Abstract
Double-stranded short interfering RNAs (siRNA) induce post-transcriptional silencing in a variety of biological systems. In the present study we have investigated the structural requirements of chemically synthesised siRNAs to mediate efficient gene silencing in mammalian cells. In contrast to studies with Drosophila extracts, we found that synthetic, double-stranded siRNAs without specific nucleotide overhangs are highly efficient in gene silencing. Blocking of the 5′-hydroxyl terminus of the antisense strand leads to a dramatic loss of RNA interference activity, whereas blocking of the 3′ terminus or blocking of the termini of the sense strand had no negative effect. We further demonstrate that synthetic siRNA molecules with internal 2′-O-methyl modification, but not molecules with terminal modifications, are protected against serum-derived nucleases. Finally, we analysed different sets of siRNA molecules with various 2′-O-methyl modifications for stability and activity. We demonstrate that 2′-O-methyl modifications at specific positions in the molecule improve stability of siRNAs in serum and are tolerated without significant loss of RNA interference activity. These second generation siRNAs will be better suited for potential therapeutic application of synthetic siRNAs in vivo.
INTRODUCTION
The term RNA-mediated interference (RNAi) was initially introduced by Fire and co-workers (1) to describe the observation that injection of double-stranded RNA (dsRNA) into the nematode Caenorhabditis elegans can block expression of genes highly homologous in sequence to the delivered dsRNA. RNAi is evolutionarily conserved among eukaryotes and it appears to have an essential role in protecting the genome against invasion by pathogens such as viruses, which generate dsRNA molecules upon activation and replication (2). Most recently two independent studies demonstrated that the yeast RNAi machinery is required for the formation and maintenance of heterochromatin during mitosis and meiosis, indicating additional functions for RNAi (3,4). In the past, repression of genes by long dsRNAs has been less successful in mammalian cells with the exception of embryonic cells (5,6). Use of short (<30 nt) synthetic dsRNAs allowed for sequence-specific gene silencing yet avoided the non-selective toxic effects of long dsRNAs in differentiated mammalian cells (7). This discovery triggered a much wider interest in the RNAi phenomenon since it provides a new avenue for loss of function studies in somatic cells of vertebrates. Initial studies with these small interfering RNAs (siRNAs) demonstrated that the duplex must have 2 or 3 nt overhanging 3′ ends for efficient cleavage destruction of the target mRNA (8). The 3′ overhangs can be generated by cleavage of long dsRNA into siRNAs by a multidomain RNase III-like enzyme, known as Dicer (9). In a subsequent step the siRNA associates with the RNAi-induced silencing complex (RISC), which is then guided to catalyse the sequence-specific degradation of the mRNA (9–11).
RNAi induced by synthetic siRNAs is transient, and in mammalian cell culture systems re-expression of the target mRNA usually occurs after a few days (12,13). Therefore an improvement in the intracellular and extracellular stability of the effector molecules will be one crucial aspect for the successful in vivo application of synthetic siRNAs. A variety of chemical modifications, including terminal and internal modifications (e.g. 2′-O-modification or phosphorothioate linkages), have been tested to see whether they influence RNAi inducing activity (7,8,13–15). However, these studies did not adequately assess the effect of the different modifications on sensitivity of these molecules to siRNA-degrading serum-derived nucleases.
In the present work we define the minimal structural requirements of siRNAs in mammalian cells and test a series of chemical modifications to improve the stability of siRNAs for future in vivo applications. To determine differences in potency, we measured the inhibition of expression of several targets, including PTEN, p110β and Akt1, which are all members of the phosphatidylinositol (PI) 3-kinase pathway (16,17). Surprisingly, we found no overhang dependence of the siRNA duplexes in HeLa cells, which is in contrast to observations made in the Drosophila system (15). We have established the minimal required size for functional siRNAs in HeLa cells and have developed functionally active siRNA molecules with increased nuclease resistance. Our results provide a basis for the further development of synthetic siRNA molecules with improved characteristics, including higher resistance to serum-derived endonucleases.
MATERIALS AND METHODS
Synthetic siRNAs and GeneBlocs
Synthetic siRNAs were purchased from BioSpring (Frankfurt, Germany). The oligoribonucleotides were resuspended in RNase-free TE to a final concentration of 50 µM. In the case of bimolecular siRNA molecules, equal aliquots (100 µM) were combined to a final concentration of 50 µM. For the formation of duplexes the siRNAs were incubated at 50°C for 2 min in annealing buffer (25 mM NaCl, 5 mM MgCl2) and were cooled down to room temperature. The PTEN-specific GeneBlocs used have the schematic structure cap-nnnnnnNNNNNNNNnnnnnn-cap, as published previously (16), where cap represents an inverted deoxy abasic modification, n stands for 2′-O-methyl ribonucleotides (A, G, U or C) and N represents phosphorothioate-linked deoxyribonucleotides (A, G, T or C).
Cell culture and transfections
The particular HeLa cell line used in the experiments presented was a gift from M. Gossen (MDC, Berlin, Germany) and was grown in Eagle’s minimum essential medium with 2 mM l-glutamine, Earle’s balanced salt solution, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids and 10% fetal bovine serum (FBS). Synthetic siRNA and antisense GeneBloc transfections were carried out in 96-well or 10 cm plates (at 30–50% confluency) by using cationic lipids such as Oligofectamine (Invitrogen, Carlsbad, CA) or NC388 (Atugen, Berlin, Germany) as reported previously (16). HeLa cells were transfected by adding a pre-formed 5× concentrated complex of siRNAs and lipid in serum-free medium to cells in complete medium. The total transfection volume was 100 µl for cells plated in 96-wells and 10 ml for cells in 10 cm plates. The final lipid concentration was 0.8–1.2 µg/ml depending on cell density; the siRNA concentration is indicated in each experiment.
Antibodies and immunoblotting
Cell lysates were prepared and aliquots of the cell extracts containing equal amounts of protein were analysed by immunoblotting as described previously (16,17). The murine monoclonal anti-p110α antibody has been described (18). Rabbit polyclonal anti-Akt and anti-phospho-Akt (S473) antibodies were obtained from Cell Signalling Technology. The murine monoclonal anti-PTEN antibody was from Santa Cruz Biotechnology.
Quantitation of mRNA by Taqman analysis
The RNA of cells transfected in 96-wells was isolated and purified using the Invisorb RNA HTS 96 kit (InVitek GmbH, Berlin, Germany). Inhibition of targeted mRNA expression was detected by real time RT–PCR (Taqman) analysis using 300 nM 5′ forward primer, 300 nM 3′ reverse primer and 100 nM Fam-Tamra-labelled Taqman probe. The gene-specific primer sequences can be obtained on request. The reaction was carried out in 50 µl and assayed in an ABI PRISM 7700 Sequence detector (Applied Biosystems) according to the manufacturer’s instructions under the following conditions: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C.
Nuclease resistance assay
For the stability assay 5 µl of (2.5 µM) siRNA was incubated in 50 µl of fetal bovine serum for 15 or 120 min at 37°C. The solution was extracted with phenol and siRNA was precipitated with ethanol and separated on 10% polyacrylamide gels followed by ethidium bromide staining. Equal amounts of siRNA before serum incubation (0 min) was extracted with phenol in parallel and loaded as an input control.
RESULTS
Comparison of synthetic siRNAs and antisense molecules in HeLa cells
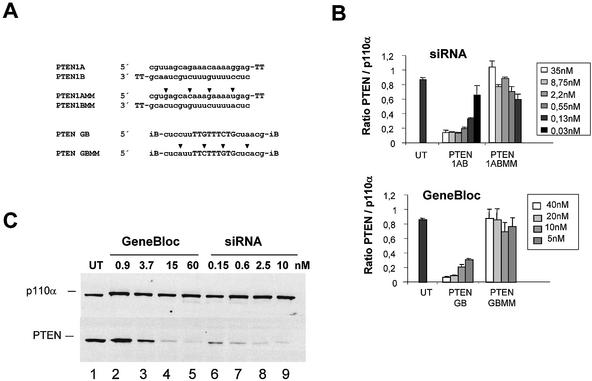
As most published studies with siRNA in mammalian cells have involved the knock-down of ectopically or abundantly expressed genes, we first set out to demonstrate that synthetic siRNAs can be efficient tools in reducing endogenous mRNA and protein levels. For this purpose we designed and analysed siRNA molecules specific for the tumour suppressor PTEN. For comparison and as a positive knock-down control, we employed in parallel conventional antisense molecules containing the same nucleotide sequences. The antisense molecules, here called GeneBlocs, consisted of nine internal deoxyribonucleotides for activation of RNase H flanked by six 2′-O-methyl-modified ribonucleotides, whereas the siRNAs used consisted of two 21mer ribonucleotides with two deoxythymidine nucleotides at the 3′ termini (Fig. 1A). The double-stranded siRNA as well as the single-stranded GeneBlocs are identical in length (21mer + terminal modifications). To control for non-specific transfection effects, we included mismatch sequences as negative controls. HeLa cells were transfected with increasing amounts of these molecules and the PTEN mRNA level was analysed 24 h later by real time PCR (Taqman). The maximal reduction of PTEN mRNA normalised to the mRNA of p110α, one of the catalytic subunits of PI 3-kinase, was reached with 2.2 nM siRNA and 20 nM GeneBloc (Fig. 1B). To verify the result on the mRNA level, we analysed protein reduction induced by antisense GeneBloc and siRNA transfection in a second set of experiments. HeLa cells were transfected with increasing amounts of these molecules and cell lysates were analysed 48 h later by immunoblot analysis using PTEN-specific antibodies. p110α protein level served as a loading control (Fig. 1C). Maximal PTEN protein knock-down was detected with 2.5 nM siRNA and 15 nM GeneBloc. These experiments demonstrate that siRNA molecules can efficiently reduce mRNA and protein levels of endogenous genes. Furthermore, these siRNAs can be more efficient in mediating mRNA reduction when compared to conventional antisense molecules directed against the same target sequence. The observed differences in efficacy may be due to different mechanisms of target recognition and/or degradation and may reflect the involvement of more efficient catalytic steps in the case of RNAi.
Figure 1.
mRNA and protein knock-down of endogenous PTEN and induced by transfection of siRNA or GeneBloc (GB, antisense) in HeLa cells. (A) The sense (A) and antisense (B) strands of siRNA targeting human PTEN mRNA are shown in comparison to the corresponding GeneBloc (antisense molecules). siRNAs were synthesised with 2 nt deoxythymidine (TT) 3′ overhangs. GeneBlocs representing the third generation of antisense oligonucleotides with inverted abasic (iB) end modifications (see Materials and Methods). The sequences of the respective mismatch controls (MM) containing 4 nt changes are shown. (B) Reduction of PTEN mRNA expression in siRNA- and GeneBloc-transfected HeLa cells. HeLa cells were transfected with the indicated amounts of siRNA and GeneBloc as described. After 24 h, RNA was prepared and subjected to real time RT–PCR (Taqman) analysis to determine PTEN mRNA levels relative to p110α mRNA levels. Each bar represents triplicate transfections (± SD). (C) Inhibition of PTEN protein expression analysed by immunoblot. The cells were harvested 48 h after transfection of the indicated amounts of GeneBlocs or siRNAs. Cell lysates were separated by SDS–PAGE and analysed by immunoblotting using anti-PTEN and anti-p110α antibody. The amount of p110α, a catalytic subunit of PI 3-kinase, was used as a loading control. Control cell extract from untransfected HeLa cells (UT) were loaded in the left lane.
No overhang requirements of siRNA duplexes in mammalian cells
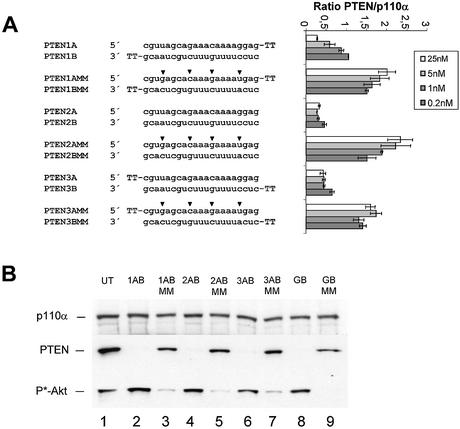
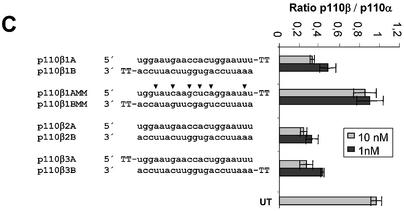
The structural–functional relationship of siRNAs has been extensively studied biochemically using Drosophila melanogaster embryo lysates (7,15). Using this system it has been demonstrated that duplexes with 2 nt 3′ overhangs were the most efficient triggers of mRNA degradation (8,15,19). To test whether a similar dependence is true for mammalian cells, we transfected PTEN siRNA molecules with 3′ or 5′ overhangs or without overhangs into HeLa cells. Surprisingly, we did not observe a more efficient knock-down of target RNA with siRNAs containing 3′ overhangs when compared to blunt molecules or those with 5′ overhangs (Fig. 2A). Similar results were obtained for the second target p110β (Fig. 2C). To verify the observed mRNA knock-down, we performed an immunoblot analysis using PTEN-specific antibodies (Fig. 2B). In these experiments the reduction of PTEN protein expression as well as the downstream phosphorylation of Akt1 kinase, a consequence of PTEN protein inhibition (16,17), was very similar with the different siRNA duplexes. We concluded from these data that 3′ overhangs on synthetic siRNAs are not essential for RNAi in HeLa cells.
Figure 2.
siRNA duplexes with a 3′ or 5′ overhang or without overhang (blunt) are equally potent in mediating gene silencing in HeLa cells. (A) Inhibition of PTEN mRNA expression in HeLa cells transfected with the indicated amounts of siRNA molecules. The sequences and different terminal structures of the siRNAs molecules are shown on the left. Mutations in the mismatch molecules are indicated by arrowheads. Samples were analysed in parallel for the level of PTEN mRNA expression 24 h after transfection by real time RT–PCR (Taqman) analysis. PTEN mRNA levels are shown relative to the mRNA levels of p110α, which served as internal reference. Each bar represents triplicate transfections (± SD). (B) Inhibition of PTEN protein expression by use of siRNAs with different terminal structures. The cells were harvested 48 h after transfection of the indicated siRNAs (30 nM) (lanes 2–7) or GeneBlocs (30 nM) (lanes 8 and 9). Cell extracts were separated by SDS–PAGE and analysed by immunoblotting using anti-p110α, anti-PTEN or anti-phospho-Akt antibody. The amount of p110α was used as a loading control and control cell extracts from untransfected HeLa cells (UT) were loaded in lane 1. (C) Inhibition of p110β mRNA expression in HeLa cells transfected with the indicated amounts of siRNA molecules. p110β mRNA levels are shown relative to the mRNA levels of p110α, which served as internal reference. The ratio of p110β/p110α mRNA of untransfected HeLa cells is shown at the bottom (UT). Each bar represents triplicate transfections (± SD).
Duplex length requirements of siRNA duplexes in mammalian cells
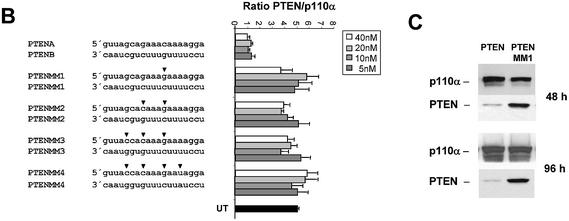
We next examined the effects of duplex length variations on siRNA activity. For these experiments we used the Akt1 kinase as a target. Duplexes of 19 nt length were highly efficient in reducing Akt1 mRNA levels independent of the nature (deoxyribonucleotides or ribonucleotides) of the 3′ overhang (Fig. 3A, compare molecules 1AB, 2AB, 3AB and 4AB). This result is consistent with our observation that a 3′ overhang appears not to be crucial for siRNA function in HeLa cells. Next we reduced the duplex length to 17 nt (Fig. 3A, molecule 5AB). This molecule did show a dramatically reduced silencing activity, suggesting that active siRNA duplexes must have a minimal length (∼19 nt), which is in agreement with experiments assessing activity of siRNA molecules with different duplex lengths in Drosophila extracts (15). This minimal duplex length for active siRNA molecules might be explained mechanistically by two different requirements. First, a minimal base pairing between the antisense siRNA and the target mRNA may be obligatory or, second, incorporation into the RISC requires a minimal length of the siRNA duplex. To address this question we synthesised and transfected 19 nt long siRNA duplex molecules with one and two terminal mutations (CG and UA inversion) relative to the wild-type sequence (Fig. 3A, molecules 6AB and 7AB). Both molecules, even the molecule with a stretch of only 15 nt base pairing to the target mRNA, were functional in reducing the Akt1 mRNA level. We concluded from this result that the duplex length itself, but not the base pairing of the antisense siRNA with the target mRNA, seems to determine the minimal length of functional siRNAs. These data suggest that the length of the double-stranded helix is an important determinant for incorporation into the RISC complex. Since the introduced mismatches at the terminal ends of the siRNA duplexes had little effect on RNAi, we wanted to analyse the effect of mismatches located in the centre of the molecule (Fig. 3B). For these experiments we used 19 nt long blunt siRNAs directed against PTEN mRNA. The sequence changes in one siRNA strand were compensated by complementary changes in the other strand to avoid disrupting duplex formation. A siRNA with only one point mutation in the centre of the molecule was severely compromised in its ability to reduce mRNA and protein expression levels (Fig. 3B and C). This result indicates that the RNAi machinery is highly discriminative between perfect and imperfect base pairing between target mRNA and siRNA in the centre of the duplex. The dependence on a perfect complementarity between target and siRNA has been investigated for RNAi before (13–15,19).
Figure 3.
Duplex length requirement and tolerance for mutation in siRNAs in HeLa cells. (A) Inhibition of Akt1 mRNA expression in HeLa cells transfected with the indicated amounts of siRNA molecules. The sequences, lengths and different terminal structures (3′ deoxyribonucleotides in upper case letters) of the siRNA molecules are shown on the left. The nucleotide changes in the mismatch siRNA molecule are indicated by arrowheads. Samples were analysed in parallel for the level of Akt1 and p110α mRNA expression 24 h after transfection by real time RT–PCR (Taqman) analysis. The mRNA levels of p110α served as internal reference. Each bar represents triplicate transfections (± SD). (B) Inhibition of PTEN mRNA expression in HeLa cells transfected with the indicated amounts of siRNA molecules. The sequences of the corresponding siRNA molecules are shown on the left. (C) Inhibition of PTEN protein expression analysed by immunoblot. The cells were harvested 48 or 96 h after transfection of the indicated siRNAs (30 nM). Cell extracts were separated by SDS–PAGE and analysed by immunoblotting as described previously. The positions of PTEN and p110α, which was used as a loading control, are indicated on the left.
Effects on RNAi and siRNA stability of 5′ and 3′ terminal modifications
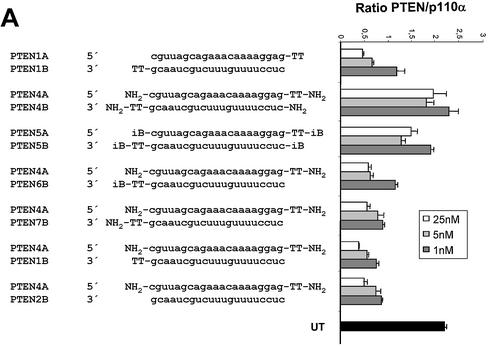
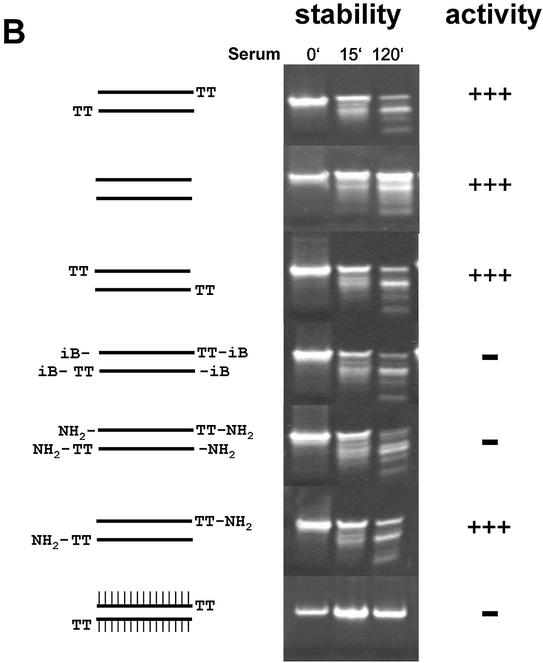
Having established the minimal structural requirements of siRNA duplexes, we wanted to test whether chemical end modifications can lead to more stabilised, active siRNA molecules. As a starting point we have used an inverted deoxy abasic (iB) modification (for details and structure see 16) and an amino end modification (NH2) (amino group with 6-carbon linker at the terminal phosphate; for details see 20). Similar end modifications have been successfully used to stabilise conventional antisense molecules and ribozymes (21) and are generally considered to protect therapeutic nucleic acids against serum-derived exonuclease activities (22). RNAi induction by siRNA duplexes with an NH2 or iB modification was dramatically reduced when all four termini were modified (Fig. 4A, compare molecule 1AB with 4AB and 5A5B). However, siRNA molecules which were modified only at the terminus of the sense strand showed no reduction in gene silencing activity (Fig. 4A, molecule 2B4A). Even siRNA molecules, which had no end protection at the 5′-end of the antisense strand but on all other termini, showed no reduction in RNAi activity (Fig. 4A, molecules 6B4A and 7B4A). This result is in agreement with recently published results demonstrating the importance of the 5′-hydroxyl group of the antisense strand for RNAi (20,23). In order to test whether the terminal modifications were capable of increasing stability in serum, we incubated the different siRNA duplexes at 37°C in calf serum, followed by separation on 10% polyacrylamide gels. Surprisingly we were not able to detect any increase in stability or nuclease resistance (Fig. 4B). Neither the NH2 nor the iB end modification stabilised the siRNA duplexes significantly. This result suggests that siRNA molecules are degraded predominantly by serum-derived endonucleases. To test this hypothesis we synthesised siRNA duplex containing internal 2′-O-methyl ribonucleotides at all positions, a commonly used modification to increase stability of RNA-containing molecules (24). This internally modified molecule showed extreme resistance against degradation in our serum incubation assay (Fig. 4B, siRNA molecule at the bottom), and therefore might have better pharmacodynamic properties in vivo.
Figure 4.
RNAi activity and stability in serum of modified siRNA molecules. (A) Activity of siRNA with modified termini. 19mer duplex siRNAs specific for PTEN mRNA were synthesised with 2 nt deoxythymidine (TT) 3′ overhangs or without overhangs. iB represents inverted deoxy abasic end modifications, NH2 represents end protection with amino-C6 linker at the terminal phosphates. Inhibition of PTEN mRNA expression in HeLa cells transfected with the indicated amounts of modified siRNA molecules was determined by Taqman analysis as described before. (B) Stability assay of siRNAs with different chemical modifications. The structure of the modified siRNA molecules are schematically shown on the left. The siRNA molecule at the bottom was synthesized with 2′-O-methyl ribonucleotides (A, G, U and C) at all positions, indicated by IIIIII. Polyacrylamide gel (10%) electrophoresis of the indicated siRNA molecules after incubation in serum as described in Materials and Methods.
Synthetic siRNA molecules with specific internal 2′-O-methyl modification mediate RNAi and have increased stability in serum
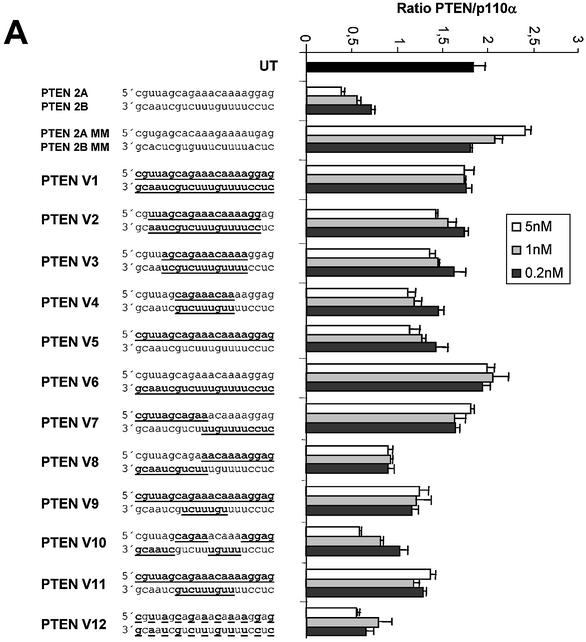
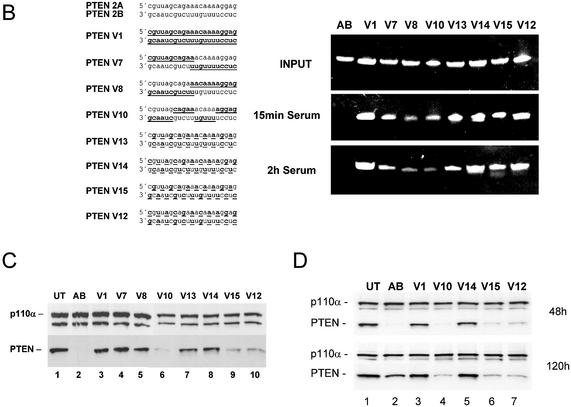
To identify synthetic siRNA molecules which have increased stability, but are also able to efficiently induce RNAi, we tested a series of molecules with 2′-O-methyl residues at different positions (Fig. 5). siRNA molecules with either one or both strands consisting of 2′-O-methyl residues were not able to induce RNAi in our mammalian system (Fig. 5A, molecules V2, V5 and V6). Similar results were observed with 2′-O-methyl-modified siRNAs in Drosophila lysates employing a luciferase-based RNAi assay (15). However, the decrease in activity was less pronounced when only parts of the strands were modified. This result is in agreement with a recent study employing siRNAs, which were not modified in the core but contained different internal modifications at the terminal nucleotides of both strands (14). Interestingly, a molecule with an unmodified antisense strand (lower) and a completely modified sense strand was significantly more active when compared to the reversed version (Fig. 5A, compare molecules V5 and V6). This result suggests again that the antisense strand of the siRNA seems to be more critical and sensitive to modification. The most efficient molecules in reducing PTEN mRNA had only stretches of modification leaving the 5′-end unmodified or were modified on alternating positions on both strands (Fig. 5A, molecules V10 and V12). At this point it was crucial to analyse the stability and activity of partially 2′-O-methyl-modified siRNA molecules in more detail. To demonstrate nuclease resistance we incubated the different siRNA versions in serum followed by polyacrylamide gel electrophoresis. As shown before, blunt-ended siRNA molecules with unmodified ribonucleotides were very rapidly degraded whereas a complete substitution with 2′-O-methyl nucleotides mediated resistance against serum-derived nucleases (Fig. 5B, compare molecule AB with V1). siRNA molecules with partial 2′-O-methyl modification also showed an increased stability when compared to unmodified siRNAs. Especially, molecules with alternating modifications on both strands showed a significant improvement in stability (Fig. 5B, molecules V13, V14, V15 and V12). More importantly, transfection of three of these molecules into HeLa cells did result in a significant down-regulation of PTEN protein expression (Fig. 5C, lanes 6, 9 and 10). Detection of p110α protein by immunoblot was used as a loading control. In this RNAi activity assay we observed an unexpected preference for molecules which were modified at every second nucleotide beginning with the most 5′ terminal nucleotide of the antisense strand (molecules V15 and V12). Molecules which contained modifications beginning with the second nucleotide at the 5′ end of the antisense strand were more stable but had a strongly reduced activity in gene silencing (molecules V13 and V14). This result points towards highly specific interactions between the involved enzymes and precise nucleotides in the siRNA duplex. Taken together our data demonstrate that 2′-O-methyl modifications at particularly selected positions in the siRNA duplex can increase nuclease resistance and do not necessarily abolish RNAi completely.
Figure 5.
(Opposite) siRNA molecules with distinct 2′-O-methyl ribonucleotide modifications show increased stability in serum and mediate protein knock-down in HeLa cells. (A) RNAi activity of siRNA molecules with various 2′-O-methyl ribonucleotide modifications. Inhibition of PTEN mRNA expression in HeLa cells transfected with the indicated amounts of modified siRNA molecules. 2′-O-methyl ribonucleotide modifications are underlined and indicated by bold letters in the sequence on the left. Samples were analysed in parallel for the level of PTEN mRNA as described before. (B) Polyacrylamide gel electrophoresis of modified and unmodified siRNA molecules after incubation in serum. The PTEN siRNA sequence and the position of the 2′-O-methyl ribonucleotide modifications (underlined and bold letters) are shown on the left. (C) Inhibition of PTEN protein expression analysed by immunoblot. The cells were harvested 48 h after transfection of the indicated siRNAs (30 nM). Cell extracts were separated by SDS–PAGE and analysed by immunoblotting as described previously. The positions of PTEN and p110α, which was used as a loading control, are indicated at left. (D) siRNA molecules with distinct 2′-O-methyl ribonucleotide modifications mediate a prolonged protein knock-down. Inhibition of PTEN protein expression analysed by immunoblot. The cells were harvested 48 or 120 h after transfection of the indicated siRNAs (30 nM). Cell extracts were separated by SDS–PAGE and analysed by immunoblotting as described previously. The positions of PTEN and p110α, which was used as a loading control, are indicated on the left.
An increased stability of synthetic siRNAs should primarily have implications for in vivo application, e.g. mouse models or in therapeutic applications of siRNA. Nevertheless we wanted to analyse whether the introduced modification can also lead to an extended protein knock-down in cell culture systems. In order to demonstrate this effect, we transfected HeLa cells transiently for 6 h using our cationic lipids with different versions of PTEN-specific siRNAs. The lipid siRNA complex was then washed away and the PTEN protein knock-down was analysed 48 and 120 h later. Generally knock-down experiments without continuous transfection of siRNAs are complicated due to rapid growth of untransfected cells in this time period resulting in a very transient knock-down (13). However, here we were able to demonstrate a prolonged PTEN protein knock-down with siRNA molecules stabilised by the described 2′-O-methyl modifications. At 48 h post-transfection the unmodified siRNA (AB) shows the biggest reduction in PTEN protein levels; however, at 120 h post-transfection the reduction in PTEN protein expression is superior with the siRNAs stabilised by alternating 2′-O-methyl modifications (Fig. 5D, compare lane 2 with lanes 4, 6 and 7). At this point we do not know whether this increase in stability will also improve the pharmacodynamic properties of these molecules, and it is very likely that further modifications and improvements in delivery are necessary to enable the use of synthetic siRNAs in vivo.
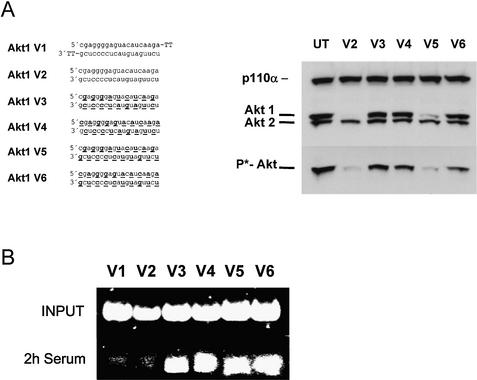
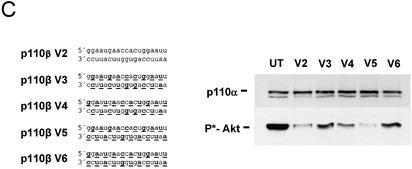
It was puzzling that not all siRNA molecules with alternating 2′-O-methyl modifications on both strands were functional. As mentioned above, the starting nucleotide position of the alternating modification seems to be important. To test this preference in more detail, we synthesised and tested for functionality two additional series of siRNAs, one specific for the kinase Akt1, the other specific for p110β, one of the two catalytic subunits of PI 3-kinase. Here we used shorter, only 19 nt long, siRNA duplexes either without any or with 2′-O-methyl modification on every second nucleotide (Fig. 6). Using Akt1 as a target we observed an efficient protein knock-down as well as a dramatic reduction in phospho-Akt levels with blunt, unmodified siRNAs (Fig. 6A, right panel). From the different versions of molecules with modifications on every second nucleotide, only one efficiently mediated RNAi (Fig. 6A, molecule V5). This siRNA molecule contained an antisense strand which was modified at the most terminal 5′ and 3′ nucleotides. The sense strand started with the unmodified nucleotides at the most terminal positions, resulting in a structure in which the modified and unmodified ribonucleotides of both strands are facing each other. As expected, this molecule was also protected against serum-derived nucleases (Fig. 6B, molecule V5). Interestingly, a very similar 19 nt long siRNA molecule (V4) with modifications beginning at the second nucleotide of the antisense strand showed no RNAi activation in our assay. Molecule V6, in which the modified nucleotides of the antisense strand face modified nucleotides on the sense strand, was also inactive in this experiment. An identical series of 19 nt long siRNA molecules specific for p110β confirmed these observations (Fig. 6C). Again, the similarly modified siRNA molecule (V5) was the most active, as indicated by reducing Akt phosphorylation, which is indicative of a reduced PI 3-kinase activity due to reduced p110β levels (16,17). The reduced activity of the V6 molecule might be explained by reduced duplex stability since the same structure was active in the PTEN knock-down experiment with 21mer siRNAs. It is well documented that 2′-O-methyl modification on both strands facing each other will reduce the stability of nucleic acid duplexes (25). However, the difference between the activity of siRNA molecules V4 and V5 (Fig. 6B and C) in these experiments is probably not due to differences in duplex stability since the number of base pairings of modified and unmodified nucleotides is identical. This difference in activity might be due to specific requirements of the interacting proteins involved in the degradation of the target mRNA. Interestingly, our data demonstrate that the most terminal nucleotides of the antisense strand can be modified at the 2′-hydroxyl group without significant loss of silencing activity.
Figure 6.
siRNA molecules with distinct 2′-O-methyl ribonucleotide modifications specific for Akt1 and p110β mRNA show increased stability in serum and mediate protein knock-down in HeLa cells. (A) The Akt1-specific siRNA sequence, the position of the 2′-O-methyl modifications (underlined and bold letters) and the integrity of the indicated siRNA molecules after incubation in serum are shown. (B) Inhibition of Akt1 protein expression as well as Akt phosphorylation analysed by immunoblot after transfection of the indicated siRNAs (30 nM). (C) Inhibition of the phosphorylation of the downstream kinase Akt1 by 2′-O-methyl-containing p110β-specific siRNAs. The positions of the phoshorylated Akt1 and p110α, which was used as a loading control, are indicated on the left.
Taken together the presented differences in the potency of different siRNA chemistries suggest that not only an alternating design principle but also the specific nucleotide positions of the internal 2′-O-alkyl modifications in the siRNA duplex are crucial to balance beneficial stabilising effects with activity reducing effects.
DISCUSSION
siRNAs represent a new tool for functional knock-down studies in mammalian cells (7). In contrast to long dsRNA, siRNAs do not activate the dsRNA-dependent protein kinase response (8,26) and are thus better suited to phenotypic loss of function studies. In order to facilitate the usage of this new class of gene knock-down tools as potential therapeutic nucleic acids, we have analysed the minimal structural requirements of siRNA function in HeLa cells. A comprehensive study on the functional anatomy of siRNAs in Drosophila has been published by Elbashir and colleagues (15). In this study an in vitro RNAi assay with D.melanogaster embryo lysates was used to measure the silencing activity of different types of siRNAs. Here we have used cationic delivery vehicles to transfect different siRNA molecules of various chemistries into HeLa cells and have quantitated the gene silencing activity by Taqman analysis. Our analysis reveals some striking differences in the structural requirements for siRNAs between the Drosophila system and the mammalian system. For instance, in Drosophila extracts it seems to be obligatory to use siRNAs with 3′ overhangs (15) whereas no particular overhang requirement was observed in the HeLa cell system (Fig. 2). Previously, Drosophila microinjection experiments with phosphorylated siRNAs, lacking 3′ overhangs, showed moderate activity at high concentrations (19). At this point we cannot exclude the formation of a 3′ overhang by an intracellular nuclease activity such as Dicer in HeLa cells. However, a potential processing of these 19 nt long siRNAs would lead to molecules which are most likely too short to be functional (see below). Regarding the minimal length for functional siRNA duplexes in mammalian cells, we concluded that efficient RNAi can be achieved with blunt 19 nt long siRNA duplexes in mammalian cells. There are at least three different factors which may influence the minimal length requirement: (i) a minimal length of the double-stranded siRNA duplex which has to be recognised by the RISC complex or other protein complexes involved; (ii) a minimal stretch of homologous base pairing between the target mRNA and the antisense strand of the siRNA duplex to ensure specific recognition; (iii) a minimal siRNA length to guarantee stable duplex formation. It seems very likely that factors (ii) and (iii) will be dependent on the particular sequence composition (e.g. GC content). However, from our data we conclude that the duplex length itself represents one critical factor below a duplex length of 19 bp since a 17 bp siRNA duplex was inactive, even in the presence of additional target-specific overhangs (Fig. 3A). A 19 bp long siRNA duplex with up to four mismatches (with respect to the target) at the terminal positions still mediated efficient gene silencing, suggesting that the base pairing with the target mRNA is not the most decisive factor for determining the minimal length. The result that 19 nt long siRNA with only a 15 nt target-specific stretch (two mismatches on either side) was able to mediate a significant knock-down may have general implications for target specificity and off-target effects of siRNAs. In contrast, a single nucleotide inversion in the centre of the siRNA duplex resulted in a complete loss of gene silencing, suggesting that a longer undisrupted stretch of base pairing with the target mRNA is necessary. Several reports published on the issue of sequence specificity of target recognition have shown that single mutations within the centre of a siRNA duplex are more discriminating than mutations located at the 5′- and 3′-ends (13–15). More recently it has been suggested that even single-stranded antisense siRNA molecules, preferably phosphorylated at the 5′-hydroxyl, can be sufficient to mediate RNAi in vitro as well as in cell culture systems (14,27,28). We have attempted to repeat these studies in HeLa cells using our siRNA transfection protocol followed by Taqman or immunoblot analysis. So far we have not been able to demonstrate significant knock-down with single-stranded siRNA molecules either with or without phosphorylated 5′ termini (data not shown). Currently we do not know whether this discrepancy may be due to a difference in cell lines or transfection protocols used in these experiments.
One major drawback of the usage of siRNAs is its transient nature owing to the limited stability and availability of the synthetic RNA-based molecules. Using conventional transient transfection protocols gene expression is inhibited for a maximum period of 2–4 days (14,29). Therefore, we reasoned that it will be beneficial to develop more stable siRNA molecules, especially for potential in vivo application of siRNAs, where sustained delivery will be one major obstacle. Our data on siRNA stability in serum suggest that unmodified siRNAs are for the most part substrates for endonucleases and to a lesser extent substrates for exonucleases. Consequently, chemical modifications at the termini of the RNA strands which are known to abolish exonuclease activity do not significantly improve stability of siRNAs in serum (Fig. 4B). Initial experiments with internal 2′-O-alkyl modifications have demonstrated that complete substitution of all nucleotides abolished RNAi activity completely (15). Partial substitution with up to a total of six modifications in the form of either methylation or thiolation of the backbone has recently been reported to reduce silencing activity only marginally in HaCat cells (14). According to our data it is essential to balance the effects of 2′-O-alkyl modifications on stability against the negative effects on activity, which makes it important to be aware of various design principles. First, modifications at the 5′-hydoxyl group of the antisense strand should be avoided since end modifications (-NH2 or inverted abasic) at this position abolish RNAi inducing activity (Fig. 4A). This result is consistent with a proposed in vivo phosphorylation step of the 5′ terminus necessary for siRNA activity (23). Second, alternating 2′-O-alkyl modification but not blocks of 2′-O-alkyl modification lead to significant resistance against serum-derived nucleases without significant loss of RNAi activity (Fig. 5). Third, 2′-O-methyl-modified siRNAs are more active when the incorporated modifications are on every second nucleotide, facing a non-modified nucleotide. This may indicate critical changes in duplex stability since it is known that 2′-O-methyl modifications stabilise base pairing with non-modified nucleotides but can destabilise the duplex when incorporated on both strands (25). A fourth feature arises from the observation that siRNAs in which the alternating modifications at the 2′-hydroxyl group start with the most terminal nucleotides of the antisense strand are more efficient than siRNAs which begin this modification at the second nucleotide of the antisense strand (Fig. 6, compare molecules V3 and V5). Taken together the results suggest that the specific structural requirements for RNAi inducing activity are at least partially distinct from the substrate requirements of serum-derived endonucleases. In summary, we conclude that specific internal modifications at the 2′-hydroxyl group are suitable to improve the stability of synthetic siRNAs without significant loss of activity, and thus may also help to advance the use of siRNAs in in vivo experiments.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the members of the Atugen team for many helpful comments during the experiments. We are grateful to Manfred Gossen and Katharina Ahrens for critical reading of the manuscript. This study was supported by a grant from the Bundesministerium für Wirtschaft und Technologie (no. 1078/02).
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 3.Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 4.Hall I.M., Shankaranarayana,G.D., Noma,K., Ayoub,N., Cohen,A. and Grewal,S.I. (2002) Establishment and maintenance of a heterochromatin domain. Science, 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- 5.Billy E., Brondani,V., Zhang,H., Muller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paddison P.J., Caudy,A.A. and Hannon,G.J. (2002) Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 10.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 11.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 12.Tuschl T. and Borkhardt,A. (2002) Small interfering RNAs: a revolutionary tool for the analysis of gene function and gene therapy. Mol. Interv., 2, 158–167. [DOI] [PubMed] [Google Scholar]
- 13.Holen T., Amarzguioui,M., Wiiger,M.T., Babaie,E. and Prydz,H. (2002) Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res., 30, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amarzguioui M., Holen,T., Babaie,E. and Prydz,H. (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res., 31, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sternberger M., Schmiedeknecht,A., Kretschmer,A., Gebhardt,F., Leenders,F., Czauderna,F., Von Carlowitz,I., Engle,M., Giese,K., Beigelman,L. et al. (2002) GeneBlocs are powerful tools to study and delineate signal transduction processes that regulate cell growth and transformation. Antisense Nucleic Acid Drug Dev., 12, 131–143. [DOI] [PubMed] [Google Scholar]
- 17.Czauderna F., Fechtner,M., Aygun,H., Arnold,W., Klippel,A., Giese,K. and Kaufmann,J. (2003) Functional studies of the PI(3)-kinase signalling pathway employing synthetic and expressed siRNA. Nucleic Acids Res., 31, 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klippel A., Escobedo,J.A., Hirano,M. and Williams,L.T. (1994) The interaction of small domains between the subunits of phosphatidylinositol 3-kinase determines enzyme activity. Mol. Cell. Biol., 14, 2675–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 20.Chiu Y.L. and Rana,T.M. (2002) RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell, 10, 549–561. [DOI] [PubMed] [Google Scholar]
- 21.Beigelman L., McSwiggen,J.A., Draper,K.G., Gonzalez,C., Jensen,K., Karpeisky,A.M., Modak,A.S., Matulic-Adamic,J., DiRenzo,A.B., Haeberli,P. et al. (1995) Chemical modification of hammerhead ribozymes. Catalytic activity and nuclease resistance. J. Biol. Chem., 270, 25702–25708. [DOI] [PubMed] [Google Scholar]
- 22.Crooke R.M., Graham,M.J., Martin,M.J., Lemonidis,K.M., Wyrzykiewiecz,T. and Cummins,L.L. (2000) Metabolism of antisense oligonucleotides in rat liver homogenates. J. Pharmacol. Exp. Ther., 292, 140–149. [PubMed] [Google Scholar]
- 23.Nykanen A., Haley,B. and Zamore,P.D. (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321. [DOI] [PubMed] [Google Scholar]
- 24.Lubini P., Zurcher,W. and Egli,M. (1994) Stabilizing effects of the RNA 2′-substituent: crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem. Biol., 1, 39–45. [DOI] [PubMed] [Google Scholar]
- 25.Blackburn G.M. and Gait.,M.J. (1996) Nucleic Acids in Chemistry and Biology, 2nd Edn. Oxford University Press, Oxford and New York.
- 26.Clemens M.J. (1997) PKR—a protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol., 29, 945–949. [DOI] [PubMed] [Google Scholar]
- 27.Martinez J., Patkaniowska,A., Urlaub,H., Luhrmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 28.Schwarz D.S., Hutvagner,G., Haley,B. and Zamore,P.D. (2002) Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell, 10, 537–548. [DOI] [PubMed] [Google Scholar]
- 29.Tuschl T. (2002) Expanding small RNA interference. Nat. Biotechnol., 20, 446–448. [DOI] [PubMed] [Google Scholar]