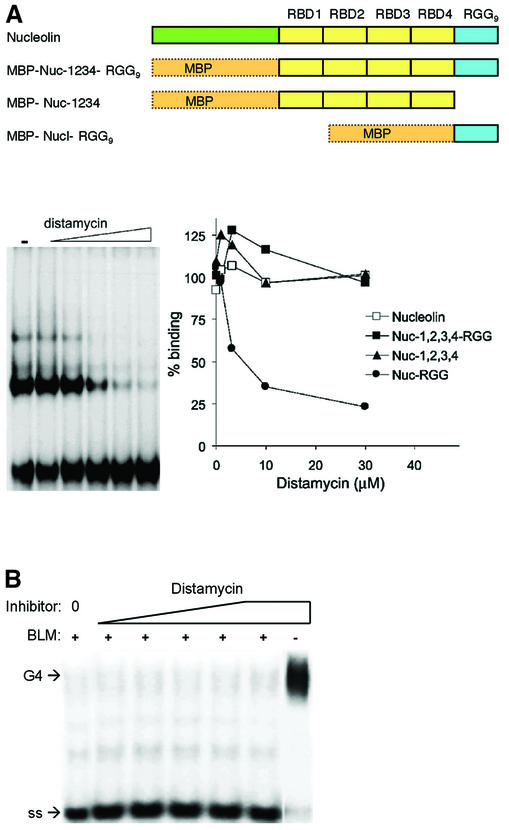
G4 DNA was formed from the oligonucleotide TGGACCAGACCTAGCAGCTATGGGGGAGCTGGGGAAGGTGGGAATGTGA and the structure verified by gel electrophoresis and DMS footprinting, as previously described (14). Full length murine nucleolin, recombinant nucleolin containing amino acid residues 281–709 but lacking the acidic N-terminus (Nuc-1,2,3,4-RGG) and recombinant nucleolin containing only the four RBD domains (Nuc-1,2,3,4) or the RGG domain (Nuc-RGG) were cloned and overexpressed as maltose binding protein (MBP) fusion proteins (Fig. 2A) and assayed for binding as described (15). Recombinant full length BLM helicase was expressed in yeast and purified as a hexahistidine tagged polypeptide (35) and DNA unwinding activity assayed as previously described (21), in reactions containing 5 ng of BLM and 10 fmol 32P-5′-end-labeled G4 DNA. For assays of binding in the presence of distamycin, G4 DNA was preincubated for 30 min at 25°C with distamycin (Sigma) aliquoted from a 1 mM stock solution in DMSO prior to addition of protein. Protein–G4 DNA complex formation and G4 DNA unwinding were quantitated by phosphorimager analysis of dried gels.
Figure 2.
Effect of distamycin on G4 DNA binding and unwinding. (A) (Top) Diagram of recombinant derivatives of nucleolin assayed as fusions to MBP. (Bottom) Effect of distamycin on G4 DNA binding by nucleolin and recombinant nucleolin derivatives. (Left) Distamycin inhibition of binding of Nuc-RGG to G4 DNA. [32P]G4 DNA was preincubated with 0, 0.12, 0.37, 1.11, 3.33 and 10 µM distamycin and then binding to 1 nM Nuc-RGG9 was assayed by gel mobility shift. (Right) Quantitation of binding assays carried out with purified murine nucleolin (Nucleolin, open square) and recombinant nucleolin carrying the RBDs and RGG9 (Nuc-1,2,3,4-RGG9, filled square), the nucleolin RBDs (Nuc-1,2,3,4, filled triangle) and the RGG9 domain of nucleolin (Nuc-RGG9, filled circle). (B) Effect of distamycin on BLM helicase G4 DNA unwinding activity. Unwinding assays were carried out in 0–125 µM distamycin. Control reactions contained distamycin, but no protein (right). Positions of G4 DNA substrate and single-stranded product (ss) are indicated.
Preparation of NMR samples
DNA samples were synthesized at the Yale University School of Medicine Keck Center, purified using a C18 column, lyophilized and dissolved directly into the sample buffer (150 mM KCl, 25 mM KH2PO4/K2HPO4, pH 7.0, 2 mM d-EDTA). G4 DNA molecules analyzed were formed from three sequences: [TAGGGTTA]4, [TAGGGGTT]4 and [TAGGGGGT]4. DNA containing a run of three guanines spontaneously formed G4 DNA with the correct register. DNA containing runs of four or more guanines was heated to 85°C for 10 min prior to structural studies or the addition of ligand.
Distamycin was first dissolved into a small volume of ethanol and diluted with water to a final concentration of 10 mM. For titration of [TAGGGTTA]4, aliquots of distamycin were added to a 0.2 mM DNA solution; the final ethanol concentration was <2%. For two-dimensional (2D) NMR analysis, an aliquot of the stock distamycin solution was added to a 2 mM strand (0.5 mM tetramer) DNA preparation and the sample was then lyophilized and resuspended to the original volume of the DNA portion, maintaining the original DNA and sample buffer concentrations. It should be noted that in some cases a small amount of distamycin did not redissolve when the sample was resuspended. The final stoichiometry was determined by integration of DNA and ligand proton NMR signals.
NMR experiments
NOESY spectra with 125, 200 and 300 ms mixing times were collected at 35°C on a Varian Unity 500 MHz NMR spectrometer. G4 DNA was initially confirmed by tracking NOE connectivities between G imino protons resistant to fast exchange with water. DNA alone and ratios of 2:1, 4:1 and 8:1 distamycin:G4 DNA were examined in both H2O and D2O. Water elimination was achieved using a modified WATERGATE (36,37). Matrix sizes were 2048 × 256 complex points, zero filled to 4096 × 512 and apodized using a Gaussian window of 10 Hz. Data were processed with NMRpipe (36) and analyzed using Sparky (37).
Measurement of diffusion constants by NMR
NMR diffusion measurements were made at 35°C in a Varian Inova 500 MHz wide bore NMR spectrometer. The pulse sequence used for these experiments was essentially that of Lapham et al. (38), based on the study of Tanner (39). These experiments were performed on a wide bore spectrometer using a narrow bore probe suspended in air within the magnet bore using a plastic ring well below the probe RF coil. The resulting distance between the probe housing and the shim set was 1.8 cm, rather than being in contact as in a conventional narrow bore magnet. Measurements of signal recovery time after a strong gradient pulse showed that the shim set eddy current relaxation, which for older probes typically requires a 1–2 ms delay before acquisition, was not apparent in the wide bore spectrometer. This allowed for the reduction of the final delay te from 2 ms to 100 µs. For our probes, use of the wide bore spectrometer for diffusion measurements affords improved sensitivity and phase properties of the spectra. However, we note that for newer probes, manufacturers have improved signal recovery following gradient pulses through better shielding.
RESULTS
Distamycin inhibits G4 DNA binding by the RGG domain of nucleolin
We assayed the effect of distamycin on the G4 DNA binding activity of nucleolin and nucleolin derivatives shown previously to bind G4 DNA (15). These were expressed as MBP fusions and contained the RBDs and RGG9 domain (Nuc-1,2,3,4-RGG9) or the RGG9 (Nuc-RGG9) domain (Fig. 2A). Distamycin inhibited binding of G4 DNA by Nuc-RGG9. In contrast, distamycin had no effect on G4 DNA binding by full length murine nucleolin (Nucleolin), recombinant nucleolin containing amino acid residues 281–709 but lacking the acidic N-terminus (Nuc-1,2,3,4-RGG9) or recombinant nucleolin containing only the four RBD domains (Nuc-1,2,3,4). The distamycin concentration at which binding was reduced to 50% of untreated levels (the IC50) was estimated to be 3 µM. Binding inhibition by distamycin was not dominant, as distamycin did not inhibit binding by Nuc-1,2,3,4-RGG9, which contains the RBDs and the RGG9 domain.
Others have shown that distamycin does inhibit unwinding of AT-rich duplex DNA by BLM helicase (40). However, assays of G4 DNA unwinding by BLM in the presence of distamycin showed no effect of this inhibitor (Fig. 2B), although inhibition was readily apparent at low concentrations of the anionic porphyrin derivative N-methyl mesoporphyrin IX (NMM), as we have previously reported (21,22). Distamycin is therefore a specific inhibitor of duplex DNA unwinding by BLM (and probably other RecQ helicases).
NMR characterization of the distamycin-[TAGGGTTA]4 complex
These data suggested that there are at least two separate modes of protein–G4 DNA recognition which can be distinguished by sensitivity to distamycin, and led us to ask how distamycin interacts with G4 DNA. G4 DNA is very stable in vitro and has proven to be an excellent candidate for structural studies by NMR (41–46). Three studies have demonstrated binding of RGG domains from nucleolin and FMRP to over 20 different substrates containing G-quadruplex structures (15,24,25). These RGG domains do not bind the same DNA when it is in the single-strand form, establishing the generic quadruplex structure as the target of protein RGG domain recognition. Because the gel-shift DNA substrate was too large to study by NMR, we chose a DNA sequence that has been a useful substrate in deriving structural models for quadruplex-specific ligands, [TAGGGTTA]4 (46). This substrate was permuted to increase the groove length since the prevailing theory was that distamycin bound in the groove of G-quadruplex DNA. We felt that the demonstrated general binding of RGG domains to quadruplex was sufficient justification to allow us a reductionist approach in the NMR characterization.
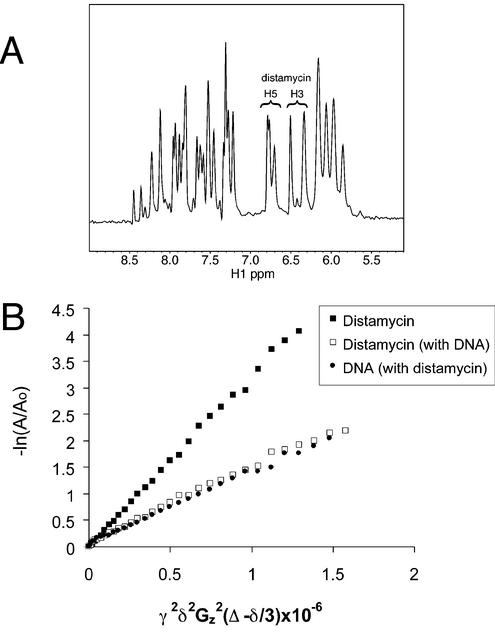
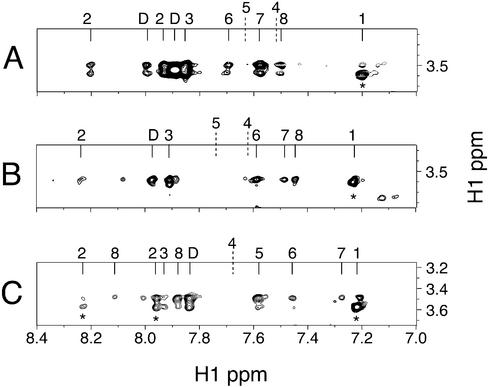
For initial characterization of the G4 DNA-distamycin complex, we titrated distamycin into a parallel G4 [TAGGGTTA]4 DNA sample and collected one-dimensional (1D) NMR spectra (not shown). Distamycin pyrrole H3 and H5 protons resonate in a region where there are no DNA signals (Fig. 3A) and allow for easy monitoring by 1D NMR and interpretation of NOEs in 2D NOESY experiments. Only one set of distamycin signals was apparent at all concentrations up to 8 equivalents per quadruplex. These signals changed only slightly with increasing ligand, sharpening and uniformly shifting downfield by 0.1–0.2 p.p.m. Although distamycin peaks sharpened slightly during the titration, they remained broader than the DNA base signals. The existence of only one set of signals requires the ligand to be in fast exchange among all bound conformations.
Figure 3.
NMR characterization of the distamycin-[TAGGGTTA]4 complex. (A) 1D proton NMR spectra of the [TAGGGTTA]4 G4 DNA with 4 equivalents of distamycin. The distamycin signals marked are the pyrrole H5 and H3 protons. (B) NMR diffusion measurements. Distamycin alone (filled square), parallel G4 [TAGGGTTA]4 DNA in complex with distamycin (open square) and distamycin in the presence of DNA (filled circle) (4:1 distamycin:G4 DNA molecule). The diffusion coefficient is the slope; γ, gyromagnetic ratio (hydrogen); δ, duration of gradient pulse; Gz, gradient amplitude; Δ, delay during which molecule diffuses; A, peak volume; Ao, initial peak volume. Samples were prepared in D2O, 150 mM KCl, 25 mM potassium phosphate, pH 7.0, 2 mM d-EDTA and the NMR data were collected at 35°C.
Because nucleolin RGG binding to G4 DNA (assayed at 1 nM protein and 67 pM DNA) was inhibited by an excess of distamycin (3 µM), we studied the complex under conditions in which the ligand was in excess in order to characterize all of the potential binding sites. Diffusion measurements using NMR gradient methods have been shown to be useful in determining ligand binding to DNA and proteins and to serve as a spectral editing tool based on molecule size. In the experiments presented here, diffusion measurements were made on the [TAGGGTTA]4-distamycin NMR preparation with a molar ratio of 4:1 distamycin to quadruplex to assay the extent of association of distamycin with parallel G4 DNA. We found that distamycin alone has a diffusion coefficient (D) of 3.08 × 10–6 cm2/s (Fig. 3B) in the buffer used to produce G4 DNA. However, in the presence of DNA, the distamycin diffused with the DNA molecule (distamycin D = 1.50 × 10–6 cm2/s; G4 DNA D = 1.43 × 10–6 cm2/s), indicating a strong or frequent interaction. The value of D for DNA alone was 1.45 × 10–6 cm2/s. The diffusion measurements indicate that the majority of the ligand is associated with the DNA, even when present in 4-fold molar excess as in our NOESY experiments.
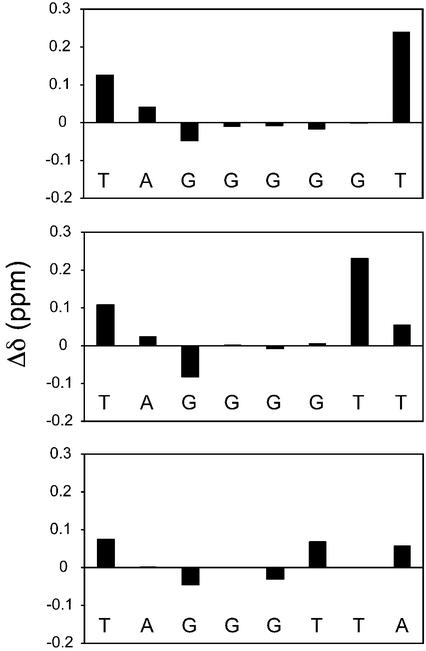
The structure of the [TAGGGTTA]4 G4 DNA was confirmed with NOESY experiments using published assignments (46). Comparison of chemical shifts (Fig. 4, bottom panel) showed that, upon addition of distamycin, all of the resonances shifted; however, within the central G-plane, the G4 H8 signal shifted the least. Specific contacts between distamycin and [TAGGGTTA]4 are listed in Table 1. Notably, there were no detectable NOEs from distamycin to any part of the interior G4 plane, and this plane therefore appears to have no contact with the ligand. However, there were NOEs arising between distamycin methyls and multiple positions in both the G3 and G5 base planes. The ligand contacts the entire face of each terminal G-plane as evidenced by NOEs to DNA G H1 protons. The ligand methyl groups resonate in a region of the NMR spectrum mostly devoid of DNA peaks (∼3.5 p.p.m.); NOEs from the distamycin methyls to the DNA H6/H8 protons are shown in Figure 5C. Based on observed NOEs, the ligand pyrrole H3, H5 and methyl protons are positioned near the G3 and G5 planes but also populate conformations in proximity to flanking bases. Weak NOEs from the distamycin pyrrole H3 and H5 protons to deoxyribose H1′ signals on both sides of the G-stack and additional flanking deoxyribose NOEs require that the pyrrole rings also be in proximity to the sugar portions of the backbone. The formyl group (HF) at one end of distamycin only gave rise to NOEs to the flanking A and T bases. Thus, although this part of the ligand does interact with DNA, it does not appear to be constrained to the G-planes.
Figure 4.
Difference in chemical shifts of DNA H8 or H6 protons upon distamycin binding (distamycin complex minus DNA alone). From top to bottom, the panels correspond to [TAGGGGGT]4, [TAGGGGTT]4 and [TAGGGTTA]4.
Table 1. NOEs between distamycin and [TAGGGTTA]4 DNAa.
| Distamycin group | δ (p.p.m.) | NOEs to [TAGGGTTA]4b |
|---|---|---|
| Pyrrole CH3-1 | 3.51 | A2 H2, A2 H8, |
| G5 H1, G5 H1′, A8 H2, T6 CH3 | ||
| Pyrrole CH3-2 | 3.49 | T1 H1′, |
| G5 H1, G5 H8, T6 H6, T7 H1′, T7 H6 | ||
| Pyrrole CH3-3 | 3.59 | G3 H8, |
| G5 H1′, G5 H8, A8 H2, T7 CH3 | ||
| Pyrrole H3-1, H3-2 | 6.34 | T1 CH3, |
| T6 CH3, T6 H1′, T7 H1′ | ||
| Pyrrole H3-3 | 6.51 | T1 CH3, |
| G5 H1′, T6 CH3, T6 H1′, T6 H2′, T7 H1′ | ||
| Pyrrole H5-1 | 6.77 | T1 CH3, |
| T6 CH3, T6 H2′ | ||
| Pyrrole H5-2 | 6.71 | T6 CH3, T7 CH3 |
| Pyrrole H5-3 | 6.80 | T6 H1′, T7 CH3 |
| Formyl HF | 7.84 | T1 CH3, |
| T6 CH3 |
aNOESY τm = 250 ms, 35°C, D2O, molar ratio of 4:1 distamycin to G4 DNA molecule.
bFour additional weak NOEs exist between distamycin H3 and H5 protons and overlapping DNA deoxyribose protons.
Figure 5.
Sections from NOESY spectra (τm = 250 ms, 35°C) of distamycin-DNA complexes. The NOEs between distamycin methyls (∼3.5 p.p.m.) and G4 DNA are the strongest in the NOESY spectra. NOE cross-peaks directly above * are intra-DNA; D, distamycin. (A) Distamycin-[TAGGGGGT]4 (H2O). Cross-peaks from left to right, A2 H8, DHF, A2 H2, DNH (unassigned), G3 H8, G6 H8, G7 H8, T8 H6 and T1 H6. The resonance positions of G4 H8 and G5 H8 are also indicated. (B) Distamycin-[TAGGGGTT]4 (D2O). Cross-peaks from left to right, A2 H8, DHF, G3 H8, G6 H8, T7 H6, T8 H6 and T1 H6. The resonance positions of G4 H8 and G5 H8 are also indicated. (C) Distamycin-[TAGGGTTA]4 (D2O). Cross-peaks are from left to right, A2 H8, A8 H8, A2 H2, A8 H2, DHF, G5 H8, T6 H6, T7 H6 and T1 H6. The resonance position of G4 H8 is also indicated.
Distamycin binding is independent of length of G-runs and 3′ flanking region
The effect of extending the run of guanines (and thereby the length of the grooves) and truncating the flanking region 3′ of the G-plane was assessed by analysis of distamycin-[TAGGGGTT]4 and distamycin-[TAGGGGGT]4 DNA complexes. DNA peak assignments of these complexes were made based on sequential H1′ to H8 and H1 to H1 NOEs. As with [TAGGGTTA]4, chemical shift changes of the DNA H6/H8 signals induced by the ligand were restricted to the terminal planes and flanking bases (Fig. 4, middle and top panels). The intermolecular NOE patterns detected in these two complexes were essentially identical to those of the distamycin-[TAGGGTTA]4 complex (Fig. 5A and B), with the obvious exception of the missing 3′ base(s). One difference was noted in the distamycin-[TAGGGGGT]4 complex, which contains only a single base 3′ of the G-planes: an additional NOE between the distamycin methyl resonances and the DNA G6 H8 protons appeared. This is unlikely to be an intercalation as there are no NOEs from internal G6 H1 protons to any part of distamycin and the chemical shifts of G6 and G7 protons were not significantly influenced by ligand binding. In both examples of DNA with longer grooves, again, no NOEs between ligand and other internal planes were detected. Distamycin-G4 DNA contacts are therefore effectively independent of the length of the G-run or number of 3′ flanking bases.
DISCUSSION
We have shown that distamycin associates with four-stranded parallel G4 DNA via stacking interactions with the terminal guanine residues at the ends of G-runs and contacts to the bases that flank the G-runs. As diagrammed in Figure 6A, two molecules of distamycin lie flat on each of the terminal G-quartets at the end of a G4-stack. Distamycin pyrrole rings are separated by amides that could potentially hydrogen bond to another ligand. In this model, flanking DNA bases would sample conformations on top of the distamycin ligands.
Figure 6.
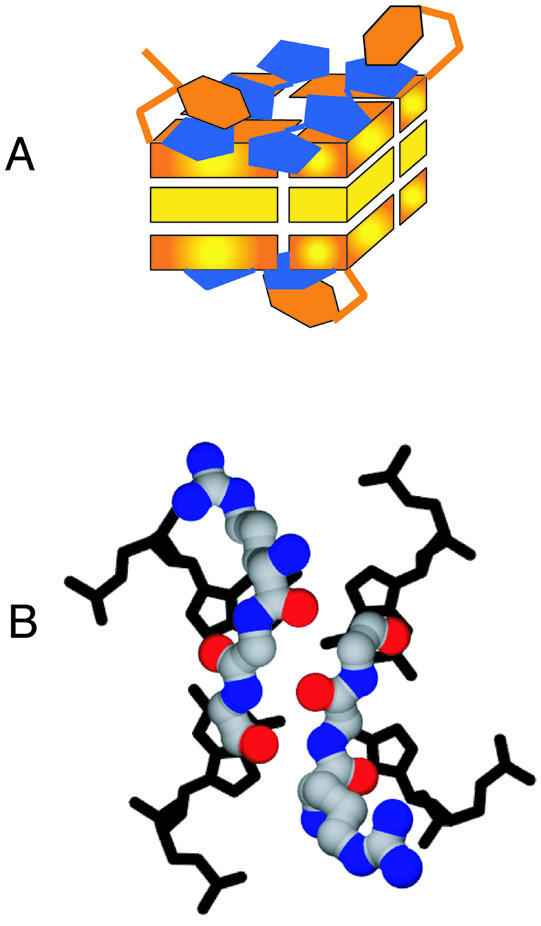
Models for interaction of distamycin and RGG repeats with G4 DNA. (A) A binding mode which allows two distamycin molecules to dock onto each terminal G-plane. Distamycin bound as shown would inhibit protein binding by occluding the terminal G-planes. Unfortunately, owing to the symmetry of the DNA and because the ligand is in fast exchange between multiple conformations with relatively few NOEs, the data are insufficient to derive a single structural model for binding. This is one model consistent with the NMR data. (B) Speculative model for the interaction of two protein RGG units with G4 DNA. The two RGGs appear in a short β-sheet conformation with the arginines positioned to form salt bridges with DNA phosphate groups.
Distamycin interactions with duplex DNA involve the minor groove of AT-rich regions (34). It had been speculated that distamycin binds to the groove of G4 DNA, just as it binds to the minor groove of duplex DNA (47). This is not the case. In fact, the binding mode we identify actually resembles more closely that used by porphyrins and tri-substituted acridines bound to G4 DNA (48,49). The planar G-quartets provide an ideal platform for the stacking of aromatic rings presented by distamycin. In binding to the DNA this way, the distamycin terminal cationic group could salt bridge to the DNA phosphate backbone, and distamycin could easily fit onto the G-plane in multiple conformations and satisfy the NMR constraints determined here.
Distamycin is intriguing in its ability to bind G-quadruplex and duplex DNA using very different modes. The four strands of parallel G4 DNA are separated by four identical grooves which are comparable in width (measured as distance between phosphate backbones) to the minor groove of AT-rich B-form DNA. Despite similarities in groove width, we find no evidence of interaction of distamycin with the groove of G4 DNA. This is not surprising since G-G grooves present only one potential hydrogen bond partner to this ligand, in contrast to the two provided by AT minor grooves in B-form DNA. In G4 DNA, the absence of suitable hydrogen bonding partners in the G-G groove is compounded by the steric impediment created by the protruding G-NH2 functionality. An analogous explanation has been put forward to account for the inability of distamycin to bind to GC-rich duplex DNA (34).
Distamycin inhibits G4 DNA binding by the RGGs of nucleolin, but not by the RBDs. These conserved domains therefore recognize distinct determinants in G4 DNA. Distamycin also does not inhibit G4 DNA unwinding by the RecQ family helicase BLM. In contrast, NMM is a potent inhibitor of G4 DNA unwinding by BLM, but it does not prevent helicase binding to G4 DNA (21). Distamycin, NMM and other G4 DNA ligands have been shown to influence protein activities on G4 DNA in the micromolar range (49–61), comparable to the inhibition concentration we find for distamycin and nucleolin RGG binding. Like distamycin, porphyrin derivatives probably bind to the quartets at the ends of G-runs (52). However, NMM is a covalently closed ring that could effectively seal the ends of the G4 DNA structure, preventing the contact with the bases that is necessary for a helicase to destabilize DNA structure. In contrast, each bound distamycin molecule bridges only two of the four strands, leaving open the possibility for a helicase to separate the strands of the DNA structure.
A possible model for the interactions of RGG domains with G4 DNA which is consistent with the NMR and binding inhibition data is shown in Figure 6B. The model assumes that the RGG domain makes contact with the region of G4 DNA that is occluded by distamycin. The protein conformation is shown as two short antiparallel β-strand segments, in which the two RGG units span one of the terminal G-planes, with the arginines positioned to form salt bridges with the DNA phosphate backbone. The glycines in the motif generate a β-sheet without protruding side chains and allow for tight turns around the DNA. The secondary structure of the RGG domain free in solution contains very small amounts of short β-turns that are stabilized at high salt concentrations (62). It is possible that the conformation of the RGG domain changes significantly upon binding to G4 DNA, and this is a question we are continuing to explore.
The ability of telomeric repeats to form G4 DNA has suggested that small molecules that bind this structure might have therapeutic potential (see, for example, 58–61). Interestingly, a distamycin derivative was recently found to inhibit telomerase activity (61). The fact that distamycin can inhibit binding to G4 DNA by a specific conserved polypeptide motif suggests that this ligand, or its derivatives, might be useful in the design of therapeutics targeted against specific proteins in vivo.
SUPPLEMENTARY MATERIAL
Tables of chemical shifts of three DNA complexes, alone and in the presence of distamycin, are available as Supplementary Material at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Gauri Dhavan for assessing the purity of NMR DNA samples by gel electrophoresis, Jim Prestegard for advice on NMR gradient pulses, and Don Crothers, Peter Moore and Scott Strobel for comments on the manuscript. This work was supported by NIH grants R01 GM39799 and R01 GM65988 to N.M.
REFERENCES
- 1.Gellert M., Lipsett,M.N. and Davies,D.R. (1962) Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA, 48, 2014–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D. and Gilbert,W. (1988) Formation of parallel four-stranded complexes by guanine rich motifs in DNA and its implications for meiosis. Nature, 334, 364–366. [DOI] [PubMed] [Google Scholar]
- 3.Sen D. and Gilbert,W. (1990) A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature, 344, 410–414. [DOI] [PubMed] [Google Scholar]
- 4.Sen D. and Gilbert,W. (1992) Novel DNA superstructures formed by telomere-like oligomers. Biochemistry, 31, 65–70. [DOI] [PubMed] [Google Scholar]
- 5.Sundquist W.L. and Klug,A. (1989) Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature, 342, 825–829. [DOI] [PubMed] [Google Scholar]
- 6.Sundquist W.I. (1993) Conducting the G quartet. Curr. Biol., 3, 893–895. [DOI] [PubMed] [Google Scholar]
- 7.Kim J., Cheong,C. and Moore,P.B. (1991) Tetramerization of an RNA oligonucleotide containing a GGGG sequence. Nature, 351, 331–332. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert D.E. and Feigon,J. (1999) Multistranded DNA structures. Curr. Opin. Struct. Biol., 9, 305–314. [DOI] [PubMed] [Google Scholar]
- 9.Giraldo R. and Rhodes,D. (1994) The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J., 13, 2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frantz J.D. and Gilbert,W. (1995) A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem., 270, 20692–20697. [DOI] [PubMed] [Google Scholar]
- 11.Frantz J.D. and Gilbert,W. (1995) A yeast gene product, G4p2, with a specific affinity for quadruplex nucleic acids. J. Biol. Chem., 270, 9413–9419. [DOI] [PubMed] [Google Scholar]
- 12.Sarig G., Weisman-Shomer,P., Erlitzki,R. and Fry,M. (1997) Purification and characterization of qTBP42, a new single-stranded and quadruplex telomeric DNA-binding protein from rat hepatocytes. J. Biol. Chem., 272, 4474–4482. [DOI] [PubMed] [Google Scholar]
- 13.Sun H., Karow,J.K., Hickson,I.D. and Maizels,N. (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem., 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- 14.Sun H., Bennett,R.J. and Maizels,N. (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res., 27, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanakahi L.A., Sun,H. and Maizels,N. (1999) High affinity interactions of nucleolin with G-G paired rDNA. J. Biol. Chem., 274, 15908–15912. [DOI] [PubMed] [Google Scholar]
- 16.Dempsey L.A., Sun,H., Hanakhai,L.A. and Maizels,N. (1999) G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D. J. Biol. Chem., 274, 1066–1071. [DOI] [PubMed] [Google Scholar]
- 17.Fry M. and Loeb,L.A. (1999) Human werner syndrome DNA helicase unwinds tetrahelical sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 18.Muniyappa K., Anuradha,S. and Byers,B. (2000) Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol., 20, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eversole A. and Maizels,N. (2000) In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol., 20, 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H., Yabuki,A. and Maizels,N. (2001) A human nuclease specific for G4 DNA. Proc. Natl Acad. Sci. USA, 98, 12444–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber M.D., Lee,D.C. and Maizels,N. (2002) G4 DNA unwinding by BLM and Sgs1p: substrate-specificity and substrate-specific inhibition. Nucleic Acids Res., 30, 3954–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X. and Maizels,N. (2001) Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res., 29, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darnell J.C., Jensen,K.B., Jin,P., Brown,V., Warren,S.T. and Darnell,R.B. (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell, 107, 489–499. [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer C., Bardoni,B., Mandel,J.L., Ehresmann,B., Ehresmann,C. and Moine,H. (2001) The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J., 20, 4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown V., Jin,P., Ceman,S., Darnell,J.C., O’Donnell,W.T., Tenenbaum,S.A., Jin,X., Feng,Y., Wilkinson,K.D., Keene,J.D. et al. (2001) Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell, 107, 477–487. [DOI] [PubMed] [Google Scholar]
- 26.Oliver A.W., Bogdarina,I., Schroeder,E., Taylor,I.A. and Kneale,G.G. (2000) Preferential binding of fd gene 5 protein to tetraplex nucleic acid structures. J. Mol. Biol., 301, 575–584. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson G.N., Lee,M.P. and Neidle,S. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature, 417, 876–880. [DOI] [PubMed] [Google Scholar]
- 28.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 29.Ginisty H., Sicard,H., Roger,B. and Bouvet,P. (1999) Structure and functions of nucleolin. J. Cell Sci., 112, 761–772. [DOI] [PubMed] [Google Scholar]
- 30.LaBranche H., Dupuis,S., Ben-David,Y., Bani,M.-R., Wellinger,R.J. and Chabot,B. (1998) Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nature Genet., 19, 1–4. [DOI] [PubMed] [Google Scholar]
- 31.Kaytor M.D. and Orr,H.T. (2001) RNA targets of the fragile X protein. Cell, 107, 591–603. [DOI] [PubMed] [Google Scholar]
- 32.Mohaghegh P., Karow,J.K., Brosh,R.M.,Jr, Bohr,V.A.,Jr and Hickson,I.D. (2001) The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res., 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wemmer D.E. (2000) Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct., 29, 439–461. [DOI] [PubMed] [Google Scholar]
- 34.Kopka M.L., Yoon,C., Goodsell,D., Pjura,P. and Dickerson,R.E. (1985) The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc. Natl Acad. Sci. USA, 82, 1376–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 37.Goddard T.D. and Kneller,D.G. SPARKY 3. University of California, San Francisco, CA.
- 38.Lapham J., Rife,J.P., Moore,P.B. and Crothers,D.M. (1997) Measurement of diffusion constants for nucleic acids by NMR. J. Biomol. NMR, 10, 255–262. [DOI] [PubMed] [Google Scholar]
- 39.Tanner J.E. (1970) Use of the stimulated echo in NMR diffusion studies. J. Chem. Phys., 52, 2523–2526. [Google Scholar]
- 40.Brosh R.M. Jr, Li,J.L., Kenny,M.K., Karow,J.K., Cooper,M.P., Kureekattil,R.P., Hickson,I.D. and Bohr,V.A. (2000) Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y. and Patel,D.J. (1993) Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure, 1, 263–282. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y. and Patel,D.J. (1993) Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol., 234, 1171–1183. [DOI] [PubMed] [Google Scholar]
- 43.Wang K.Y., McCurdy,S., Shea,R.G., Swaminathan,S. and Bolton,P.H. (1993) A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry, 32, 1899–1904. [DOI] [PubMed] [Google Scholar]
- 44.Feigon J., Koshlap,K.M. and Smith,F.W. (1995) 1H NMR spectroscopy of DNA triplexes and quadruplexes. Methods Enzymol., 261, 225–255. [DOI] [PubMed] [Google Scholar]
- 45.Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 164–168. [DOI] [PubMed] [Google Scholar]
- 46.Fedoroff O.Y., Salazar,M., Han,H., Chemeris,V.V., Kerwin,S.M. and Hurley,L.H. (1998) NMR-based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry, 37, 12367–12374. [DOI] [PubMed] [Google Scholar]
- 47.Randazzo A., Galeone,A., Esposito,V., Varra,M. and Mayol,L. (2002) Interaction of distamycin A and netropsin with quadruplex and duplex structures: a comparative 1H-NMR study. Nucl. Nucl., 21, 535–545. [DOI] [PubMed] [Google Scholar]
- 48.Han H., Langley,D.R., Rangan,A. and Hurley,L.H. (2001) Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc., 123, 8902–8913. [DOI] [PubMed] [Google Scholar]
- 49.Read M., Harrison,R.J., Romagnoli,B., Tanious,F.A., Gowan,S.H., Reszka,A.P., Wilson,W.D., Kelland,L.R. and Neidle,S. (2001) Structure-based design of selective and potent G quadruplex-mediated telomerase inhibitors. Proc. Natl Acad. Sci. USA, 98, 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wemmer D.E. (2000) Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct., 29, 439–461. [DOI] [PubMed] [Google Scholar]
- 51.Pelton J.G. and Wemmer,D.E. (1990) Binding modes of distamycin A with d(CGCAAATTTGCG)2 determined by two-dimensional NMR. J. Am. Chem. Soc., 112, 1393–1399. [Google Scholar]
- 52.Anantha N.V., Azam,M. and Sheardy,R.D. (1998) Porphyrin binding to quadruplexed T4G4. Biochemistry, 37, 2709–2714. [DOI] [PubMed] [Google Scholar]
- 53.Ren J. and Chaires,J.B. (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry, 38, 16067–16075. [DOI] [PubMed] [Google Scholar]
- 54.Han H., Hurley,L.H. and Salazar,M. (1999) A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res., 27, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li J.L., Harrison,R.J., Reszka,A.P., Brosh,R.M.,Jr, Bohr,V.A., Neidle,S. and Hickson,I.D. (2001) Inhibition of the Bloom’s and Werner’s syndrome helicases by G-quadruplex interacting ligands. Biochemistry, 40, 15194–15202. [DOI] [PubMed] [Google Scholar]
- 56.Riou J.F., Guittat,L., Mailliet,P., Laoui,A., Renou,E., Petitgenet,O., Megnin-Chanet,F., Helene,C. and Mergny,J.L. (2002) Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl Acad. Sci. USA, 99, 2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David W.M., Brodbelt,J., Kerwin,S.M. and Thomas,P.W. (2002) Investigation of quadruplex oligonucleotide-drug interactions by electrospray ionization mass spectrometry. Anal. Chem., 74, 2029–2033. [DOI] [PubMed] [Google Scholar]
- 58.Bears D.J., Hurley,L.H. and Von Hoff,D.D. (2000) Telomere maintenance mechanisms as a target for drug development. Oncogene, 19, 6632–6641. [DOI] [PubMed] [Google Scholar]
- 59.Mergny J.L., Lacroix,L., Teulade-Fichou,M.P., Hounsou,C., Guittat,L., Hoarau,M., Arimondo,P.B., Vigneron,J.P., Lehn,J.M., Riou,J.F. et al. (2001) Telomerase inhibitors based on quadruplex ligands selected by a fluorescence assay. Proc. Natl Acad. Sci. USA, 98, 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neidle S. and Parkinson,G. (2002) Telomere maintenance as a target for anticancer drug discovery. Nature Rev. Drug Discov., 1, 383–393. [DOI] [PubMed] [Google Scholar]
- 61.Zaffaroni N., Lualdi,S., Villa,R., Bellarosa,D., Cermele,C., Felicetti,P., Rossi,C., Orlandi,L. and Daidone,M.G. (2002) Inhibition of telomerase activity by a distamycin derivative: effects on cell proliferation and induction of apoptosis in human cancer cells. Eur. J. Cancer, 38, 1792–1801. [DOI] [PubMed] [Google Scholar]
- 62.Ghisolfi L., Joseph,G., Amalric,F. and Erard,M. (1992) The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem., 267, 2955–2959. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.