Abstract
The MCM8 protein from HeLa cells, a new member of the MCM family, co-isolates through several steps with MCM6 and MCM7, and MCM8 co-immunoprecipitates with MCM4, MCM6 and MCM7, proteins reportedly forming a helicase complex involved in initiation of DNA replication. MCM8 mRNA is expressed in placenta, lung and liver, but is also significantly expressed in adult heart, a tissue with a low percentage of proliferating cells. The MCM8 gene, consisting of 19 exons, is located contrapodal to a gene, consisting of 11 exons, encoding a homolog of the yeast GCD10 gene product. The region between these two transcription units, comprising as few as 62 bp, is TATA-less and highly GC-rich, containing multiple CpG units. MCM8 expression is altered in certain forms of neoplasia. In a case of choriocarcinoma MCM8 mRNA is aberrant, leading to expression of a protein lacking 16 amino acids. In several cases of colon adenocarcinoma MCM8 expression is greatly reduced relative to matched non-cancerous tissue. The potential helicase domain of MCM8 is different from those of other MCM proteins in that it is more homologous to canonical ATP-binding domains of other known helicases. Results suggest that MCM8 may interact with other MCM proteins to alter the function of the replicative MCM protein complex.
INTRODUCTION
Genes encoding the minichromosome maintenance (MCM) proteins were originally identified in Saccharomyces cerevisiae in screens to detect proteins involved in plasmid maintenance during the cell cycle (reviewed in 1,2). The six MCM proteins, MCM2, MCM3, MCM4, MCM5, MCM6 and MCM7, are all highly conserved throughout eukaryotes, and each of these six has a counterpart in all eukaryotes that corresponds closely to the yeast protein. All six of these proteins are essential for DNA replication, and complexes containing various combinations of these proteins have been implicated in licensing DNA for replication in Xenopus egg extracts (3,4). More recent reports have documented a processive DNA helicase activity of a complex consisting of MCM4, MCM6 and MCM7 (5,6). While this complex from human cells had relatively low processivity using oligonucleotide substrates annealed to plasmid DNA, it has been reported that the presence of 5′ or 3′ tails greatly enhances the processivity of the corresponding complex from Schizosaccharomyces pombe, allowing unwinding of DNA >500 bp (7). The MCM4,6,7 complex from human cells has been observed by electron microscopy to adopt a heterotrimeric ring-like structure with DNA in the center, a configuration characteristic of several known helicases (8). Despite these tantalizing reports of helicase activity, it is not known at this time whether the MCM proteins function as a primary replicative helicase in S phase. Nor is it known what role any individual MCM protein plays in the MCM contribution to replication. MCM2 or MCM3/5 have been reported to inhibit helicase activity of the MCM4,6,7 complex (9), suggesting a diversity of functions in the MCM family. In addition to the MCM2–MCM7 proteins, another MCM protein, MCM10, is reportedly essential for plasmid maintenance in yeast (10). The human homolog of this protein associates with other MCMs and with the origin recognition complex, ORC (11), but little is presently known about the function of MCM10 in replication. The essential replication protein human Cdc45 has been reported to associate with both MCM7 and the p70 subunit of DNA polymerase α (12), implicating MCM7 in a potential transition from initiation to formation of the replication fork.
We describe here a new member of the MCM family of DNA replication proteins, termed MCM8, and we characterize the genomic locus of its gene. Unlike MCM2–MCM7 or MCM10, this protein does not have a direct counterpart in yeast, although it is highly conserved in several higher eukaryotes. Of all the previously known MCMs, MCM8 is most homologous to MCM2, MCM5 and MCM7, and in the region of potential helicase motifs it is most like MCM7. Unlike MCM7, however, MCM8 possesses highly canonical Walker A and B boxes (13,14), characteristic of helicase ATP-binding regions. MCM8 associates with MCM4, MCM6 and MCM7 proteins, suggesting a role in DNA replication, but distribution of MCM8 expression is not limited to highly replicating tissues, suggesting that MCM8 may extend the range of MCM functions beyond replication. This notion is further supported by our present findings of altered expression of MCM8 in certain forms of neoplasia.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in spinner culture in Eagle’s minimal essential medium (with l-glutamine and without calcium and magnesium; Cellgro Mediatech) for suspension cultures, supplemented with 10% (v/v) fetal bovine serum (Gibco BRL) at 37°C in a humidified atmosphere of 5% CO2, 95% air (15).
PCR analysis of MCM8 and an altered MCM8 form in human tissue cDNA panels
The primers used to amplify an indicator segment of the MCM8 coding sequence were both 20mers: 5′-ccaggaattgatgtctgatg-3′ (M8EX9F) and 5′-cagcttgaatctcttggatg-3′ (M8EX10R). These primers are from exons 9 and 10, respectively, of the MCM8 genomic sequence. They are not homologous to any other human gene, including MCM genes, and they encompass a region found to be missing in choriocarcinoma. These primers amplify a segment of 317 bp from normal MCM8 mRNA or cDNA and a segment of 269 bp from the transcript containing the deletion. Since they encompass an intron in the MCM8 gene, the primers amplify a segment of 559 bp from human genomic DNA. The MCM8 sequences were amplified from samples of the Human Multiple Tissue cDNA Panel I, from the Multiple Tissue Tumor Panel and from matched pairs of various non-cancerous and tumor tissues from individuals, all obtained from Clontech.
DNA sequencing
Vectors were pOTB7 for IMAGE clone 3546350 from choriocarcinoma and for IMAGE clone 4588564 from renal cell adenocarcinoma, and pCMV-SPORT6 for IMAGE clone 4395048 from a duodenal adenocarcinoma cell line. The bacterial host (DH10B) was amplified in Luria–Bertani growth medium with chloramphenicol (pOTB7) or ampicillin (pCMV-SPORT6), and DNA was prepared using a Qiagen kit. DNA sequencing was performed using phage M13 forward and reverse DNA primers of 21 nt. Automated sequencing was performed at the Mount Sinai Medical School DNA Core Facility.
Antibody production, immunoblotting and immunoprecipitation
The following synthetic peptide, keyhole limpet hemocyanin-conjugated at the C-terminal Cys, was obtained from Research Genetics Inc. and employed to immunize mice for antibody production: GEIQSFPLPDGKYSLPTKC. This peptide corresponds to amino acids 245–263 of the MCM8 protein sequence, and it is specific for that protein. Serum obtained from a mouse boosted three times was used for immunoblotting. SDS–PAGE and immunoblot transfer of proteins to Immobilon-P membranes were performed as described previously (16,17). Detection was with the Supersignal chemiluminescence system (Pierce). Antibodies for MCM4–MCM7 were all obtained from Santa Cruz Biotechnology. Immuno precipitations were performed rapidly after cell lysis as described previously (18). Co-immunoprecipitation of proteins using goat polyclonal anti-MCM4 (K-18), anti-MCM6 (C-20) or anti-MCM7 (V-17) antibodies was performed in RIPA lysis buffer (137 mM NaCl, 2.7 mM KCl, 4.2 mM sodium phosphate, 1.5 mM potassium phosphate, pH 7.4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitors essentially as described previously (18) using aliquots of 107 HeLa cells. Primary antibody was incubated with lysates for 1.5 h at 4°C, and agarose beads conjugated to protein G (Santa Cruz Biotechnology) were then incubated with the mixture overnight at 4°C. Precipitates were obtained by centrifugation at 1000 g, rinsed with RIPA buffer four times and redissolved in SDS sample buffer. Samples were heated to 98°C for 5 min and subjected to SDS–PAGE on 6% gels prior to immunoblotting.
Glycerol gradient centrifugation
A complex containing MCM4, MCM6 and MCM7 was prepared from a HeLa cell lysate by DEAE cellulose column chromatography essentially as described previously (6). Fractions containing the MCM proteins, eluted from the column at 0.2 M NaCl, were concentrated using a Centricon YM-10 (Millipore), dialyzed against buffer containing 0.15 M NaCl, 0.5 mM EDTA, 1.0 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 0.01% Triton X-100 and 20 mM Tris–HCl, pH 7.5, and an aliquot of 0.5 ml loaded onto a 10–35% glycerol gradient (38 ml) containing the same buffer. A parallel gradient was prepared with a 0.5 ml aliquot of a HeLa cell lysate in the same buffer. This lysate had previously been cleared by centrifugation at 12 500 g for 30 min. Glycerol gradients were subjected to centrifugation at 25 000 r.p.m. in a SW28 rotor (Beckman) for 24 h. Fractions were eluted and to each was added an equal volume of 10% trichloroacetic acid. Precipitates were collected by centrifugation at 13 000 r.p.m. for 5 min in an Eppendorf microfuge. Each precipitate was vacuum dessicated and dissolved in 10 µl of 1 M Tris–HCl, pH 8.8, and SDS gel sample buffer was added to each. After electrophoresis on a 6% polyacrylamide gel, samples were immunoblotted and incubated with antibodies to different MCM proteins as described above. Detection was with the Supersignal chemiluminescence system (Pierce). Band intensities on exposed film were quantitated using histogram counts on the gray channel with Adobe Photoshop 7.0.
RESULTS
Contrapodal genomic organization of human genes encoding MCM8 and GCD10
By searching the dBEST nucleotide database of sequence tags from expressed mRNAs with the protein sequence of MCM7 using the tBLASTn algorithm (19), we identified several clones containing ESTs from a sequence similar, but not identical, to MCM7. Segments of such sequences could be arranged contiguously to form an open reading frame (ORF). Selected IMAGE cDNA clones (GenBank accession nos BE278386, BG023796 and BG422937) were then ordered from the ATCC and Incyte Genomics for sequencing to confirm limits of the ORF and to identify sequences for additional searches. The resulting deduced ORF is bounded at the 5′ end by an ATG start codon upstream of which are TAA or TAG stop codons in all three reading frames. It is bounded at the 3′ end by a TAA stop codon, 200 nt downstream of which is an AATAAA potential polyadenylation signal. Further searching the HTGS database of human genomic sequences with unique sequences from this ORF using the tBLASTn algorithm revealed a BAC clone containing a unique gene encoding the new ORF at chromosome band 20p12.3–13 (GenBank accession no. AL035461). This gene is composed of 19 exons, comprising 43 796 genomic bp, and it encodes precisely the ORF deduced by our sequencing of ESTs. A complete mRNA sequence encoding this protein is available. Due to protein homologies to be described in this article, this protein is considered a member of the MCM protein family. Although no data yet link this protein to minichromosome maintenance, the name MCM is given the new protein, and the new gene, since it reflects family sequence similarities. Family members MCM2–MCM7 have been identified in humans, as has protein MCM10. Since the name MCM8 has not yet been used, the new family member is given the name MCM8. While this manuscript was undergoing initial review, a paper by a separate group reported the same protein, also terming it MCM8 (20). That paper did not report on the MCM8 genomic organization.
This new MCM family member has certain aspects unique among MCM proteins. Notably, it has no clear homolog in yeast. The N-terminal 250 amino acids of MCM8 are quite different from the N-termini of other MCMs. When a database search is initiated with the N-terminal sequences of MCM8, significant homologs are found in Drosophila, roundworms and the plant Arabadopsis, but no homologs are detected in yeast or lower organisms. This suggests that MCM8 evolved, perhaps from another MCM family member, at some point in metazoan development. The central region of MCM8 is very similar to those of all the other MCMs, especially that of MCM7. Further aspects of this MCM8 region will be considered below.
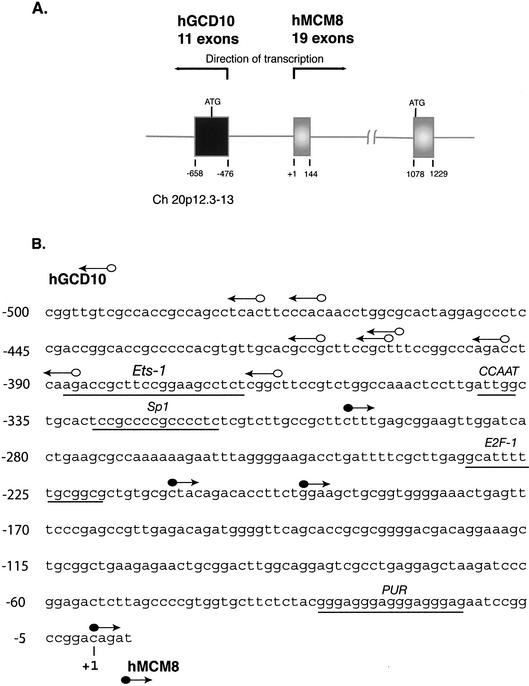
Intriguingly, the MCM8 gene is oriented head-to-head with another gene in the same BAC (GenBank accession no. AL035461; Fig. 1A). This gene consists of 11 exons comprising 12 698 genomic bp and encoding two alternative splice forms of mRNA, as determined by comparison to multiple recorded ESTs. The protein encoded by this gene is homologous to yeast protein P41814, termed the Gcd10p protein. Gcd10 from humans has not yet been described, but in S.cerevisiae the protein is a RNA-binding protein involved in mediating mRNA translation (21). Based on the significant homology, the new human coding sequence located contrapodal to MCM8 at 20p12.3–13 is named hGCD10. The start points of several extended cDNAs representing mRNAs for MCM8 and GCD10 are indicated by circles with arrows in Figure 1B. The 5′ start points, sequenced by us, include that for the EST from choriocarcinoma lacking codons for 16 amino acids, AY158211 (hMCM8, submitted by us 3 October 2002). Of the start points indicated in Figure 1B, that for hMCM8 at +1 is represented in two clones, and that for hGCD10 at –476 is found in three clones, making these at present the most common start points for the two genes. It is not known whether there are tissue or developmental specifications for start points. We have recently reported that certain other genes located in head-to-head fashion are each characterized by an array of transcriptional start points (22). Another GenBank entry for the MCM8 transcript, AJ439063, has an apparent start point located at –364. It can be seen that the entire sequence separating the transcription units represented in Figure 1B is highly GC-rich and that it contains no obvious candidates for TATA boxes for either gene. It does possess one potential CCAAT element, located less than 30 bp upstream of one hGCD10 start point. The region contains potential elements for binding transcription factors Sp1, Ets-1, E2F-1 and Purα, as indicated in Figure 1B. At this time there is no evidence that any of these transcriptional control elements is functional in the depicted region. The region separating the transcription units also contains multiple CpG units. In fact, the ratio of CpG to GpC in the 500 bp shown in Figure 1B is 1.07, a value high for mammalian GC-rich sequences and one that qualifies this region as a CpG island. The configuration of TATA-less promoters containing Sp1 elements is characteristic of several genes recently reported to be oriented in contrapodal fashion (22–26). No further characterization of the hGCD10 gene or its encoded protein was undertaken for the present report, which focuses on expression and preliminary functional characterization of MCM8.
Figure 1.
(A) Genomic organization of human MCM8 and GCD10 genes at chromosome band 20p12.3–13. Arrows indicate direction of transcription of the two genes. Gray boxes represent MCM8 exons, black boxes GCD10 exons. Translational start codons are indicated by ATG. MCM8 and GCD10 exon positioning was determined from the genomic sequence, GenBank accession no. AL035461. (B) Genomic DNA sequences in the region of multiple 5′ transcription start points of mRNAs for hMCM8 and hGCD10. Arrowed lines indicate the direction of transcription. Open circles indicate start points for hGCD10, filled circles for hMCM8. Certain potential cis-regulatory regions are underlined as indicated. The start point for hMCM8 at +1 is represented in two clones, and that for hGCD10 at –476 is found in three clones. These are at present the most common start points. The closest two start points are separated by 62 bp which are highly GC-rich.
An aberrant MCM8 mRNA form in choriocarcinoma
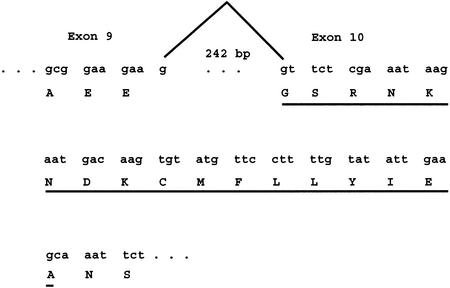
Sequencing of IMAGE clone 3546350 (GenBank accession no. BE278386) by us revealed an anomaly in the encoded MCM8 ORF in the form of a deletion of codons 342–357, encoding 16 amino acids, present in many reported ESTs as well as in the genomic sequence of the ORF. This IMAGE clone is a cDNA from an mRNA expressed in tumor tissue in a case of placental choriocarcinoma. In the genomic sequence of BAC RP5-967N21 (GenBank accession no. AL035461), obtained from non-cancerous cells, the codons missing in the choriocarcinoma clone are present in exon 10. This anomaly could represent either a genomic deletion in the tumor tissue DNA, a polymorphism found in non-cancerous or tumor DNA or aberrant processing of the mRNA in the tumor tissue. An examination of splicing in MCM8 mRNA, as exemplified by comparison of a full-length cDNA clone with genomic DNA, offers clues as to the apparent anomaly in choriocarcinoma. In Figure 2, which represents genomic DNA, the nucleotides missing in the choriocarcinoma mRNA are underlined. In the usual case …gaa g of exon 9 is a splice donor site to gt tct… of exon 10, the splice acceptor site. These sequences are standard for splice donor and acceptor sites. The splice forms the codons for amino acids EGS, beginning with codon 342. In the choriocarcinoma case it is exactly as though the …gaa g of exon 9 is spliced to the sequence ca aat…, occurring 48 nt downstream of the beginning of the usual exon 10 splice acceptor site. In this case the resulting nucleotide sequence encodes amino acids EAN. The ca aat… is not a canonical splice acceptor site. Nevertheless, it is quite conceivable that it has acted as a cryptic splice acceptor site in this case. At this time, however, the alternative explanation that the mRNA deletion may result from a genomic DNA deletion cannot presently be ruled out. This cDNA may be a valuable mutant in studies of MCM8 function.
Figure 2.
Missing codons in an MCM8 transcript from a case of choriocarcinoma. Genomic DNA in the region of exons 9 and 10 is depicted showing codon triplets. Normal splicing, employing canonical donor and acceptor sites, removes an intron of 242 bp, as shown by the peaked line at the top. Missing codons, representing 16 amino acids, are underlined.
Distribution of expression of MCM8 mRNA in non-cancerous and neoplastic human tissues
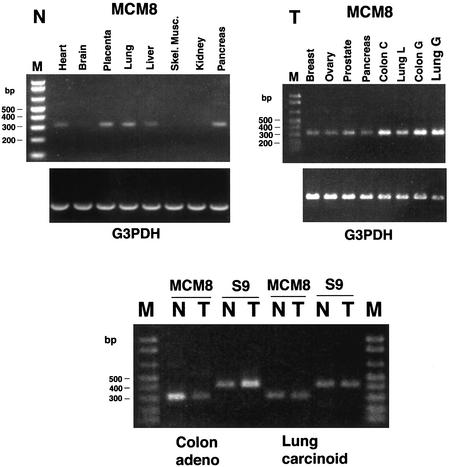
Tissue distribution of MCM8 mRNA expression was examined using a cDNA panel of multiple adult human tissues, as shown in Figure 3 (N, non-cancerous). The G3PDH controls show that overall mRNA representation is approximately the same in each lane. The amplified MCM8 bands (top) reveal considerable variation in expression in different tissues. In addition, only a band of 317 bp is seen in each lane, indicating that only appropriately spliced exons 9 and 10 are expressed in each tissue. Note that the samples in this panel, obtained from Clontech, are from non-cancerous tissues. Highest levels of MCM8 expression are seen in the placenta, lung and pancreas. Skeletal muscle and kidney lanes have little or no amplified MCM8 bands. There is a faint, but visible, MCM8 band in the brain sample lane. The levels of MCM8 expression do not necessarily correlate with levels of cell proliferation, i.e. with expected levels of DNA replication. For example, MCM8 amplification is significant in the heart sample lane, although adult heart generally does not have a high percentage of proliferating cells. Caution should be exercised, however, in interpreting this type of distribution study since differences in exsanguination of tissues could influence levels of proliferating cells. It is interesting that placenta, with relatively high levels of MCM8 expression, has only the 317 bp band and not the 269 bp band that would be generated by the aberrant mRNA. The aberrant mRNA described was from a choriocarcinoma obtained from the placenta.
Figure 3.
Tissue distribution of expression of MCM8 mRNA. (N, non-cancerous) PCR amplification of an indicator segment comprising 317 bp of MCM8 mRNA or a 269 bp segment comprising the region with a deletion in eight different non-cancerous tissue types. G3PDH housekeeping gene expression is shown as a positive control. PCR amplification is shown at 35 cycles using 1.0 ng first strand cDNA per sample based on reported normalized concentration (Clontech). (T, tumor) PCR amplification of the MCM8 mRNA indicator segment described above in a panel of eight different tumors. G3PDH expression in the tumors is shown as a positive control. PCR products derived from the use of cDNAs from two different colon adenocarcinomas (CX-1 and GI-112) and two different lung carcinomas (LX-1 and GI-117) are included. Total PCR cycles were 34, and 1.0 ng of each first strand cDNA was used for each sample based on reported normalized concentration. (N/T) Expression of MCM8 and ribosomal protein S9 control mRNAs in matched non-cancerous and tumor pairs from the same patient. PCR amplification of the 317 bp MCM8 indicator segment and the 431 bp S9 control using matched human colon adenoid (colon adenocarcinoma) and lung carcinoid (lung malignant carcinoid) non-cancerous and tumor tissue pairs is shown. Specimens were adjusted to 1.0 ng first strand cDNA per sample based on reported initial normalized concentration. PCR amplification is shown at 35 cycles. Left and right markers contained 2 and 4 µg marker DNA, respectively.
In a panel of cDNAs representing mRNAs expressed in various tumor tissues MCM8 was detected by PCR in every sample (Fig. 3, T, tumor). There were only slight variations in overall expression level, although colon and lung tumor samples were relatively high. There is variability in expression levels in the lung tumor samples, but the significance of this can only be evaluated upon comparison of matched non-cancerous and tumor samples from the same individual. One notable result of this tumor tissue study is that the 269 bp aberrant mRNA found in the choriocarcinoma is not detected in these other tumors. It remains to be determined whether other alterations that would affect MCM8 gene expression occur in different neoplastic tissues or stages.
MCM8 mRNA expression was compared in matched non-cancerous (N) and tumor (T) samples from selected individuals (Fig. 3, bottom panel). The tumors examined were a metastatic colon adenocarcinoma and a malignant lung carcinoid tumor. These cDNA samples were normalized as to total mRNA amounts. MCM8 expression in the lung tumor (T) showed no change relative to matched non-cancerous lung tissue (N), as seen by comparison to control expression of ribosomal protein S9 in each tissue. In contrast, MCM8 expression in the colon tumor was significantly reduced relative to non-cancerous tissue, as compared with S9 expression. S9 expression was increased in the tumor tissue. Previous reports have indicated enhanced expression of ribosomal protein S9 mRNA in a minor percentage of cases of colon cancer (27). MCM8 expression is several-fold lower relative to its level in non-cancerous tissue (N) and relative to the level of S9 expression. These comparisons, shown in Figure 3 (bottom panel), at 34 PCR cycles, were essentially the same over a range of different cycles of amplification.
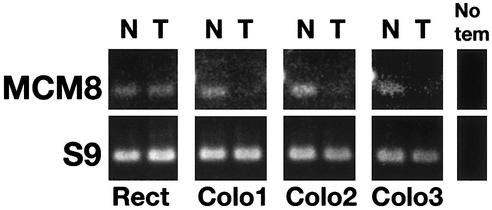
To further examine the link between reduced MCM8 gene expression and colon cancer, mRNAs from three additional cases of colon adenocarcinoma and one case of rectal carcinoma were analyzed. All samples were commercially obtained (Clontech) together with matched non-cancerous colon tissue samples and all had been prepared for PCR with first strand cDNA synthesis. Control S9 ribosomal protein mRNA was assayed to standardize mRNA amounts. The comparisons of non-cancerous (N) and tumor (T) MCM8 amplified DNA and of control S9 DNA are shown in Figure 4. There is no change in MCM8 mRNA levels between non-cancerous and tumor rectal tissue. It can be seen that in each case of colon adenocarcinoma MCM8 gene expression is several-fold lower than in matched non-cancerous tissue. In contrast, there is no significant difference in S9 mRNA levels in non-cancerous and tumor tissue. Densitometry of the bands obtained from all four patients analyzed reveals that the average level of MCM8 mRNA in the colon tumor tissue is only 0.153 ± 0.044 that of the level in the matched non-cancerous colon tissue.
Figure 4.
Reduced levels of MCM8 mRNA in colon adenocarcinoma. All samples of matched non-cancerous (N) and tumor (T) mRNAs were commercially prepared with first strand cDNA synthesis. PCR was performed as described in Figure 3. Samples for electrophoresis were at 35 and 36 cycles for S9 and MCM8, respectively. A control lane with no template is shown for each set of primers. Densitometry on bands was performed using Photoshop 7.0.
Association of MCM8 with other MCM proteins involved in initiation of DNA replication
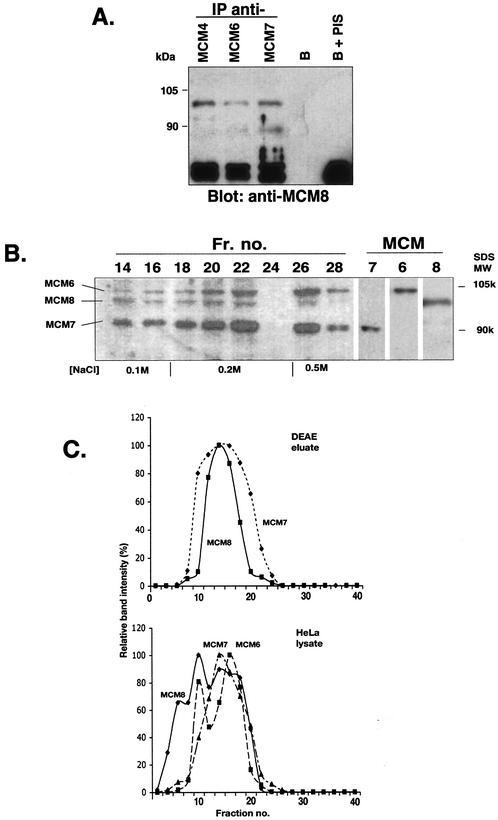
All known MCM proteins have been detected in association with the DNA replication apparatus in various organisms (1,2). MCM2–MCM7 can be isolated as a distinct complex, and among these, MCM4, MCM6 and MCM7 from mammalian cells have been reported to function as a helicase complex (6,7). These proteins co-isolate through several purification steps culminating in DEAE cellulose chromatography (6). We sought to determine whether MCM8 associates with other MCM proteins, particularly MCM4, MCM6 and MCM7. Figure 5A shows that MCM8 can be co-immunoprecipitated with either anti-MCM6, anti-MCM4 or anti-MCM7. Figure 5B shows that MCM8 co-isolates with MCM6 and MCM7 through DEAE cellulose column chromatography. MCM8 is visualized upon SDS–PAGE and staining with anti-MCM8 antibody as a band of slightly less than 100 kDa, migrating slightly more rapidly than MCM6 and slightly retarded relative to MCM7. MCM8 has a molecular weight of 92 kDa, based on its length of 840 amino acids, but its migration at nearly 100 kDa is consistent with anomalous migration of several of the MCM proteins.
Figure 5.
(Next page) Co-isolation and co-immunoprecipitation of MCM8 with other MCM proteins in a potential helicase complex. (A) Co-immunoprecipitation of MCM8 from a HeLa cell lysate using antibodies against either MCM4, MCM6 or MCM7 and agarose beads conjugated to protein G. Antiserum against MCM8 was used in analysis of the blot obtained after gel electrophoresis of the immunoprecipitated samples. Controls are shown to the right: beads without primary antibody (B) and beads with preimmune serum replacing the primary antibody (PIS). (B) Co-isolation of MCM8 with MCM6 and MCM7 after protein purification steps. Overlapping radiographs from western blot analysis following SDS–PAGE indicate the presence of MCM8 (slightly smaller than 100 kDa) co-isolated with MCM6 and MCM7 at various salt concentrations in fractions following DEAE cellulose column chromatography. An immunoblot of the indicated DEAE chromatographic fractions was subjected separately to each of the specific anti-MCM antibodies to obtain three fluorescent radiographic images. These were then precisely superimposed and scanned to obtain the image presented. Individual detection of MCM6, MCM7 and MCM8 in fraction 20 is shown to the right under MCM. (C) Comparison of glycerol gradient centrifugation of MCM8-containing complexes in whole HeLa cell lysate and after elution from a DEAE cellulose column. Glycerol gradients were prepared and subjected to centrifugation and MCM protein levels in fractions quantitated as described in Materials and Methods. The HeLa lysate gradient is in the bottom panel. Multiple peaks for MCM8 (diamonds), two for MCM6 (squares) and one for MCM7 (triangles) can be seen. Fractions from the DEAE column of Figure 4B containing MCM6, MCM7 and MCM8 were pooled and subjected to glycerol gradient centrifugation, shown in the top panel. A single peak for MCM8 (squares) coincides with that for MCM7 (diamonds).
Glycerol gradient centrifugation helps in assessing the functional state of MCM protein complexes. In Figure 5C centrifugation of a complex isolated from the DEAE column of Figure 5B is compared with centrifugation of complexes in a HeLa cell soluble extract. In the HeLa extract, which is a whole cell lysate cleared by centrifugation, shown in Figure 5C (bottom panel), MCM8 is detected in several gradient peaks. The lowest density peak at fraction 6 does not coincide with the presence of any other MCM proteins assayed. It may represent free MCM8. It cannot be immunoprecipitated with antibodies to MCM4, MCM6 or MCM7. Another sharp peak at fraction 10 coincides precisely with a sharp peak of MCM6. MCM7 does not have a peak at this position. Another MCM8 peak at fraction 14 coincides with the sole peak of MCM7. MCM6 sediments in two relatively sharp peaks, both of which coincide or overlap considerably with MCM8 and one of which overlaps with that of MCM7. Results of centrifugation of the DEAE isolate (Fig. 5C, top panel) are straightforward. MCM8 sediments in one peak at fraction 14, a peak that is also seen in the bottom panel. MCM7 from the DEAE isolate also peaks at this position, almost exactly as it does in the bottom panel. MCM6 and MCM4 from the DEAE isolate also peak at fraction 14, although for neatness sake, only MCM7 is shown in Figure 5C (top). Thus, this peak at fraction 14 most likely represents the complex of MCM4, MCM6 and MCM7, which also at times includes a portion of MCM8. By integrating the area under the MCM8 peaks in the bottom panel it can be estimated that the maximum portion of MCM8 present in the MCM complex peaking at fraction 14, the potential MCM4,6,7 complex, is ∼32%. In contrast, in the non-synchronous HeLa lysate nearly all MCM7 is in this complex. These results also suggest a dynamic association of different MCM proteins since the highest peak of MCM8 in the HeLa lysate coincides with one of MCM6 but not of MCM7. More work would have to be done to establish that these coinciding peaks represent a complex. The results indicate that MCM8 associates with other MCM proteins that play a role in DNA replication. At this point, however, the results do not unequivocally implicate MCM8 itself in DNA replication since it is conceivable that the MCM proteins are involved in other cellular functions.
DISCUSSION
MCM8 includes ATP-binding domains characteristic of helicases
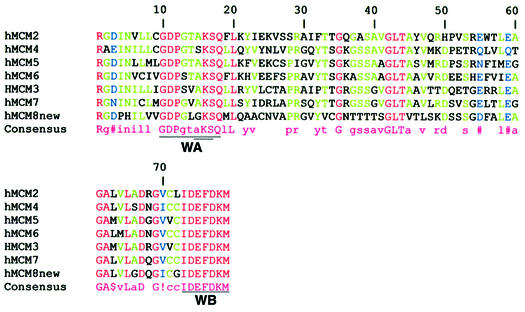
The MCM8 gene encodes a protein of 840 amino acids. According to the Multalign algorithm (28), based on the total hMCM8 amino acid sequence, hMCM8 has 12% identity with hMCM3, 12% identity with hMCM10, 19% identity with hMCM4 and 21% identity with hMCM6. hMCM8 has 24% identity with hMCM5 and hMCM7 and 25% identity with hMCM2. Human MCM8 is 88% identical to mouse MCM8. In the ATP-binding, potential helicase domain, common to all MCM proteins, hMCM8 is more homologous to hMCM7 than to any other human MCM protein. This protein region of 79 amino acids is compared using the Multalign algorithm (28) for hMCM2–hMCM8 in Figure 6. Amino acids identical among all the shown MCM proteins in this helicase domain, 30.4% overall, are displayed in red. Scrutiny reveals that this region is more similar between MCM8 and MCM7 (51.9% identity) than between MCM8 and any other MCM. The percent identities between MCM8 and MCM2–MCM6 in this helicase domain are 46.8, 46.8, 50.6, 48.1 and 48.1%, respectively. Walker A and B boxes, characteristic ATP-binding sites of virtually all helicases (13,14,29), are underlined and noted as WA and WB, respectively, in Figure 6. All of the MCM proteins save MCM8 possess an unusual A-box motif, either AKS or SKS, as indicated by double underlining of the sequence in Figure 6. MCM8, intriguingly, possesses the A-box motif GKS, which is much more prevalent among helicases (29). The A-box reportedly makes contact with an ATP phosphate (13). The B-box motif, reportedly involved in ATP phosphate binding via Mg2+ (13), is strongly conserved in all MCM proteins in Figure 6. It will be of interest to determine whether MCM8 participates, either positively or negatively, in a helicase complex or whether it possesses helicase activity itself. The 16 amino acid sequence eliminated in the choriocarcinoma clone (GSRNKNDKCMFLLYIE) is N-terminal to the conserved helicase-like region of MCM8 and may be involved in DNA binding (1). The removed amino acid sequence is itself conserved in complete MCM8 proteins as it is identical in the human and mouse MCM8. Further work will reveal whether the MCM8 variant reported here acts in a dominant negative fashion with regard to MCM8 function.
Figure 6.
Alignment of MCM protein sequences encompassing potential helicase ATP-binding domains. MCM8 is compared to MCM2–MCM7, proteins implicated in initiation of DNA replication, using the Multalign algorithm (19). Walker A (WA) and Walker B (WB) boxes (14) are underlined in the MCM consensus sequence. A component of the A-box motif, either AKS or SKS, a characteristic of MCM proteins other than MCM8, is indicated with double underlining. In MCM8 this A-box is GKS. Residues conserved for all presented sequences, consensus upper case letters, red in individual sequences. Residues conserved for 50% or more, lower case letters, green. Residues conserved for <50%, white space, black. I or V conserved positions, (!), blue. LM conserved positions, ($), black. NDQE conserved positions, (#), blue.
Parallels in contrapodal genomic orientation of MCM8-GCD10 and WRN-PURG
The contrapodal orientation of MCM8 and GCD10 in the genome, with implied closely associated or overlapping regulatory regions, is intriguing since both encoded proteins are involved in nucleic acid metabolism. This type of head-to-head organization has now been observed for several genes encoding proteins involved in DNA replication or repair, including DNA helicases. For example, the gene encoding another MCM protein, MCM4, a component of the putative MCM helicase complex, is located contrapodal to the gene encoding the catalytic subunit of DNA-dependent protein kinase, DNAPKCS (30). The gene encoding the breast cancer susceptibility protein BRCA1, thought to be involved in DNA recombination/repair (31), is located contrapodal to that encoding NBR1 (25), a protein that may be involved in cell signaling pathways (32). The gene encoding poly(ADP-ribose) polymerase 2 (PARP-2), involved in the cellular response to DNA damage, is located contrapodal to that encoding RNase P (26). Recently we have reported that the Werner syndrome helicase gene, WRN, is located contrapodal to PURG, encoding Purγ, a member of the Pur family of sequence-specific single-stranded DNA-binding proteins (22). There are certain parallels between the orientations of MCM8-GCD10 and WRN-PURG. Primary transcription start sites for WRN and PURG are separated by ∼90 bp (22). Primary start sites for MCM8 and GCD10 are ∼60–500 bp apart. The promoters for WRN and PURG are TATA-less and there are no obvious TATA boxes between the MCM8 and GCD10 genes. Instead, the regions separating each of the two sets of genes are overwhelmingly GC-rich and contain Sp1 and PUR elements. This type of transcriptional regulatory region is common among housekeeping genes and frequently comprises a CpG island in which the CpG/GpC ratio is >0.6 (33,34). The CpG/GpC ratio for the sequence comprising MCM8 and GCD10 transcription start sites is approximately 1.1, an unusually high value. In the case of WRN and PURG there is only limited shared use of critical transcriptional regulatory elements, and the pattern of tissue levels of expression is not parallel (22). That is also true of most genes oriented head-to-head that possess GC-rich regulatory regions (reviewed in 21). What advantage can be derived from contrapodal orientation of genes with overlapping promoters if the genes are not strictly co-regulated? One answer may be that at certain times it is advantageous to modulate the expression of both genes in parallel. To that effect, such processes as chromatin remodeling or DNA methylation of a shared regulatory region could either up- or down-regulate both genes together.
Tissue distribution of MCM8 expression and association of MCM8 protein with other MCMs provide insight into MCM8 functions
Our present data may be compared with those of a contemporary paper on the same protein subject (20), published while this manuscript was under review. That group did not observe co-immunoprecipitation of MCM8 with anti-MCM4 antibody whereas we did. This difference may be due to the antibodies employed but may also be due to the fact that a relatively minor percentage of MCM8 is in a complex with MCM4, as indicated by a comparison of glycerol gradient centrifugation of MCM8 isolated as a complex with MCM8 in a total HeLa cell lysate (Fig. 5C). With regard to details of cell and tissue distribution of MCM8 the two papers are generally consistent, although presence or absence of low levels in certain tissues may need further clarification.
The tissue distribution of MCM8 mRNA expression may help in assessing the functions of this new MCM protein family member. MCM8 expression levels are relatively high in tissues known to possess a significant percentage of replicating epithelial cells, i.e. placenta and lung. MCM8 expression is, however, also high in certain tissues not thought to have many replicating cells, i.e. heart and pancreas. Therefore, while a role for the MCM8 protein in DNA replication cannot be excluded, it is likely that the protein perhaps serves additional function(s) that are not related to replication. DNA repair processes, in which DNA synthesis plays a role, are one potential area of MCM protein function. If MCM8 were involved in repair, however, one might expect high levels of expression in the brain, since that organ is well known to require high DNA repair activity. MCM8 expression is relatively low in the brain. It is quite conceivable that MCM8 could function in DNA replication or some other activity only in specific cell types. In that regard, MCM8 may, at certain times in development or in the cell cycle, substitute for one of the six MCM proteins previously demonstrated to function together. MCM8 associations with other MCM proteins help illuminate this point. MCM8 was detected in co-immunoprecipitates with anti-MCM4, anti-MCM6 and anti-MCM7 and it sediments on glycerol gradients with these proteins. The results suggest that further work be done to address the possibility that MCM8 could substitute for one of the three MCMs at certain times in the MCM4,6,7 complex. Since MCM8 has helicase motifs that are unique among MCM proteins, such a substitution could add a regulatory dimension to the function of that complex.
Reduction of MCM8 expression in colon adenocarcinoma
All known MCM proteins have been ascribed a function in DNA replication, and certain of them are reported to be good markers for proliferating cells in cancer (35). MCM8 does not fit this pattern. MCM8 mRNA levels were strongly reduced in multiple colon adenocarcinoma tumors as compared to matched non-cancerous colon tissue from the same patients (Figs 3 and 4). In contrast, there were no changes in MCM8 expression in lung carcinoid or rectal tumors. The change in expression in colon tumors is dramatic, being present in all cases tested and averaging ∼85% reduction. The relation of this reduction in expression to stage of tumor development remains to be determined. Three of the four cases examined were early, non-metastatic tumors. This observation also sheds light on function, since it indicates that MCM8 does not facilitate enhanced DNA replication in neoplastic cells. The possibility that reduced MCM8 expression could be an early marker for colon cancer should be further explored.
Acknowledgments
ACKNOWLEDGEMENTS
Hong Liu provided valuable assistance with PCR. This work was supported by a Mount Sinai Dean’s Incentive Award to D.C.D. and by grants from the NCI and the NINDS to E.M.J.
DDBJ/EMBL/GenBank accession no. AY158211
REFERENCES
- 1.Tye B.K. (1999) MCM proteins in DNA replication. Annu. Rev. Biochem., 68, 649–686. [DOI] [PubMed] [Google Scholar]
- 2.Bell S.P. and Dutta,A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem., 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 3.Madine M.A., Khoo,C.Y., Mills,A.D. and Laskey,R.A. (1995) MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature, 375, 421–424. [DOI] [PubMed] [Google Scholar]
- 4.Chong J.P., Mahbubani,H.M., Khoo,C.Y. and Blow,J.J. (1995) Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature, 375, 418–421. [DOI] [PubMed] [Google Scholar]
- 5.You Z., Komamura,Y. and Ishimi,Y. (1999) Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol. Cell. Biol., 19, 8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- 7.Lee J.K. and Hurwitz,J. (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6 and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA, 98, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M., Gotow,T., You,Z., Komamura-Kohno,Y., Uchiyama,Y., Yabuta,N., Nojima,H. and Ishimi,Y. (2000) Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol., 300, 421–431. [DOI] [PubMed] [Google Scholar]
- 9.Ishimi Y., Komamura,Y., You,Z. and Kimura,H. (1998) Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem., 273, 8369–8375. [DOI] [PubMed] [Google Scholar]
- 10.Maine G.T., Sinha,P. and Tye,B.K. (1984) Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics, 106, 365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawasaki Y., Hiraga,S. and Sugino,A. (2000) Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells, 5, 975–989. [DOI] [PubMed] [Google Scholar]
- 12.Kukimoto I., Igaki,H. and Kanda,T. (1999) Human CDC45 protein binds to minichromosome maintenance 7 protein and the p70 subunit of DNA polymerase alpha. Eur. J. Biochem., 265, 936–943. [DOI] [PubMed] [Google Scholar]
- 13.Hodgman T.C. (1988) A new superfamily of replicative proteins. Nature, 333, 22–23. [DOI] [PubMed] [Google Scholar]
- 14.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karn J., Johnson,E.M., Vidali,G. and Allfrey,V.G. (1974) Differential phosphorylation and turnover of nuclear acidic proteins during the cell cycle of synchronized HeLa cells. J. Biol. Chem., 249, 667–677. [PubMed] [Google Scholar]
- 16.Barr S.M. and Johnson,E.M. (2001) Ras-induced colony formation and anchorage-independent growth inhibited by elevated expression of Puralpha in NIH3T3 cells. J. Cell. Biochem., 81, 621–638. [DOI] [PubMed] [Google Scholar]
- 17.Daniel D.C. (1999) Dual immunofluorescence labeling with cell-specific markers localizes BRCA1 in both basal and luminal epithelial cells in primary outgrowth from noncancerous mammary ductal and alveolar preparations. Cell Tissue Res., 298, 481–487. [DOI] [PubMed] [Google Scholar]
- 18.Johnson E.M., Chen,P.-L., Krachmarov,C.P., Barr,S., Ma,Z.-W. and Lee,W.-H. (1995) Association of human Purα with the retinoblastoma protein, Rb, regulates binding to the Purα single-stranded DNA recognition element. J. Biol. Chem., 270, 24352–24360. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 20.Gozuacik D., Chami,M., Lagorce,D., Faivre,J., Murakami,Y., Poch,O., Biermann,E., Knippers,R., Brechot,C. and Paterlini-Brechot,P. (2003) Identification and functional characterization of a new member of the human Mcm protein family: hMcm8. Nucleic Acids Res., 31, 570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson J., Phan,L. and Hinnebusch,A.G. (2000) The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 5173–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H. and Johnson,E.M. (2002) Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res., 30, 2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinya E. and Shimada,T. (1994) Identification of two initiator elements in the bidirectional promoter of the human dihydrofolate reductase and mismatch repair protein 1 genes. Nucleic Acids Res., 22, 2143–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito T., Matsuda,Y., Ishii,H., Watanabe,F., Mori,M., Hayashi,A., Araki,R., Fujimori,A., Fukumura,R., Morimyo,M., Tatsumi,K., Hori,T. and Abe,M. (1998) Mouse cdc21 only 0.5 kb upstream from dna-pkcs in a head-to-head organization: an implication of co-evolution of ATM family members and cell cycle regulating genes. Mamm. Genome, 9, 769–772. [DOI] [PubMed] [Google Scholar]
- 25.Dimitrov S., Brennerova,M. and Forejt,J. (2001) Expression profiles and intergenic structure of head-to-head oriented Brca1 and Nbr1 genes. Gene, 262, 89–98. [DOI] [PubMed] [Google Scholar]
- 26.Ame J.C., Schreiber,V., Fraulob,V., Dolle,P., de Murcia,G. and Niedergang,C.P. (2001) A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA. structure and expression of the mouse PARP-2 gene. J. Biol. Chem., 276, 11092–11099. [DOI] [PubMed] [Google Scholar]
- 27.Frigerio J.M., Dagorn,J.C. and Iovanna,J.L. (1995) Cloning, sequencing and expression of the L5, L21, L27a, L28, S5, S9, S10 and S29 human ribosomal protein mRNAs. Biochim. Biophys. Acta, 1262, 64–68. [DOI] [PubMed] [Google Scholar]
- 28.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonin E.V. (1993) A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res., 21, 2541–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimori A., Araki,R., Fukumura,R., Ohhata,T., Takahashi,H., Kawahara,A., Tatsumi,K. and Abe,M. (2000) Identification of four highly conserved regions in DNA-PKcs. Immunogenetics, 51, 965–973. [DOI] [PubMed] [Google Scholar]
- 31.Daniel D.C. (2002) Highlight: BRCA1 and BRCA2 proteins in breast cancer. Microsc. Res. Tech., 59, 68–83. [DOI] [PubMed] [Google Scholar]
- 32.Whitehouse C., Chambers,J., Howe,K., Cobourne,M., Sharpe,P. and Solomon,E. (2002) NBR1 interacts with fasciculation and elongation protein zeta-1 (FEZ1) and calcium and integrin binding protein (CIB) and shows developmentally restricted expression in the neural tube. Eur. J. Biochem., 269, 538–545. [DOI] [PubMed] [Google Scholar]
- 33.Razin A. and Kafri,T. (1994) DNA methylation from embryo to adult. Prog. Nucleic Acid Res. Mol. Biol., 48, 53–81. [DOI] [PubMed] [Google Scholar]
- 34.Bird A.P. (1986) CpG-rich islands and the function of DNA methylation. Nature, 321, 209–213. [DOI] [PubMed] [Google Scholar]
- 35.Freeman A., Morris,L.S., Mills,A.D., Stoeber,K., Laskey,R.A., Williams,G.H. and Coleman,N. (1999) Minichromosome maintenance proteins as biological markers of dysplasia and malignancy. Clin. Cancer Res., 5, 2121–2132. [PubMed] [Google Scholar]