Abstract
Deletions in mitochondrial DNA (mtDNA) accumulate with age in humans without overt mitochondriopathies, but relatively limited attention has been devoted to the measurement of the total number of mtDNA molecules per cell during ageing. We have developed a precise assay that determines mtDNA levels relative to nuclear DNA using a PCR-based procedure. Quantification was performed by reference to a single recombinant plasmid standard containing a copy of each target DNA sequence (mitochondrial and nuclear). Copy number of mtDNA was determined by amplifying a short region of the cytochrome b gene (although other regions of mtDNA were demonstrably useful). Nuclear DNA content was determined by amplification of a segment of the single copy β-globin gene. The copy number of mtDNA per diploid nuclear genome in myocardium was 6970 ± 920, significantly higher than that in skeletal muscle, 3650 ± 620 (P = 0.006). In both human skeletal muscle and myocardium, there was no significant change in mtDNA copy number with age (from neonates to subjects older than 80 years). This PCR-based assay not only enables accurate determination of mtDNA relative to nuclear DNA but also has the potential to quantify accurately any DNA sequence in relation to any other.
INTRODUCTION
Human mitochondrial DNA (mtDNA) is a circular 16 569 bp, multicopy genome that encodes 13 polypeptides of the electron transport chain and ATP synthase. Normal assembly and operation of the respiratory chain requires an intact and functional mitochondrial genome (1). The functional complement of mtDNA in a cell depends on both the copy number of mitochondrial genomes, which was first shown to be of the order of thousands per cultured mammalian cell (2), as well as the integrity of each mtDNA molecule. Many studies during the past decade have demonstrated somatic mtDNA mutations that accumulate with age (3). Human mtDNA is known to accumulate a variety of deletional mutations that occur in all regions of the genome (4). Various qualitative and quantitative assays have demonstrated an age-related increase in the incidence and abundance of mtDNA deletions of variable sizes and locations (2,5,6) which would be expected to reduce the complement of fully functional mtDNA. However, comparatively few studies have focused on mtDNA genome copy number, especially in human tissues.
Previous studies on various mammalian tissues have claimed that mtDNA copy number increases with age, decreases with age or remains more or less the same (7–14). Such studies (7–12) have mostly relied on Southern blotting approaches. Quantification can also be obtained using PCR (13,14). Quantitative PCR approaches have been extensively applied by us to the measurement of the abundance of the accumulated age-associated deletions, in proportion to total mtDNA, in a variety of mammalian tissues (6,15). The use of reference plasmids containing cloned segments of normal mtDNA, and those bearing specific deletions to be quantified, has been an essential feature leading to precise and reliable data (16,17).
In this study we have refined such PCR procedures to quantify total mtDNA relative to nuclear DNA as a function of age in human tissues. To ensure accurate quantification of mtDNA and nuclear DNA relative to each other, we devised a procedure based on a single reference plasmid that contains segments of both an appropriate region of mtDNA and a single copy nuclear gene encoding β-globin. We thereby used a PCR-based assay to accurately determine total mtDNA copy number relative to the diploid chromosomal DNA content. First, we demonstrated that any part of several regions tested of the mitochondrial genome could be used as a copy number marker. We then determined mtDNA copy number per diploid nuclear genome in a series of skeletal muscle and myocardial samples obtained from individuals of different ages and without overt mitochondrial disease.
MATERIALS AND METHODS
Human tissues and DNA extraction
Of the 29 human skeletal muscle samples studied here, 14 were obtained at autopsy from the quadratus femoris, deltoid and diaphragmatic muscles in individuals who had died from a variety of conditions including trauma [among those reported in Liu et al. (6) and Zhang et al. (16)]. A further 15 samples were obtained from vastus lateralis muscle during surgery of patients undergoing hip operations (Epworth Hospital, Melbourne). The age range for the whole group was between 1 h and 95 years. Samples of right atrial myocardium (n = 35) were harvested from tissue discarded during cardiopulmonary bypass cannulation of patients undergoing cardiac surgery at the Alfred Hospital, Melbourne (n = 28) and the Royal Children’s Hospital, Melbourne (n = 7). The age range in this group was between 3 weeks and 80 years. No individual had an overt mitochondrial disorder. DNA was extracted from fibroblasts of a patient with Pearson’s syndrome (kindly provided by Dr D. R. Thorburn, Murdoch Institute, Melbourne). The study protocol was approved by the Monash University Standing Committee on Ethics in Research on Humans.
Total cellular DNA was extracted from muscle samples (10–100 mg wet wt) as described previously (18). The DNA was stored in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). Absorbance readings (260 nm) of DNA extracts indicated the DNA concentration to be in the range 50–300 ng/µl.
Oligonucleotide primers
Oligonucleotide primers (Bresatec Ltd, Adelaide, South Australia) encompassing four regions spaced around human mtDNA and within the human β-globin gene are summarised in Table 1, together with PCR product sizes.
Table 1. Oligonucleotide primer positions and product lengths.
| Target | Region | Forward primer | Reverse primer | Length of PCR product (bp) |
|---|---|---|---|---|
| mtDNAa | A | L3108(20) | H3500(20) | 393 |
| B | L5223(24) | H5586(20) | 364 | |
| C | L12759(21) | H13188(22) | 430 | |
| D | L14820(23) | H15253(20) | 434 | |
| β-Globinb | (5′) 84–103 (3′) | (5′) 439–421 (3′) | 356 |
aNomenclature of primers is according to Vaillant and Nagley (19). The first number after L or H (corresponding to light and heavy chains of human mtDNA, respectively) is the sequence position (20) of the 5′ end of the primer. The number in parentheses is the length of the primer in nucleotides. Each primer pair corresponds to a particular region of mtDNA used for quantification of total mtDNA.
bSequence as reported in Lawn et al. (21), with nucleotide positions relative to 5′ end of first exon of β-globin.
Polymerase chain reaction
PCR conditions were the same for all experiments, except that total cycle number (between 17 and 30 cycles) varied between assays. Plasmid or total cellular DNA (0.5–10 µl) was added to the PCR mix, which contained 1.25 U Taq polymerase (Biotech International Ltd, Western Australia), dATP, dTTP, dCTP and dGTP (Biotech) each at 0.2 mM, PCR buffer [67 mM Tris–HCl, pH 8.8 at 25°C, 16.6 mM (NH4)2SO4, 0.45% Triton X-100, 0.2 mg/ml gelatin (Biotech), 1.25 mM MgCl2], 20 pmol of each primer and autoclaved deionized water to a final volume of 50 µl. PCR took place in temperature-controlled serial oil/water baths with automated sample transfer (Model T100; Bartlett Instruments, Victoria). PCR was initiated by heating the mixture to 95°C for 5 min (denaturation phase), followed by a 1.5 min annealing phase at 56°C, then a 2.5 min polymerisation phase at 72°C. All subsequent cycles of PCR were the same except that the denaturation phase was 1 min. After the completion of PCR, 10 µl of each PCR mix was analysed by electrophoresis on a 1.5% agarose gel, stained with ethidium bromide and subsequently viewed under UV transillumination, the image being recorded using a video camera linked to a thermal printer (Videocopy; Mitsubishi).
Plasmid pFM11
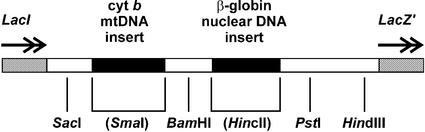
A 356 bp section of the human β-globin gene and a 434 bp sequence of mtDNA located within the cytochrome b gene (Table 1, region D) were inserted into the cleaved blunt-ended SmaI and HincII restriction sites, respectively, of the polylinker region of pUC18 (Fig. 1). The resultant plasmid pFM11 (3746 bp) was verified as having only one copy of each insert by restriction enzyme digestion and DNA sequencing. The concentration of DNA in the CsCl gradient-purified pFM11 plasmid (22) was estimated from the absorbance (260 nm) and adjusted to give a stock of 1 ng/µl. Further 10-fold serial dilutions down to a concentration of 10–8 ng/µl were prepared.
Figure 1.
Dual-insert plasmid pFM11 showing sites into which specific segments of human mtDNA and nuclear DNA have been inserted. Schematic (not to scale) indicates polylinker region of plasmid pUC18 with sequences bounded by LacI and LacZ′. Representative restriction sites are indicated including insertion sites of mtDNA (part of cytochrome b gene) and nuclear DNA (part of β-globin gene) at SmaI and HincII sites in the original pUC18 plasmid, respectively (the now-destroyed sites are indicated in parentheses).
Quantitative PCR to determine mtDNA copy number per diploid nuclear genome
Quantification required two separate PCR amplifications, each using the reference plasmid pFM11. First, region D of mtDNA from various amounts of tissue DNA extract was amplified (17 cycles of PCR), ensuring that at least one such PCR product would lie in the exponential PCR amplification range. A serially diluted input of pFM11 (see above), in separate PCR tubes but within the same PCR run, was also amplified to allow comparison of the yields of tissue-derived PCR products with those of the standard. Second, the abundance of β-globin template in the tissue DNA extracts was similarly determined using PCR (28 cycles) based on β-globin-specific primers (Table 1). Smaller inputs of pFM11 were required, consistent with lower copy numbers of β-globin templates in the tissue DNA extracts relative to those of mtDNA genomes (abundance greater by about three orders of magnitude). Abundances of the respective templates in the tissue extracts were initially expressed as nanogram plasmid equivalent. The relative copy number of mtDNA genomes per diploid nuclear genome in a given tissue extract was readily computed by taking twice the ratio of the mtDNA and β-globin plasmid equivalent values.
Statistical methods
Values given are the mean ± standard error of the mean. Comparison between groups was performed by one way analysis of variance. Linear regression analysis was used to relate two variables. To obtain the coefficient of variation (CV) of the copy number [mtDNA abundance (M)/diploid nuclear genome abundance (N)] the following formula was used: copy number CV = √(CVM2 + CVN2),where CVM and CVN are the individual coefficients of variation for the abundance of mtDNA genomes and nuclear genomes, respectively. Statistical significance was set at P < 0.05.
RESULTS
Selection of appropriate mtDNA targets for amplification by PCR
In selecting suitable primers for the quantification of total mtDNA copy number in tissue samples from subjects of different ages, attention must be paid to the following issue. That is, the possible interference in PCR-based assays by accumulated mtDNA deletions, which may be clustered preferentially in one region of the mtDNA genome and which could be of sufficiently high abundance to skew the data should the target for amplification be selected from within such a preferentially deleted region.
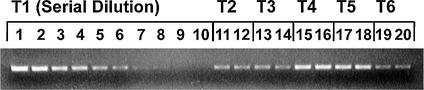
Prior to selecting any one region of mtDNA to act as a reliable marker of mtDNA copy number, four short regions (A–D) situated around the mtDNA genome (Table 1) were amplified from six skeletal muscle DNA extracts (T1–T6), previously studied in our laboratory (6,16). Of these, T1 (a skeletal muscle extract from a subject aged 1 h at autopsy) was designated as reference tissue extract because it does not contain detectable levels of mtDNA deletions (6) and, as the tissue extract from the youngest subject studied here, probably has the lowest level of mtDNA deletions. For T1 and the other tissue samples T2–T6, subject ages 5 weeks to 90 years (Table 2), the abundance of the common deletion mtDNA4977 was as follows: in T1, T2 and T3, mtDNA4977 was not detectable; in T4, T5 and T6, mtDNA4977 represented 0.0355, 0.08 and 0.257% of total mtDNA, respectively (6). Moreover, no multiple deletions of mtDNA could be detected in T1, T2 or T3, whereas T4, T5 and T6 contained various levels of such multiple deletions.
Table 2. Normalised relative abundance of different regions of mtDNA in skeletal muscle.
| Tissue | Age | Region | CV (%) | P | |||
|---|---|---|---|---|---|---|---|
| |
|
A |
B |
C |
D |
|
|
| T1 | 1 h | 1 | 1 | 1 | 1 | ||
| T2 | 5 weeks | 1.00 ± 0.05 (10) | 1.03 ± 0 (3) | 0.97 ± 0.04 (6) | 1.03 ± 0 (4) | 3 | 0.88 |
| T3 | 5 years | 0.99 ± 0.14 (8) | 1.09 ± 0.23 (5) | 1.07 ± 0.19 (4) | 0.68 ± 0 (2) | 20 | 0.68 |
| T4 | 50 years | 1.01 ± 0.07 (8) | 1.01 ± 0.04 (5) | 0.91 ± 0.03 (4) | 1.11 ± 0 (2) | 9 | 0.41 |
| T5 | 84 years | 0.98 ± 0.04 (12) | 1.12 ± 0.07 (5) | 1 ± 0 (4) | 0.93 ± 0.07 (3) | 8 | 0.23 |
| T6 | 90 years | 1.00 ± 0.08 (8) | 0.90 ± 0.15 (5) | 1.03 ± 0.28 (4) | 1.12 ± 0 (4) | 9 | 0.77 |
The mean relative abundance data for each tissue (T2–T6), region by region (Table 1), were determined relative to the intensity of the relevant PCR products from T1 (which was considered to have the lowest abundance of deletions in mtDNA of the tissue samples used here; see text). The normalised values for the reference tissue T1 are by definition unity for each region. The values for other tissues are displayed with standard errors of the mean. The numbers of repeat measurements made are displayed in parentheses. CV indicates coefficient of variation, namely the coefficient of the standard deviation divided by the mean; the mean CV for tissues T2–T6 was 9.8. P values were derived from analysis of variance for data from the four regions of each skeletal muscle sample and incorporate the multiple results that contribute to each average regional value, initially expressed in relative units (see text).
For each selected mtDNA region, replicate PCR runs of each tissue extract provided a mean relative abundance expressed as relative units (RU), derived by matching intensity of PCR product bands from T2–T6 with those from T1 (Fig. 2). We found that the absolute efficiency of amplification for each of the regions differed. The yield of PCR product for a given input of tissue extract was optimal in region D, while those of regions A, B and C were slightly smaller, giving less intense bands on the gel. Other regions tested showed much lower amplification efficiencies (data not shown). To correct for these differences in amplification efficiency, the mean relative abundance data for each region were normalised to that for region A. On this basis, for a tissue with fully intact mtDNA (or bearing a population of deletions uniformly distributed round the mtDNA genome), one would expect a value of 1 RU for each region.
Figure 2.
Representative gel of PCR products of human skeletal muscle DNA extracts used to determine relative abundance of different regions of mtDNA. Gel image shows PCR products from serially diluted inputs of reference DNA extract (1 h old skeletal muscle extract, T1, lanes 1–10) and other human skeletal muscle DNA: tissues T2 (5 weeks), T3 (5 years), T4 (50 years), T5 (84 years) and T6 (90 years). Tissues T2–T6 underwent duplicate analyses (lanes 11–20); the product intensities are similar for each pair, demonstrating a high degree of reproducibility.
Indeed, the data in Table 2 show that for each of the selected regions A–D, in each of the tissues T2–T6, the normalised abundance (in RU) was, in general, not significantly different from unity. The mean CV for the measurement of relative abundance was close to 10% (Table 2), which indicates an acceptable degree of precision. The results are also reliable in the sense that there is no one region, of those tested here, which is systematically depressed in its normalised relative abundance that would be suggestive of relatively high levels of preferentially accumulated deletions in that region. Such preferential deletions may occur but not be so abundant as to be evident by this test. In spite of the age-associated increase in accumulated mtDNA deletions (3,17), the absolute levels of mtDNA deletions in any given region of mtDNA, generally less than 1% of total mtDNA (6–8), are too low to have a significant impact on quantification of mtDNA as a whole.
The use of primer pairs specific for particular regions of mtDNA is valuable in quantifying accurately the abundance of a single deletion known to come from a particular region of mtDNA. Accordingly, we used a DNA extract prepared from a fibroblast culture derived from a patient with Pearson’s syndrome. This sample had been previously shown to harbour a single mtDNA deletion spanning the mtDNA sequence between nucleotides 10368 and 12828 and, on the basis of Southern hybridisation, accounting for about 80% of total mtDNA (23). Using this extract, we amplified and quantified a region outside the deletion (nucleotides 12759–13188; primer pair not shown in Table 1) and a region within the deletion zone (region C, Table 1). The ratio of normalised relative units obtained in this manner was used to calculate the abundance of the single mtDNA deletion harboured by the fibroblast culture, namely 77% of total mtDNA (data not shown). This result agreed well with the abundance estimated by Southern hybridisation (80%) and indicated a clear reduction in template copy number in the affected region compared to the unaffected region.
Copy number of mtDNA per nucleus in skeletal and cardiac muscle
As no evidence of regional inequality in mtDNA was found in extracts of normally ageing subjects, all relative units being close to unity, particularly those of T4–T6 (Table 2), any of the regions could serve as a marker of mtDNA genome content. We chose region D as it had greater PCR efficiency than those of the other regions. Template sequences corresponding to mtDNA region D were amplified with PCR from portions of the serially diluted reference plasmid pFM11 and, in parallel, from extracts containing total cellular DNA of muscle samples. Repeated measurements were made on such a series of tissue DNA samples and a CV of 3.5% for this technique was obtained. Amplification was then performed using the human β-globin gene-derived templates in both pFM11 and the muscle extracts. The CV for this technique was 6.1%. The overall CV for the copy number technique as a whole was close to 7%, obtained by combining the two individual values for quantification of mtDNA and nuclear DNA (see Statistical methods). As with the quantification based on PCR amplification of different regions of mtDNA, this indicates that our procedures have an acceptable degree of reproducibility.
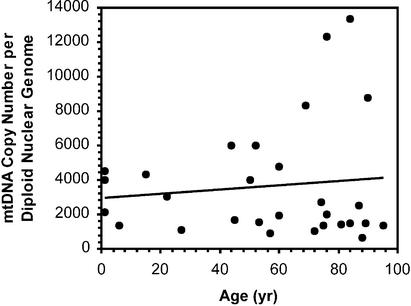
We applied these techniques to measure the mtDNA copy number in 29 DNA extracts from human skeletal muscle over the age range 1 h to 95 years (Fig. 3). The average number of mitochondrial genomes per diploid nuclear genome in these tissues was 3650 ± 620. Linear regression of copy number against age failed to show any significant age-associated change (P = 0.56). We noted that those tissue samples from patients undergoing hip operations tended to have lower mtDNA copy numbers (all but one value fell below the regression line) than those from individuals, mainly trauma subjects, from whom skeletal muscle was taken at autopsy. Whether this reflects a tendency for muscle atrophy (and hence reduced mtDNA copy number) in patients with hip pathology remains to be established.
Figure 3.
Human mtDNA copy number in skeletal muscle as a function of age (n = 29). The line represents a linear regression analysis (R2 = 0.01, P = 0.56).
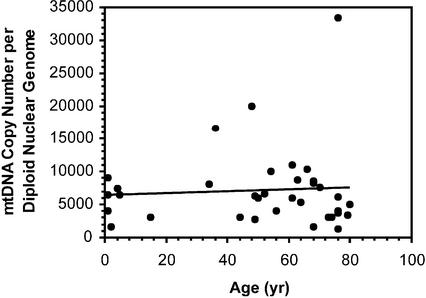
Copy number of mtDNA was also determined in 35 right atrial samples taken from hearts of patients aged between 3 weeks and 80 years. The average mtDNA copy number in this group was 6970 ± 920. This was significantly higher than that of the skeletal muscle group (P = 0.006). As in the case of skeletal muscle, we found no relationship between age and mtDNA copy number per nucleus (linear regression yielding P = 0.70) (Fig. 4). Of the 35 patients whose data are represented in Figure 4, 75% had ischaemic heart disease, and hence would not have developed significant right atrial hypertrophy (that might have elevated mtDNA copy number). Other patients had non-ischaemic cardiac conditions and, in all but three of these, no right atrial hypertrophy would have been anticipated. For the remaining three, all with congenital heart disease, right atrial hypertrophy would be expected, but their mtDNA copy number was not grossly out of the range of others of comparable ages (patients aged 3 weeks, 6 months and 36 years, with mtDNA copy numbers of 4000, 6400 and 16 700, respectively, in Fig. 4). Nevertheless, a systematic study of mtDNA copy number in hearts exhibiting cardiac hypertrophy might be warranted, given their apparent increase in mtDNA copy number with age.
Figure 4.
Human mtDNA copy number in right atrium of heart as a function of age (n = 35). In cases where multiple atrial strips were analysed (Table 3) the mean was used. The line represents a linear regression analysis (R2 = 0.005, P = 0.70).
The method of right atrial sample collection allowed two or three small strips (10 mg) to be harvested for mtDNA copy number analysis from the same sample of heart tissue. Data for 12 subjects where this was done are shown in Table 3. Where multiple strips were subjected to mtDNA copy number analysis from a single atrial appendage, the mean value was used to ascribe one value per patient for the purpose of correlating age with copy number (Fig. 4). In most cases the variation between strips from the same patient was low (Table 3). However, in several cases there was considerable variability in copy number of mtDNA between different strips from the same right atrium (e.g. P2 and P1 in Table 3). The CV was calculated for the copy number values in each patient and was found to vary between 7 and 54% with an average value of 26%. These findings are characteristic of a ‘tissue mosaic’ with respect to mtDNA abundance as a whole, previously observed at the level of mtDNA deletions (24). Such tissue ‘mosaicism’ in terms of the abundance and multiplicity of mtDNA deletions was later demonstrated at the cellular level (25) by analysis of individual cardiomyocytes.
Table 3. Copy number of mtDNA can vary in multiple samples from single atrial appendages.
| Patient code | Age (years) | Sample no. | Copy number of mtDNAa | Mean copy number of mtDNAa | CVb (%) |
|---|---|---|---|---|---|
| P1 | 34 | 1 | 6000 | 8000 | 35 |
| 2 | 10 000 | ||||
| P2 | 44 | 1 | 1867 | 3089 | 54 |
| 2 | 2400 | ||||
| 3 | 5000 | ||||
| P3 | 49 | 1 | 3200 | 2800 | 20 |
| 2 | 2401 | ||||
| P4 | 49 | 1 | 6000 | 6334 | 7 |
| 2 | 6667 | ||||
| P5 | 50 | 1 | 7500 | 6036 | 34 |
| 2 | 4571 | ||||
| P6 | 61 | 1 | 10 000 | 11 000 | 13 |
| 2 | 12 000 | ||||
| P7 | 63 | 1 | 9375 | 8688 | 11 |
| 2 | 8000 | ||||
| P8 | 66 | 1 | 10 667 | 10 334 | 5 |
| 2 | 10 000 | ||||
| P9 | 68 | 1 | 13 000 | 8222 | 51 |
| 2 | 5000 | ||||
| 3 | 6667 | ||||
| P10 | 70 | 1 | 8889 | 7570 | 25 |
| 2 | 6250 | ||||
| P11 | 73 | 1 | 2286 | 3016 | 34 |
| 2 | 3745 | ||||
| P12 | 74 | 1 | 2500 | 3125 | 28 |
| 2 | 3750 |
aExpressed as mtDNA genomes per diploid nuclear genome; the overall mean across the 12 subjects is 6390.
bThe mean CV (as defined in Table 2) was 26.
DISCUSSION
We have developed a novel, precise assay based on PCR to compare the relative abundance of mtDNA with nuclear DNA, using a dual-insert plasmid as a common external reference for amplification of representative segments of the relevant two genomes. Nuclear DNA was chosen as the internal reference for quantifying mtDNA. The β-globin gene was selected as a reference as it is an example of a single copy gene of known sequence (21) that conveniently acts as a marker of diploid genome content. Other nuclear genes, including 18S rDNA (8,11) and LINE sequences (9), have been used previously in Southern hybridisation as markers of nuclear content for mtDNA quantification. However, repetitive nuclear sequences may vary in number from one individual to another and thus are not attractive as markers of genome content. Use of a suitable single copy gene probe would require larger amounts of input DNA, but the limited amount of human tissue samples available usually precludes this approach. PCR of single copy nuclear genes is more sensitive in these terms. We used a short segment of the mitochondrial cytochrome b gene to serve as a marker of copy number for the entire mtDNA genome. The technique utilising the dual-insert plasmid as reference was then used to measure the absolute mtDNA copy number per diploid nuclear genome. The results we obtained are in the same range (of the order of thousands) as those originally observed in cultured mammalian cells (2).
Several studies have been published that have used PCR to determine mtDNA copy number. Some have used competitive PCR (14,26) which involves the introduction of standards into tissue DNA extracts. However, we have found that serially diluted standards in separate PCR tubes, but within the same PCR run, yield the same quantitative values as standards which were mixed with tissue samples (6,16). The use of external standards allows more tissue samples to be analysed in a given PCR run compared to the use of internal standards whereby many tubes, each with a different input of standard, are required to assay a single tissue. The target sequence being amplified in an internal standard must differ from the native tissue sequence to distinguish the two species of PCR product at the end of the PCR run. This is usually accomplished by the formation of either a point mutation causing an artificial restriction site (6,8,27) or a small deletion (14) or insertion (27) within the target sequence of a recombinant plasmid. However, a DNA sequence with modified length, as present in a deletion-bearing or insertion-bearing standard, may enjoy a replicative advantage or disadvantage, respectively, over the native DNA target and introduce a degree of inexactitude that would be difficult to quantify. The principles inherent in calibrating quantitative PCR using the dual-insert external reference plasmid, as described here, may also be used in setting up more automated systems such as real-time PCR. Thus, our method avoids the use of multiple beacons or standards (28).
Recently, mtDNA copy number in human tissues was measured (14) using an internal standard single plasmid containing short sections of the mtDNA genome and the multicopy 28S rRNA gene. In skeletal muscle from 16 healthy subjects aged 2–45 years a mean copy number of 1811 ± 546 per diploid nuclear genome was observed (somewhat less than what we found), and no correlation with age was found, as in the present work.
In the present study, myocardium was found to contain approximately twice the number of mtDNA genomes per diploid nucleus as skeletal muscle (6970 versus 3650, P = 0.006). This is in keeping with an earlier study (10) that used Southern hybridisation. This finding accords with a greater reliance on aerobic ATP production by the myocardium than by skeletal muscle. The mtDNA copy number in myocardium and skeletal muscle was found to remain unchanged over a 10 decade timespan in the tissues we studied (Figs 3 and 4), similar to the previous findings on rat heart (12).
The present approach can be used to quantify mtDNA with respect to nuclear DNA in a variety of settings, including ageing (as carried out here), tissue adaptation and disease. The currently described technique based on the dual-insert reference plasmid can readily be generalised for the measurement of any two sequences in relation to one another in any biological context.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the surgeons and operating room nurses at the Alfred Hospital and the Royal Children’s Hospital for assistance in the collection of discarded cardiac tissue, and those at the Epworth Hospital (Professor M. Richardson, in particular) for assistance in collection of skeletal muscle samples. We greatly appreciate the statistical advice provided by Rory Wolfe (Biostatistics Section of the Department of Epidemiology and Preventive Medicine, Monash University).
REFERENCES
- 1.Scheffler I.E. (1999) Mitochondria. Wiley-Liss, New York, NY.
- 2.Bogenhagen D. and Clayton,D.C. (1974) The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. J. Biol. Chem., 249, 7991–7995. [PubMed] [Google Scholar]
- 3.Nagley P. and Wei,Y.H. (1998) Aging and mammalian mitochondrial genetics. Trends Genet., 14, 513–517. [DOI] [PubMed] [Google Scholar]
- 4.Hayakawa M., Katsumata,K., Yoneda,M., Tanaka,M., Sugiyama,S. and Ozawa,T. (1996) Age-related extensive fragmentation of mitochondrial DNA into minicircles. Biochem. Biophys. Res. Commun., 226, 369–377. [DOI] [PubMed] [Google Scholar]
- 5.Melov S., Shoffner,J.M., Kaufman,A. and Wallace,D.C. (1995) Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res., 23, 4122–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu V.W.S., Zhang,C. and Nagley,P. (1998) Mutations in mitochondrial DNA accumulate differently in three different human tissues during ageing. Nucleic Acids Res., 26, 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadaleta M.N., Rainaldi,G., Lezza,A.M.S., Milella,F., Fracasso,F. and Cantatore,P. (1992) Mitochondrial DNA copy number and mitochondrial DNA deletion in adult and senescent rats. Mutat. Res., 275, 181–193. [DOI] [PubMed] [Google Scholar]
- 8.Simonetti S., Chen,X., DiMauro,S. and Schon,E.A. (1992) Accumulation of deletions in human mitochondrial DNA during normal aging: analysis by quantitative PCR. Biochim. Biophys. Acta, 1180, 113–122. [DOI] [PubMed] [Google Scholar]
- 9.Asano K., Nakamura,M., Sato,T., Tauchi,H. and Asano,A. (1993) Age dependency of mitochondrial DNA decrease differs in different tissues of rat. J. Biochem., 114, 303–306. [DOI] [PubMed] [Google Scholar]
- 10.Van den Bogert C., De Vries,H., Holtrop,M., Muus,P., Dekker,H.L., Van Galen,M.J.M., Bolhuis,P.A. and Taanman,J.W. (1993) Regulation of the expression of mitochondrial proteins: relationship between mtDNA copy number and cytochrome-c oxidase activity in human cells and tissues. Biochim. Biophys. Acta, 1144, 177–183. [DOI] [PubMed] [Google Scholar]
- 11.Barrientos A., Casademont,J., Cardellach,F., Estivill,X., Urbano-Marquez,A. and Nunes,V. (1997) Reduced steady-state levels of mitochondrial RNA and increased mitochondrial DNA amount in human brain with aging. Mol. Brain Res., 52, 284–289. [DOI] [PubMed] [Google Scholar]
- 12.Barazzoni R., Short,K.R. and Nair,K.S. (2000) Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver and heart. J. Biol. Chem., 275, 3343–3347. [DOI] [PubMed] [Google Scholar]
- 13.Filser N., Margue,C. and Richter,C. (1997) Quantification of wild-type mitochondrial DNA and its 4.8-kb deletion in rat organs. Biochem. Biophys. Res. Commun., 233, 102–107. [DOI] [PubMed] [Google Scholar]
- 14.Barthelemy C., Ogier de Baulny,H., Diaz,J., Cheval,M.A., Frachon,P., Romero,N., Goutieres,F., Fardeau,M. and Lombes,A. (2001) Late-onset mitochondrial DNA depletion: DNA copy number, multiple deletions and compensation. Ann. Neurol., 49, 607–617. [PubMed] [Google Scholar]
- 15.Nagley P., Zhang,C., Lim,M.L.R., Merhi,M., Needham,B.E. and Khalil,Z. (2001) Mitochondrial DNA deletions parallel age-linked decline in rat sensory nerve function. Neurobiol. Aging, 22, 635–643. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C., Liu,V.W.S., Addessi,C.L., Sheffield,D.A., Linnane,A.W. and Nagley,P. (1998) Differential occurrence of mutations in mitochondrial DNA of human skeletal muscle. Hum. Mutat., 11, 360–371. [DOI] [PubMed] [Google Scholar]
- 17.Nagley P. and Zhang,C. (1998) Mitochondrial DNA mutations in aging. In Singh,K.K. (ed.), Mitochondrial DNA Mutations in Aging, Disease and Cancer. R.G. Landes Co., Austin, TX, pp. 205–238.
- 18.Zhang C., Baumer,A., Maxwell,R.J., Linnane,A.W. and Nagley,P. (1992) Multiple mitochondrial DNA deletions in an elderly human individual. FEBS Lett., 297, 34–38. [DOI] [PubMed] [Google Scholar]
- 19.Vaillant F. and Nagley,P. (1995) Human cell mutants with very low mitochondrial DNA copy number (ρd). Hum. Mol. Genet., 4, 903–914. [DOI] [PubMed] [Google Scholar]
- 20.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H.L., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J.H., Staden,R. and Young,I.G. (1981) Sequence and organisation of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 21.Lawn R.M., Efstratiadis,A., O’Connell,C. and Maniatis,T. (1980) The nucleotide sequence of the human β-globin gene. Cell, 21, 647–651. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Kapsa R., Thompson,G.N., Thorburn,D.R., Dahl,H.H.M., Marzuki,S., Byrne,E. and Blok,R.B. (1994) A novel mtDNA deletion in an infant with Pearson syndrome. J. Inherit. Metab. Dis., 17, 521–526. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C., Lee,A., Liu,V.W., Pepe,S., Rosenfeldt,F. and Nagley,P. (1999) Mitochondrial DNA deletions in human cardiac tissue show a gross mosaic distribution. Biochem. Biophys. Res. Commun., 254, 152–157. [DOI] [PubMed] [Google Scholar]
- 25.Khrapko K., Bodyak,N., Thilly,W.G., Van Orsouw,N.J., Zhang,X., Coller,H.A., Perls,T.T., Upton,M., Vijg,J. and Wei,J.Y. (1999) Cell-by-cell scanning of whole mitochondrial genomes in aged human heart reveals a significant fraction of myocytes with clonally expanded deletions. Nucleic Acids Res., 27, 2434–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Cooney,D.A., Sreenath,A., Zhan,Q., Agbaria,R., Stowe,E.E., Fornace,A.J.,Jr and Johns,D.G. (1994) Quantitation of mitochondrial DNA in human lymphoblasts by a competitive polymerase chain reaction method: application to the study of inhibitors of mitochondrial DNA content. Mol. Pharmacol., 46, 1063–1069. [PubMed] [Google Scholar]
- 27.Zhang C., Peters,L.E., Linnane,A.W. and Nagley,P. (1996) Comparison of different quantitative PCR procedures in the analysis of the 4977-bp deletion in human mitochondrial DNA. Biochem. Biophys. Res. Commun., 223, 450–455. [DOI] [PubMed] [Google Scholar]
- 28.Gahan M.E., Miller,F., Rosenfeldt,F., Cherry,C., Lewin,S.R., Mijch,A., Hoy,J. and Wesselingh,S.L. (2001) Quantification of mitochondrial DNA using real-time PCR. J. Clin. Virol., 22, 241–247. [DOI] [PubMed] [Google Scholar]