Abstract
The transcription factor RUNX1 (AML-1, PEBP2αB and CBFA2) is essential for definitive haematopoiesis, and chromosomal translocations involving the RUNX1 gene are frequently found in acute leukaemias. The gene encoding the histone acetyltransferase MOZ is also rearranged in some acute leukaemias, resulting in the expression of MOZ fusion proteins. MOZ has recently been shown to interact directly with RUNX1, indicating that MOZ fusion proteins act by deregulating RUNX1 function. Macrophage inflammatory protein-1α (MIP-1α) is a proinflammatory cytokine that also inhibits proliferation of haematopoietic stem cells. Amongst the conserved sequence elements in the human MIP-1α promoter are two consensus RUNX sites. We have investigated the role of these RUNX sites in the regulation of the MIP-1α promoter by PMA/PHA stimulation in Jurkat T-cells. RUNX1 can specifically bind to both RUNX sites in vitro and chromatin immunoprecipitation assays demonstrated that endogenous RUNX1 is constitutively bound to the endogenous MIP-1α promoter. Mutation of the RUNX sites demonstrated that the proximal RUNX site is essential for PMA/PHA-stimulated activation of the MIP-1α promoter. Activation of the promoter can also be inhibited by heterologous expression of the repressor protein AML-1/ETO. We further demonstrate that MOZ can activate the MIP-1α promoter and that this activation is largely dependent upon the proximal RUNX site. Moreover, we show that co-expression of MOZ and RUNX1 can synergistically activate the MIP-1α promoter. The regulation of MIP-1α expression by RUNX1/MOZ is discussed in the context of MIP-1α’s role as an inhibitor of haematopoietic stem cell proliferation and its potential importance in leukaemias associated with RUNX1 or MOZ chromosomal rearrangements.
INTRODUCTION
The Runt-domain family of transcription factors are essential for a range of cellular differentiation programmes including osteogenesis, haematopoiesis, Drosophila eye development and neurogenesis (1–3). The Runt-domain is a DNA-binding domain that specifically recognises the consensus-binding site TGT/cGGT, and also mediates protein–protein interactions with its partner protein CBFβ, to form a heterodimer on DNA (4–6). The mammalian family of Runt-domain transcription factors is comprised of three members, RUNX1, 2 and 3. RUNX1 is a master regulator of definitive haematopoiesis and is essential for the development of the myeloid and lymphoid lineages, RUNX2 is required for development of the skeleton and RUNX3 has a role in gastric and neuronal development (7–11).
Chromosomal translocations in the gene encoding RUNX1 (also known as AML-1, PEBP2αB and CBFA2) are frequently found in acute leukaemias including acute myeloid leukaemia, B-cell acute lymphoblastic leukaemia and, more recently, in T-cell acute lymphoblastic leukaemia (4,12). At the molecular level, RUNX1 has been shown to regulate the transcription of a number of genes including the cytokines IL-3 and GM-CSF, the CSF-1 receptor, myeloperoxidase, p21WAF–1 and p14ARF (13–20).
RUNX1 has been shown to act as both a transcriptional activator and a repressor (4). These functions can be explained by the ability of RUNX1 to interact with both co-activator and co-repressor proteins. RUNX1 can repress transcription by recruitment of mSin3A and Groucho co-repressors, whilst transcriptional activation involves interaction with the histone acetyltransferases p300, CBP and MOZ (4,20–24). The MOZ gene is rearranged in some chromosomal translocations associated with acute myeloid leukaemia and MOZ was recently shown to be a strong co-activator of RUNX1 (24–26). However, to date, only the myeloperoxidase promoter has been shown to be regulated by RUNX1 and MOZ.
Macrophage inflammatory protein-1α (MIP-1α) is a member of the CC chemokine subfamily of cytokines (27). MIP-1α was initially characterised as a lymphocyte chemoattractant and, as such, acts as a proinflammatory cytokine (27). However, in addition to its role as a proinflammatory mediator, it also inhibits proliferation of haematopoietic stem cells and immature progenitors (27,28). MIP-1α expression is inducible in several mature haematopoietic cells, including macrophages, B-cells and T-cells (28–31). In T-cells, MIP-1α expression can be induced by stimulation with PHA and PMA (27–29). Induction of MIP-1α expression by PMA/PHA in Jurkat T-cells occurs via transcriptional activation of the MIP-1α promoter (29). DNase I footprinting analysis has identified a number of conserved sequence elements in the human MIP-1α promoter that are bound by transcription factors (29). However, only one of these elements, termed the ICK-1 element, has been characterised in T-cells (29). The ICK-1 element acts as both a negative and positive regulator of MIP-1α transcription, indicating that the mechanism of PMA/PHA stimulation of the MIP-1α promoter involves relief of repression as well as activation (29). In myeloid cells both the ICK-1 element and an IL-6 response element have been shown to regulate activation of the MIP-1α promoter (29,32).
Amongst the conserved sequence elements in the MIP-1α promoter is a consensus RUNX site, which has not previously been characterised (33). In addition, we identified a further consensus RUNX site in the MIP-1α promoter. The regulation of expression of a stem cell inhibitor such as MIP-1α by haematopoietic transcription factors, which are rearranged in leukaemias, is potentially important in the progression of such diseases. We therefore investigated the role of the RUNX sites in transcriptional regulation of the MIP-1α promoter. We show that RUNX1 can specifically bind to both RUNX sites but that only the proximal RUNX site is essential for PMA/PHA stimulation of the MIP-1α promoter in Jurkat T-cells. We also show that the endogenous MIP-1α promoter is constitutively bound by RUNX1. We further demonstrate that the histone acetyltransferase, MOZ, can activate the MIP-1α promoter in T-cells and that this activation is largely dependent upon the proximal RUNX site. Moreover, we show that co-expression of MOZ and RUNX1 can activate the MIP-1α promoter.
MATERIALS AND METHODS
Cell culture, transfection and reporter assays
Jurkat (T-cell lymphoma) and HeLa (epithelial carcinoma) cells were grown in standard conditions (20,29). Transfection of Jurkat cells was performed with Superfect (Qiagen) according to the manufacturer’s instructions. 5 × 105 cells were transfected in 24-well plates. A total of 1.5 µg of DNA was used in each transfection. All transfections contained 600 ng of reporter plasmid and 100 ng of Renilla luciferase plasmid pRLSV40 (Promega) to normalise for transfection efficiency. Twenty-four hours post transfection, the cells were washed in growth media and divided into two separate wells. One well was stimulated by addition of 50 ng/ml of phorbol 12-myristate 13-acetate (PMA) and 1 µg/ml phytohemaglutinin (PHA) final concentration. The PMA/PHA vehicle solution (PBS/DMSO) was added to the other well as an unstimulated control. Cells were harvested and assayed for luciferase activity 24 h after stimulation. All transfections were performed in triplicate.
Transfection of HeLa cells was performed using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. A total of 2 µg of DNA was used in each transfection. All transfections contained 700 ng of reporter plasmid and 100 ng of Renilla luciferase plasmid pRLSV40 (Promega) to normalise for transfection efficiency. Cells were transfected at 60–80% confluency in 35 mm 6-well plates and harvested 24 h after transfection. All transfections were carried out in triplicate. Promoter activity was determined using the Dual-Luciferase™ Reporter Assay System (Promega) according to the manufacturer’s recommendations.
Plasmid constructs
The MIP-1α promoter was cloned by nested PCR from genomic DNA derived from Jurkat T-cells using the following oligonucleotides: first amplification, 5′-TCCTCCAGCTTTCATTCAGTTC-3′ and 5′-GGTGACGGAATGTGGGCTCG-3′; second amplification 5′-TCGCGAGCTCTTTGCCTCTGGGAGGAGGAAG-3′ and 5′TCGCAAGCTTGAGTGTCAGCAGAGCCAAGAA-3′. The sequences were obtained from the published human MIP-1α promoter sequence (31). The resulting 499 bp fragment (Fig. 1A) was inserted into the SacI and HindIII sites of the pGL3-Basic luciferase reporter vector (Promega).
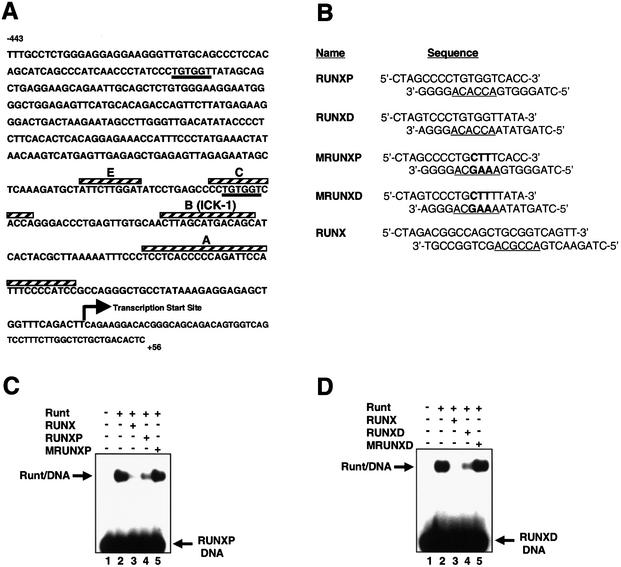
Figure 1.
The MIP-1α promoter contains two consensus RUNX1-binding sites. (A) The sequence of the human MIP-1α promoter sequence (31). The underlined sequences are the proximal and distal RUNX sites located at –132 to –137 bps and –375 to –380 bps, respectively. Sequence elements (A–E) that have been identified by DNase I footprinting in Jurkat T-cells are overlined (hatched) (29). (B) Sequences of the double-stranded oligonucleotides used in the EMSA experiments throughout this study. The RUNX sites are underlined and mutations are shown in bold. RUNXP and RUNXD encompass the proximal and distal sites respectively from the MIP-1α promoter. RUNX is a known RUNX1-binding site (40). MRUNXP and MRUNXD contain mutations of the RUNX sequence in RUNXP and RUNXD respectively. (C) EMSA, demonstrating specific binding of the RUNX1 Runt-domain to the RUNXP site. The Runt-domain was incubated with the radiolabelled RUNXP DNA in the presence or absence of 100-fold molar excess of competitor DNA as indicated above the lanes. The free DNA and Runt/DNA complexes are indicated. (D) EMSA, demonstrating specific binding of the RUNX1 Runt-domain to the RUNXD site. The Runt-domain was incubated with the radiolabelled RUNXD DNA in the presence or absence of 100-fold molar excess of competitor DNA as indicated above the lanes. The free DNA and Runt/DNA complexes are indicated.
Site-directed mutagenesis of the RUNX sites within the MIP-1α promoter was carried out using the QuikChange Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. In both cases the TGTGGT RUNX site was mutated to TGCTTT using oligos 5′-GATATCCTGAGCCCCTGCTTTCACCAGGGACCCTGA-3′ and 5′-TCAGGGTCCCTGGTGAAAGCAGGGGCTCAGGATATC-3′ for the proximal site (MRUNXP) and 5′-CATCAACCCTATCCCTGCTTTTATAGCAGCTGAGGA-3′ and 5′-TCCTCAGCTGCTATAAAAGCAGGGATAGGGTTGATG-3′ for the distal site (MRUNXD). The double mutant (MRUNXDP) was constructed by mutating the RUNXD site in the plasmid containing the proximal RUNX site mutation (MRUNXP). All DNA inserts were verified by sequencing and contain no additional mutations. The MOZ expression plasmid, pLNCX-HA-MOZ has been described previously (24). RUNX1 was expressed using pCMV5-AML-1B (34), CBFβ was expressed using pCMV5-CBFβ (34) and AML-1/ETO was expressed using pCMV-5-AML-1/ETO (20).
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed as described previously (35). Approximately 100 ng of purified Runt-domain or 2 µl of nuclear extract was used in each reaction. The anti-RUNX1 antibody and RUNX1 competitor peptide used in supershift assays were supplied by Geneka Biotechnology and the assays were performed according to the manufacturer’s protocol. Synthetic oligonucleotides were radiolabelled by incorporation of [α-32P]dCTP, using the Klenow fragment of DNA polymerase I according to standard protocols (36). The sequences of pairs of complimentary oligonucleotides used to make radiolabelled probes are shown in Figure 1B. All DNA-binding sites were purified on 10% non-denaturing polyacrylamide gels. In standard reactions, proteins and labelled DNA were incubated in a total of 10 or 12 µl volumes. Reactions were incubated at room temperature for 20 min prior to loading onto a 5% non-denaturing polyacrylamide gel. Gels were fixed, dried and visualised by autoradiography.
Protein purification and nuclear extract preparation
The Runt-domain of RUNX1 was expressed in Escherichia coli as a fusion protein with glutathione S-transferase from the plasmid pGEX-Runt (34). The Runt-domain was purified as described previously for the Elk-1 ETS-domain (37). Nuclear extracts from Jurkat cells were prepared as described previously (38).
Chromatin immunopreicpitation
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit (Upstate Biotechnology) according to the manufacturer’s protocol, except that formaldehyde cross-linking was performed for 30 min in Jurkat T-cells. Three microlitres of anti-RUNX1 antibody (Geneka Biotechnology) or 5 µl (25 µg) of the control anti-HA antibody (Roche) was used in the immunoprecipitation. The MIP-1α promoter sequence was detected with PCR primers –251FWD, 5′-CTCTTCACACTCACAGGAGA-3′ and –26REV, 5′-TAGGCAGCCCTGGCGGAT-3′. Cycling parameters were 99°C for 3 min, and 30 cycles at 95°C for 1 min, 50°C for 1 min and 72°C for 1 min. For the anti-HA ChIP, 35 cycles were performed at 55°C. The region of the MIP-1α gene downstream of the promoter (1110 to 1326 bps) was amplified using the PCR primers 1110FWD, 5′-TTGGAGCTGGAGTGAGGCAT-3′ and 1326REV, 5′-CCCACCATGGCCCCACCATT-3′. Cycling parameters were 99°C for 3 min, and 35 cycles at 95°C for 1 min, 55°C for 1 min and 72°C for 1 min. PCR was performed using Pfu polymerase (Stratagene) and the products were resolved on a 1.5% agarose gel.
RESULTS
RUNX1 binds the consensus RUNX sites in the human MIP-1α promoter
Visual inspection of the MIP-1α promoter revealed the presence of two consensus RUNX-binding sites. The positions of the proximal site (RUNXP) and the distal site (RUNXD) are shown in Figure 1A. DNase I footprinting of the MIP-1α promoter in Jurkat cells has previously shown that the proximal RUNX site is bound by a nuclear factor (Fig. 1A, footprint C) (29). However, the sequence was not recognised as a RUNX site and the identity of the factor binding to it had not previously been established. Having recognised that there are two consensus RUNX sites in the MIP-1α promoter, we therefore used the EMSA to establish whether the RUNX sites in the MIP-1α promoter are able to recruit RUNX transcription factors. Double-stranded oligonucleotides encompassing either the RUNXP or the RUNXD sites were used as radiolabelled probes (Fig. 1B). When the RUNXP site was incubated with the purified recombinant DNA-binding domain of RUNX1 (Runt), a specific retarded complex was observed (Fig. 1C, lane 2). However, in the presence of 100-fold molar excess of a known RUNX1-binding site (RUNX), complex formation was abrogated (Fig. 1C, lane 3). In contrast, competition with a mutant RUNXP site (MRUNXP) did not affect the formation of the complex (Fig. 1C, lane 5), whereas competition with the wild-type RUNXP site considerably reduced binding (Fig. 1C, lane 4). Almost identical results were also obtained with the RUNXD site (Fig. 1D). These data demonstrate that the RUNX sites in the MIP-1α promoter can be specifically recognised by the RUNX1 DNA-binding domain.
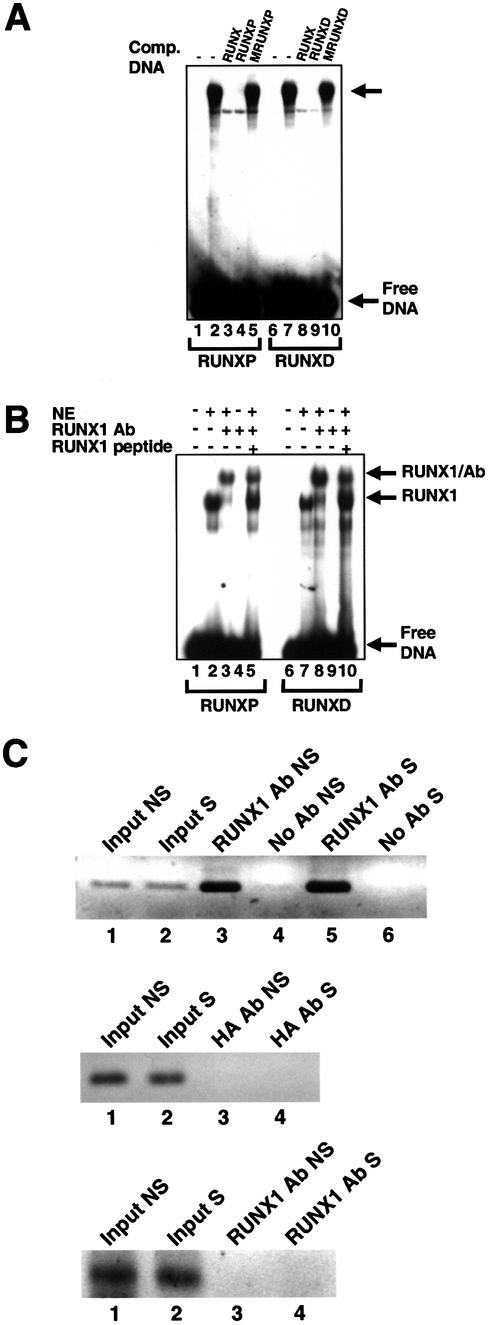
To establish whether endogenous proteins from Jurkat cells are able to bind to the RUNX sites in the MIP-1α promoter, nuclear extracts were incubated with the RUNX sites in the presence of the competitor sequences. Incubation of nuclear extracts with the RUNXP site produced a specific retarded complex (Fig. 2A, lane 2), which was completely abolished in the presence of either the RUNX or RUNXP competitors (Fig. 2A, lanes 3 and 4). In contrast the MRUNXP competitor did not affect the formation of the complex (Fig. 2A, lane 5). The same results were obtained with the RUNXD site (Fig. 2A, lanes 6–10).
Figure 2.
RUNX1 binds to the MIP-1α promoter in Jurkat cells. (A) EMSA demonstrating specific binding of a nuclear factor to the RUNX sites of the MIP-1α promoter. Nuclear extracts from Jurkat T-cells were incubated with radiolabelled RUNXP (lanes 1–5) or RUNXD (lanes 6–10) in the presence or absence of 100-fold molar excess of competitor DNA as indicated above the lanes. Lanes 1 and 6 contain radiolabelled probe only. The arrow indicates specific complexes. (B) EMSA demonstrating specific supershift of the RUNX1/DNA complexes in Jurkat T-cell nuclear extracts. Nuclear extracts (NE) from Jurkat T-cells were incubated with radiolabelled RUNXP (lanes 1–5) or RUNXD (lanes 6–10) in the presence or absence of RUNX1-specific antibody or the RUNX1 peptide competitor as indicated above the lanes. The identity of each of the complexes is indicated. (C) ChIP assays demonstrating specific binding of RUNX1 to the endogenous MIP-1α promoter. The MIP-1α promoter or an internal MIP-1α gene fragment was amplified from chromatin derived from either non-stimulated (NS) or PMA/PHA-stimulated (S) Jurkat cells as indicated above the lanes. The identity of the input chromatin or immunoprecipitated chromatin is also indicated above the lanes. Upper panel, primers specific for the MIP-1α promoter were used to amplify DNA from complexes immunoprecipitated with the anti-RUNX1 antibody (lanes 3 and 5). Lanes 4 and 6 show no-antibody controls. Middle panel, primers specific for the MIP-1α promoter were used to amplify DNA from complexes immunoprecipitated with the anti-HA antibody (lanes 3 and 4). Lower panel, primers specific for the internal MIP-1α gene fragment (+1110 to +1326 bps) were used to amplify DNA from complexes immunoprecipitated with the anti-RUNX1 antibody (lanes 3 and 4).
The identity of the RUNX protein in the Jurkat nuclear extracts was determined using a RUNX1-specific polyclonal antibody in a supershift assay. In the presence of a RUNX1-specific antibody, the specific complex was almost entirely abolished and an additional supershifted complex appeared (Fig. 2B, compare lanes 2 and 3). Addition of antibody alone did not form a complex (Fig. 2B, lane 4). Furthermore when the extracts were incubated in the presence of RUNX1-specific antibody and competitor peptide, to which the antibody was raised, the RUNX1 complex reappeared and the supershifted complex was diminished (Fig. 2B, lane 5). Almost identical results were obtained with the RUNXD site (Fig. 2B, lanes 6–10). Taken together, the data clearly demonstrate that both of the RUNX sites in the MIP-1α promoter can be specifically bound by endogenous RUNX1.
RUNX-1 binds the endogenous MIP-1α promoter
The ability of RUNX1 to bind to the MIP-1α promoter in EMSA experiments suggested that MIP-1α is a direct RUNX1 target gene. We therefore performed ChIP assays using non-stimulated and PMA/PHA-stimulated Jurkat cells. Primers were designed to amplify a 225 bp fragment of the MIP-1α promoter encompassing the proximal RUNX1 site. The MIP-1α promoter could be amplified from anti-RUNX1 complexes immunoprecipitated from either non-stimulated or stimulated cells (Fig. 2C, upper panel). In contrast, complexes were not amplified from immunoprecipitations using the control anti-HA antibody (Fig. 2C, middle panel). To establish that PCR amplification of the MIP-1α promoter was specific for RUNX1-bound DNA, internal control primers were designed to amplify a region remote from the promoter within the MIP-1α gene (+1110 to +1326 bps). The internal primers did not amplify DNA from anti-RUNX1 complexes immunoprecipitated from either non-stimulated or stimulated cells but were able to amplify a fragment from the input DNA prior to immunoprecipitation (Fig. 2C, lower panel). These data clearly demonstrate that the endogenous MIP-1α gene is a direct target of RUNX1. Furthermore RUNX1 is bound to the MIP-1α promoter prior to stimulation, and after stimulation with PMA/PHA.
The proximal RUNX1-binding site is essential for activation of the MIP-1α promoter
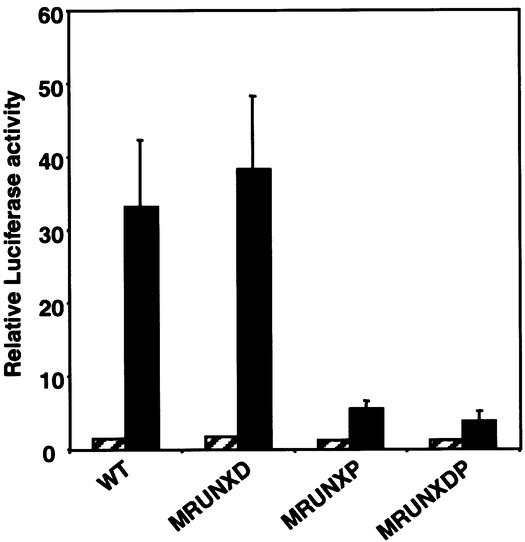
Previous studies have demonstrated that both endogenous MIP-1α expression and MIP-1α promoter reporter constructs are induced by PMA/PHA stimulation, suggesting that the response of MIP-1α promoter reporters to PMA/PHA stimulation reflects the response of the endogenous gene (29). To determine whether the RUNX-binding sites are required for transcriptional activation, mutations that abolish RUNX1 binding were introduced into the promoter. The MIP-1α promoter (Fig. 1A, +56 to –443 bps) was ligated upstream of the luciferase cDNA in the reporter plasmid pGL3-basic and the activity of the wild-type and mutant promoters was determined in Jurkat cells (Fig. 3). Previous reports have established that the MIP-1α promoter is activated in Jurkat cells by stimulation with PMA and PHA (29). The activity of the wild-type and mutant promoters in response to PMA/PHA stimulation is shown in Figure 3. PMA/PHA treatment stimulated the wild-type promoter 25-fold (Fig. 3, WT). Mutation of the distal RUNX site had little effect on the activity of the promoter as stimulation with PMA/PHA activated the promoter 23-fold (Fig. 3, MRUNXD). In contrast, when the proximal RUNX site was mutated only 5-fold activation was observed (Fig. 3, MRUNXP). When both sites were mutated, activation of the promoter was reduced to 3-fold (Fig. 3, MRUNXDP). These data clearly demonstrate that the proximal RUNX site is essential for full activation of the MIP-1α promoter and that the distal RUNX site does not make a significant contribution.
Figure 3.
The proximal RUNX site is essential for transcriptional activation of the MIP-1α promoter in Jurkat T-cells. Jurkat T-cells were transfected with the luciferase reporter plasmid containing, either the wild type promoter (WT), the distal RUNX site mutant (MRUNXD), the proximal RUNX site mutant (MRUNXP), the double RUNX site mutant (MRUNXDP) or the empty plasmid pGL3-Basic. The total amount of DNA was kept constant by the addition of pCMV-5. Transfected cells were left unstimulated (hatched) or stimulated with PMA/PHA (black). Luciferase activities were determined from triplicate experiments (mean ± standard error). All values are relative to the activity of the pGL3-Basic reporter.
A dominant negative form of RUNX1 represses PMA/PHA activation
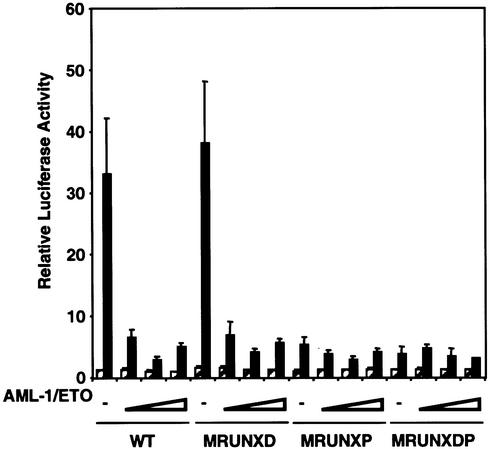
Heterologous expression of RUNX1, even with its dimerization partner CBFβ, did not further activate the MIP-1α promoter in Jurkat cells (data not shown). However, this was not surprising since the high level of expression of endogenous RUNX1 in Jurkat cells is probably already saturating. In addition, the MIP-1α promoter contains a previously characterised negative regulatory element, ICK-1, which represses promoter activity in the absence of PMA/PHA stimulation (29). We therefore used the AML-1/ETO fusion protein as a dominant negative form of RUNX1 to inhibit PMA/PHA stimulation of the MIP-1α promoter (Fig. 4). AML-1/ETO is a repressive form of RUNX1 arising from the t(8;21) translocation and is often used to inhibit transcriptional activation via RUNX sites (4,14,20,39). When the wild-type MIP-1α promoter was co-transfected with the AML-1/ETO-expressing construct in Jurkat cells, transcriptional activation of the promoter was inhibited, with a maximum 11.5-fold inhibition observed with 400 ng of AML-1/ETO expression plasmid (Fig. 4, WT). When AML-1/ETO was co-expressed with the MIP-1α promoter containing the mutated distal RUNX site, an almost identical pattern of inhibition was observed at all levels of AML-1/ETO expression, suggesting that AML-1/ETO was inhibiting activation via the intact proximal RUNX site (Fig. 4, MRUNXD). Indeed, when the proximal RUNX site was mutated we observed maximum inhibition of only 2-fold (Fig. 4, MRUNXP). AML-1/ETO therefore requires the intact proximal RUNX site to exert most of its inhibitory effects. However, we cannot discount a minor role for the distal site because when both RUNX sites were mutated, AML-1/ETO was unable to inhibit activation (Fig. 4, MRUNXDP). These data clearly demonstrate that a dominant interfering form of RUNX1 can inhibit activation of the promoter. The greatest inhibition was observed when the proximal RUNX site was intact, suggesting that AML-1/ETO acts predominantly via this site.
Figure 4.
AML-1/ETO represses PMA/PHA activation of the MIP-1α promoter. Jurkat T-cells were transfected with the luciferase reporter plasmid containing, either the wild-type promoter (WT), the distal RUNX site mutant (MRUNXD), the proximal RUNX site mutant (MRUNXP), the double RUNX site mutant (MRUNXDP) or the empty plasmid pGL3-Basic. Increasing amounts of the AML-1/ETO expression plasmid (0, 100, 400 and 800 ng) were transfected with each of the reporter plasmids as indicated on the x-axis. The total amount of DNA was kept constant by the addition of pCMV-5. Transfected cells were left unstimulated (hatched) or stimulated with PMA/PHA (black). Luciferase activities were determined from triplicate experiments (mean ± standard error). All values are relative to the activity of the pGL3-Basic reporter.
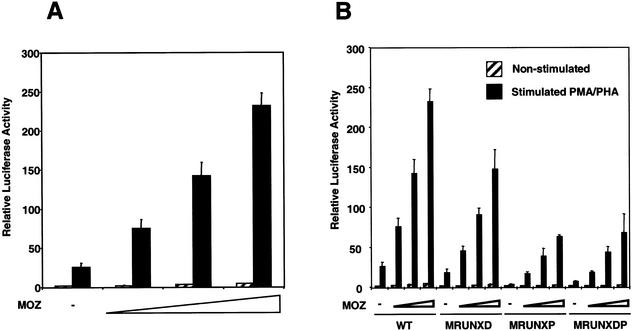
Activation of MIP-1α promoter by the acetyltransferase MOZ
RUNX1 has been shown to activate transcription by recruiting co-activators (4–6,23,24). We therefore investigated the ability of the histone acetyltransferase MOZ, which is a strong RUNX1 co-activator, to activate the MIP-1α promoter. To determine whether MOZ can activate the MIP-1α promoter in Jurkat T-cells, increasing amounts of a MOZ expression plasmid were co-transfected with the MIP-1α promoter plasmid. In the absence of PMA/PHA stimulation, no significant increase in MIP-1α promoter activity was observed. However, upon stimulation with PMA/PHA, the wild-type promoter activity increased substantially with increasing amounts of MOZ (Fig. 5A). To investigate whether the RUNX sites were required for MOZ-dependent activation of the MIP-1α promoter, increasing amounts of MOZ were co-transfected with the MIP-1α promoter plasmids containing RUNX-site mutations (Fig. 5B). MOZ enhanced activation of the wild-type and mutant promoters at every level (Fig. 5B). However, whilst the level of MOZ-dependent activation was only moderately inhibited on the distal RUNX site mutant (MRUNXD), it was severely inhibited when the proximal RUNX site was mutated (MRUNXP). Mutation of both RUNX sites resulted in a similar level of activation to that observed with the mutation of the proximal site alone (Fig. 5B, MRUNXDP). Taken together, these data clearly demonstrate that MOZ can activate the MIP-1α promoter in Jurkat T-cells and that the proximal RUNX site is required for maximum MOZ-dependent activation.
Figure 5.
MOZ activates the MIP-1α promoter in Jurkat T-cells. (A) Increasing amounts of MOZ activates the MIP-1α promoter. Jurkat T-cells were co-transfected with the wild type MIP-1α promoter reporter and increasing amounts of a MOZ expression plasmid (0, 100, 300 and 800 ng) as indicated on the x-axis. The total amount of DNA was kept constant by the addition of the empty expression plasmid pCMV-5. Transfected cells were left unstimulated (hatched) or stimulated with PMA/PHA (black). Luciferase activities were determined from triplicate experiments (mean ± standard error). All values are relative to the activity of the pGL3-Basic reporter. (B) Effect of RUNX site mutations on MOZ-dependent activation. Jurkat T-cells were co-transfected with increasing amounts of a MOZ expression plasmid (0, 100, 300 and 800 ng) and MIP-1α promoter reporter plasmids containing RUNX-site mutations as indicated on the x-axis. Transfected cells were left unstimulated (hatched) or stimulated with PMA/PHA (black). Luciferase activities were determined from triplicate experiments (mean ± standard error). All values are relative to the activity of the pGL3-Basic reporter.
RUNX1 and MOZ can cooperate to activate the MIP-1α promoter
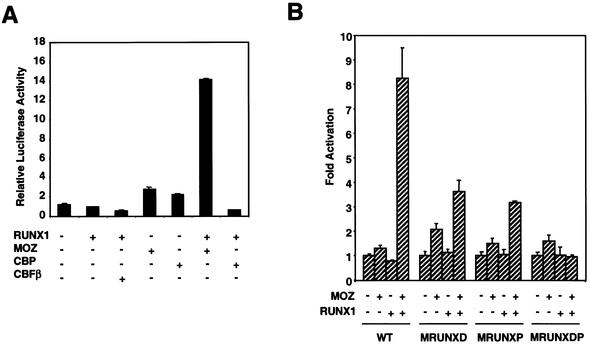
Whilst a large proportion of the MOZ-dependent activation required the intact proximal RUNX site, we noted that MOZ could still activate the MIP-1α promoter even in the absence of both RUNX sites. To determine unequivocally whether MOZ can activate the MIP-1α promoter in a RUNX1-dependent manner, we tested the ability of RUNX1 and MOZ to directly activate the MIP-1α promoter in a RUNX1 negative cell line. We therefore used HeLa cells, which do not express RUNX transcription factors and have previously been used to demonstrate the effects of heterologously expressed RUNX1 on the activity of promoters (20). Expression of RUNX1 alone in HeLa cells was unable to activate the MIP-1α promoter (Fig. 6A). However, when RUNX1 and MOZ were co-expressed, significant activation of the promoter was observed (14-fold, Fig. 6A). In contrast, only 2-fold activation was observed in the presence of MOZ alone (Fig. 6A). Co-expression of RUNX1 with either CBP or CBFβ did not activate the promoter (Fig. 6A). These results clearly demonstrate that MOZ can activate the MIP-1α promoter in a RUNX1-dependent manner.
Figure 6.
RUNX1 and MOZ activate the MIP-1α promoter. (A) Transcriptional activition of the MIP-1α promoter by RUNX1 and MOZ. HeLa cells were co-transfected with the MIP-1α promoter reporter and RUNX1, MOZ, CBP or CBFβ expression plasmids (400 ng) as indicated. Luciferase activities were determined from triplicate experiments (mean ± standard error). (B) HeLa cells were co-transfected with either the MIP-1α promoter or the RUNX site mutant derivatives and the RUNX1 and MOZ expressing plasmid as indicated. The total amount of DNA was kept constant by the addition of pCMV-5. Luciferase activities were determined from triplicate experiments (mean ± standard error). Values are expressed as fold activation relative to the basal activity of the reporters, which was set to 1.
To determine whether the ability of RUNX1 and MOZ to synergistically activate the MIP-1α promoter was dependent upon the RUNX sites, we performed co-transfections with the mutant RUNX site reporters (Fig. 6B). Mutation of either the distal or the proximal RUNX sites reduced RUNX1/MOZ-dependent transcription by >50% (Fig. 6B; compare WT with MRUNXD and MRUNXP). Moreover, when both sites were mutated, activation by RUNX1 and MOZ was completely abolished (Fig. 6B, MRUNXDP). Taken together these data demonstrate that either of the RUNX sites is potentially able to mediate MOZ-dependent activation but in T-cells the proximal RUNX site is the most important.
DISCUSSION
We have clearly demonstrated that the human MIP-1α promoter contains two consensus RUNX sites that are able to bind to RUNX1. We have also shown that endogenous RUNX1 is bound to the endogenous MIP-1α promoter in Jurkat T-cells. However, only the proximal RUNX site is essential for activation of the MIP-1α promoter in T-cells. In addition we have demonstrated that RUNX1 and the histone acetyltransferase MOZ can synergistically activate the MIP-1α promoter. RUNX1/MOZ-dependent activation in HeLa cells required at least one of the RUNX sites to be intact, suggesting that both sites can mediate this activation. However, in T-cells, MOZ-dependent activation was largely mediated via the proximal RUNX site. These findings indicate that whilst both sites are able to recruit RUNX1 and contribute to activation of the MIP-1α promoter, only the proximal RUNX site makes a significant contribution to MIP-1α promoter activation in T-cells.
A previous DNase I footprinting analysis of the MIP-1α promoter identified a number of sequence elements that are occupied by nuclear factors in Jurkat cells (Fig. 1A) (29). The proximal RUNX site is one of the sequences identified in the footprinting analysis (Fig. 1A, footprint C) (29). However, previous deletion analysis of the MIP-1α promoter did not identify the RUNX site as an essential element because the MIP-1α promoter is rendered inactive in T-cells by deletion of sequences upstream of the proximal RUNX site (29). However, site-directed mutation of the proximal RUNX site, in the context of the whole promoter, has enabled us to define it as an essential element for PMA/PHA activation of the MIP-1α promoter in T-cells. Taken together with the fact that RUNX1 in Jurkat cells is the major nuclear factor binding to the RUNX sites and that the endogenous MIP-1α promoter is bound by RUNX1, our data clearly indicate that RUNX1 has an essential role in mediating PMA/PHA activation of the MIP-1α promoter in Jurkat T-cells.
The ICK-1 element (Fig. 1A) is also essential for transcriptional activation of the MIP-1α promoter in Jurkat cells (29). This element acts as both a positive and negative regulator of transcription (29). Its role as a negative regulator in non-stimulated T-cells could explain why we did not observe activation of the MIP-1α promoter by the heterologous expression of RUNX1 in Jurkat cells. In addition, the high level of expression of endogenous RUNX1 in Jurkat cells suggests that it is already saturating. We therefore investigated the effect of RUNX1 on the MIP-1α promoter in HeLa cells in which expression of RUNX1 is not detectable (20). In the absence of any co-activators, RUNX1 is a relatively weak transcriptional regulator but cooperates effectively with other transcription factors such as Ets-1 and c-Myb, or with co-activators such as MOZ (4,5,24). Consistent with this, we found that RUNX1 alone was unable to activate the MIP1-α promoter. However, the promoter was strongly activated when MOZ was expressed with RUNX1 and this activation was dependent upon the RUNX sites. In T-cells, we demonstrated that MOZ activation of the MIP-1α promoter was largely dependent upon the proximal RUNX site. MOZ has recently been shown to interact directly with RUNX1 and strongly stimulate RUNX1-mediated transcriptional activation (24). It is therefore likely that RUNX1 directly recruits MOZ to the MIP-1α promoter via the proximal RUNX site to activate transcription. We also note that although the proximal RUNX site is required for maximum MOZ-dependent activation, MOZ can still activate the MIP-1α promoter even when both RUNX sites are mutated. This suggests that MOZ can also contribute to activation of the MIP-1α promoter in the absence of the consensus RUNX sites, possibly via non-consensus RUNX sites or alternative transcription factors.
In addition to IL-3 and GM-CSF, MIP-1α should now be considered as a RUNX1-regulated cytokine gene. Our findings therefore support the notion that cytokine genes are major targets of RUNX1 in T-cells. In addition to regulating haematopoietic genes, RUNX1 regulates transcription of the cell cycle inhibitors p14ARF and p21WAF–1 (19,20). The function of RUNX1, as a master regulator of haematopoiesis, therefore probably reflects its role as a regulator of cytokine and cell cycle genes.
Several chromosomal rearrangements involving the RUNX1 gene are associated with leukaemias and give rise to fusion proteins, which deregulate the transcriptional regulatory function of RUNX1 (4,24,26). Some of these rearrangements transform RUNX1 into a transcriptional repressor, for example, AML-l/ETO acts as a constitutive transcriptional repressor and disrupts normal differentiation of haematopoietic cells (4). Of particular note is the recently described T-cell lymphoblastic leukaemia associated with t(4;21) in which RUNX1 is translocated (12). Although the fusion protein product of this rearrangement has not yet been identified, our finding that RUNX1 regulates the MIP-1α promoter in Jurkat T-cells suggests that deregulated MIP-1α expression may occur in leukaemic T-cells carrying the t(4;21) translocation. MIP-1α can reversibly inhibit the proliferation of particular haematopoietic stem cells (27,28). Deregulation of MIP-1α expression may therefore contribute to the progression of leukaemia by perturbing haematopoietic stem cell proliferation.
Chromosomal rearrangements involving the gene encoding MOZ are associated with acute myeloid leukaemia (25,26). The t(8;16) translocation gives rise to the fusion protein MOZ-CBP which has been shown to inhibit RUNX1-dependent transcriptional activation, and is thought to contribute to the development of leukaemia via its effects on RUNX1 activity (24). Although the proximal RUNX site does not appear to be necessary for activation of the MIP-1α promoter by PMA/PHA stimulation in myeloid cells, RUNX1 fusion proteins in myeloid leukaemias may target the site and deregulate MIP-1α expression. Having established the role of RUNX1 and MOZ in activating the MIP-1α promoter in Jurkat T-cells, it will be interesting to investigate the effects of oncogenic RUNX1 and MOZ fusion proteins on the MIP-1α promoter in myeloid cells.
Since MIP-1α is able to negatively regulate haematopoietic stem cell proliferation, our finding that RUNX1 and MOZ contribute to transcriptional regulation of the MIP-1α promoter indicates that at least part of the mechanism by which RUNX1 and MOZ regulates haematopoiesis is by regulating the expression of MIP-1α. Chromosomal rearrangements in the RUNX1 and MOZ genes may therefore contribute to the development of leukaemia by alteration of MIP-1α expression.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Paul Brandwood for doing the EMSAs and Larisa Logunova for excellent technical assistance. We also thank Scott Hiebert and Dong-Er Zhang for kindly providing plasmids. We are grateful to Claire Inman for critically reading the manuscript. This work was supported by a Wellcome Trust Career Development Fellowship (P.S.), the Royal Society and a BBSRC studentship (C.B.).
REFERENCES
- 1.Karsenty G. (2000) Role of Cbfa1 in osteoblast differentiation and function. Semin. Cell Dev. Biol., 11, 343–346. [DOI] [PubMed] [Google Scholar]
- 2.Tracey W.D. and Speck,N.A. (2000) Potential roles for RUNX1 and its orthologs in determining hematopoietic cell fate. Semin. Cell Dev. Biol., 11, 337–342. [DOI] [PubMed] [Google Scholar]
- 3.Canon J. and Banerjee,U. (2000) Runt and Lozenge function in Drosophila development. Semin. Cell Dev. Biol., 11, 327–336. [DOI] [PubMed] [Google Scholar]
- 4.Lutterbach B. and Hiebert,S.W. (2000) Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene, 245, 223–235. [DOI] [PubMed] [Google Scholar]
- 5.Wheeler J.C., Shigesada,K., Gergen,J.P. and Ito,Y. (2000) Mechanisms of transcriptional regulation by Runt domain proteins. Semin. Cell Dev. Biol., 11, 369–375. [DOI] [PubMed] [Google Scholar]
- 6.Adya N., Castilla,L.H. and Liu,P.P. (2000) Function of CBFbeta/Bro proteins. Semin. Cell Dev. Biol., 11, 361–368. [DOI] [PubMed] [Google Scholar]
- 7.Okuda T., van Deursen,J., Hiebert,S.W., Grosveld,G. and Downing,J.R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell, 84, 321–330. [DOI] [PubMed] [Google Scholar]
- 8.Otto F., Thornell,A.P., Crompton,T., Denzel,A., Gilmour,K.C., Rosewell,I.R., Stamp,G.W., Beddington,R.S., Mundlos,S., Olsen,B.R., Selby,P.B. and Owen,M.J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell, 89, 765–771. [DOI] [PubMed] [Google Scholar]
- 9.Komori T., Yagi,H., Nomura,S., Yamaguchi,A., Sasaki,K., Deguchi,K., Shimizu,Y., Bronson,R.T., Gao,Y.H., Inada,M., Sato,M., Okamoto,R., Kitamura,Y., Yoshiki,S. and Kishimoto,T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 89, 755–764. [DOI] [PubMed] [Google Scholar]
- 10.Downing J.R., Higuchi,M., Lenny,N. and Yeoh,A.E. (2000) Alterations of the AML1 transcription factor in human leukemia. Semin. Cell Dev. Biol., 11, 347–360. [DOI] [PubMed] [Google Scholar]
- 11.Mundlos S., Otto,F., Mundlos,C., Mulliken,J.B., Aylsworth,A.S., Albright,S., Lindhout,D., Cole,W.G., Henn,W., Knoll,J.H., Owen,M.J., Mertelsmann,R., Zabel,B.U. and Olsen,B.R. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell, 89, 773–779. [DOI] [PubMed] [Google Scholar]
- 12.Mikhail F.M., Serry,K.A., Hatem,N., Mourad,Z.I., Farawela,H.M., El Kaffash,D.M., Coignet,L. and Nucifora,G. (2002) A new translocation that rearranges the AML1 gene in a patient with T-cell acute lymphoblastic leukemia. Cancer Genet. Cytogenet., 135, 96–100. [DOI] [PubMed] [Google Scholar]
- 13.Taylor D.S., Laubach,J.P., Nathan,D.G. and Mathey-Prevot,B. (1996) Cooperation between core binding factor and adjacent promoter elements contributes to the tissue-specific expression of interleukin-3. J. Biol. Chem., 271, 14020–14027. [DOI] [PubMed] [Google Scholar]
- 14.Uchida H., Zhang,J. and Nimer,S.D. (1997) AML1A and AML1B can transactivate the human IL-3 promoter. J. Immunol., 158, 2251–2258. [PubMed] [Google Scholar]
- 15.Takahashi A., Satake,M., Yamaguchi-Iwai,Y., Bae,S.C., Lu,J., Maruyama,M., Zhang,Y.W., Oka,H., Arai,N., Arai,K. et al. (1995) Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood, 86, 607–616. [PubMed] [Google Scholar]
- 16.Cockerill P.N., Osborne,C.S., Bert,A.G. and Grotto,R.J. (1996) Regulation of GM-CSF gene transcription by core-binding factor. Cell Growth Differ., 7, 917–922. [PubMed] [Google Scholar]
- 17.Zhang D.E., Fujioka,K., Hetherington,C.J., Shapiro,L.H., Chen,H.M., Look,A.T. and Tenen,D.G. (1994) Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol. Cell. Biol., 14, 8085–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuchprayoon I., Meyers,S., Scott,L.M., Suzow,J., Hiebert,S. and Friedman,A.D. (1994) PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2 beta/CBF beta proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol. Cell. Biol., 14, 5558–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutterbach B., Westendorf,J.J., Linggi,B., Isaac,S., Seto,E. and Hiebert,S.W. (2000) A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem., 275, 651–656. [DOI] [PubMed] [Google Scholar]
- 20.Linggi B., Muller-Tidow,C., van de Locht,L., Hu,M., Nip,J., Serve,H., Berdel,W.E., van der Reijden,B., Quelle,D.E., Rowley,J.D., Cleveland,J., Jansen,J.H., Pandolfi,P.P. and Hiebert,S.W. (2002) The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nature Med., 8, 743–750. [DOI] [PubMed] [Google Scholar]
- 21.Fisher A.L. and Caudy,M. (1998) Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev., 12, 1931–1940. [DOI] [PubMed] [Google Scholar]
- 22.Javed A., Guo,B., Hiebert,S., Choi,J.Y., Green,J., Zhao,S.C., Osborne,M.A., Stifani,S., Stein,J.L., Lian,J.B., van Wijnen,A.J. and Stein,G.S. (2000) Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J. Cell Sci., 113, 2221–2231. [DOI] [PubMed] [Google Scholar]
- 23.Kitabayashi I., Yokoyama,A., Shimizu,K. and Ohki,M. (1998) Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J., 17, 2994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitabayashi I., Aikawa,Y., Nguyen,L.A., Yokoyama,A. and Ohki,M. (2001) Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ-CBP fusion protein. EMBO J., 20, 7184–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borrow J., Stanton,V.P.,Jr, Andresen,J.M., Becher,R., Behm,F.G., Chaganti,R.S., Civin,C.I., Disteche,C., Dube,I., Frischauf,A.M., Horsman,D., Mitelman,F., Volinia,S., Watmore,A.E. and Housman,D.E. (1996) The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature Genet., 14, 33–41. [DOI] [PubMed] [Google Scholar]
- 26.Liang J., Prouty,L., Williams,B.J., Dayton,M.A. and Blanchard,K.L. (1998) Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood, 92, 2118–2122. [PubMed] [Google Scholar]
- 27.Menten P., Wuyts,A. and Van Damme,J. (2002) Macrophage inflammatory protein-1. Cytokine Growth Factor Rev., 13, 455–481. [DOI] [PubMed] [Google Scholar]
- 28.Graham G.J., Wright,E.G., Hewick,R., Wolpe,S.D., Wilkie,N.M., Donaldson,D., Lorimore,S. and Pragnell,I.B. (1990) Identification and characterization of an inhibitor of haemopoietic stem cell proliferation. Nature, 344, 442–444. [DOI] [PubMed] [Google Scholar]
- 29.Nomiyama H., Hieshima,K., Hirokawa,K., Hattori,T., Takatsuki,K. and Miura,R. (1993) Characterization of cytokine LD78 gene promoters: positive and negative transcriptional factors bind to a negative regulatory element common to LD78, interleukin-3, and granulocyte-macrophage colony-stimulating factor gene promoters, Mol. Cell. Biol., 13, 2787–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Widmer U., Manogue,K.R., Cerami,A. and Sherry,B. (1993) Genomic cloning and promoter analysis of macrophage inflammatory protein (MIP)-2, MIP-1 alpha, and MIP-1 beta, members of the chemokine superfamily of proinflammatory cytokines. J. Immunol., 150, 4996–5012. [PubMed] [Google Scholar]
- 31.Nakao M., Nomiyama,H. and Shimada,K. (1990) Structures of human genes coding for cytokine LD78 and their expression. Mol. Cell. Biol., 10, 3646–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto M., Sakao,Y. and Akira,S. (1998) Inducible expression of nuclear factor IL-6 increases endogenous gene expression of macrophage inflammatory protein-1 alpha, osteopontin and CD14 in a monocytic leukemia cell line. Int. Immunol., 10, 1825–1835. [DOI] [PubMed] [Google Scholar]
- 33.Widmer U., Yang,Z., van Deventer,S., Manogue,K.R., Sherry,B. and Cerami,A. (1991) Genomic structure of murine macrophage inflammatory protein-1 alpha and conservation of potential regulatory sequences with a human homolog, LD78. J. Immunol., 146, 4031–4040. [PubMed] [Google Scholar]
- 34.Zhang D.E., Hetherington,C.J., Meyers,S., Rhoades,K.L., Larson,C.J., Chen,H.M., Hiebert,S.W. and Tenen,D.G. (1996) CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol., 16, 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagoshima H., Akamatsu,Y., Ito,Y. and Shigesada,K. (1996) Functional dissection of the alpha and beta subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J. Biol. Chem., 271, 33074–33082. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Shore P., Bisset,L., Lakey,J., Waltho,J.P., Virden,R. and Sharrocks,A.D. (1995) Characterization of the Elk-1 ETS DNA-binding domain. J. Biol. Chem., 270, 5805–5811. [DOI] [PubMed] [Google Scholar]
- 38.Shore P., Dietrich,W. and Corcoran,L.M. (2002) Oct-2 regulates CD36 gene expression via a consensus octamer, which excludes the co-activator OBF-1. Nucleic Acids Res., 30, 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers S., Lenny,N. and Hiebert,S.W. (1995) The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol., 15, 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bae S.C., Ogawa,E., Maruyama,M., Oka,H., Satake,M., Shigesada,K., Jenkins,N.A., Gilbert,D.J., Copeland,N.G. and Ito,Y. (1994) PEBP2 alpha B/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol. Cell. Biol., 14, 3242–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]