Abstract
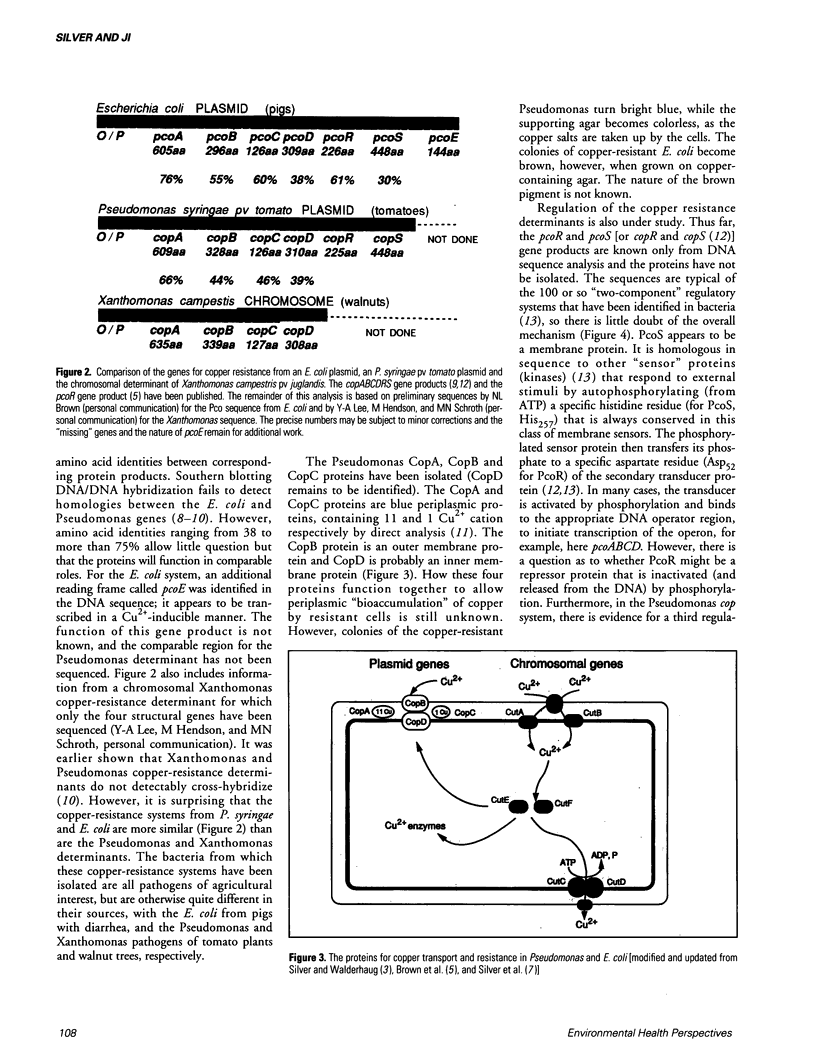
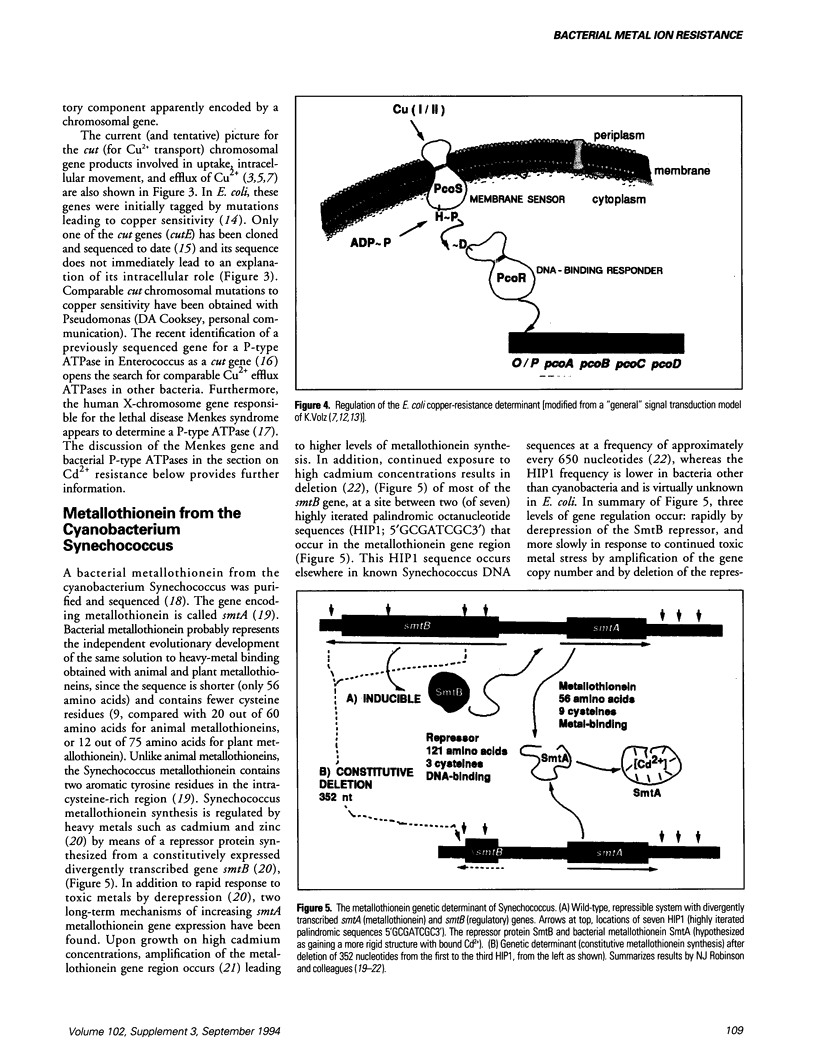
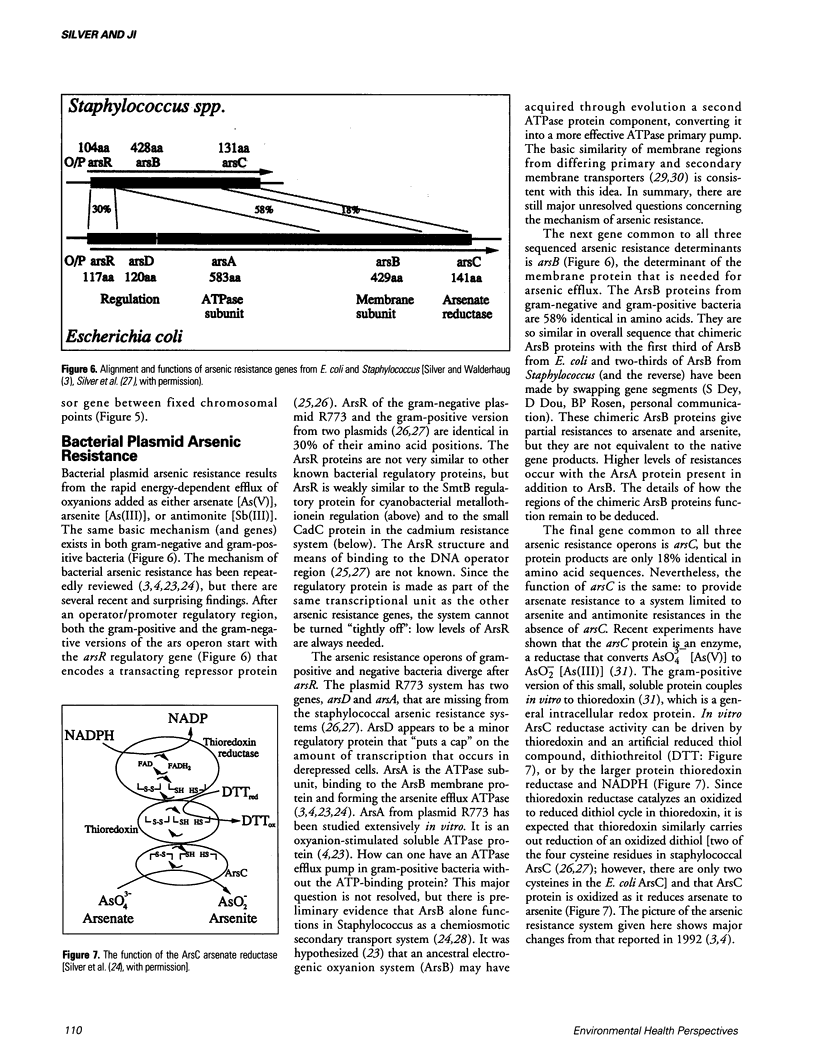
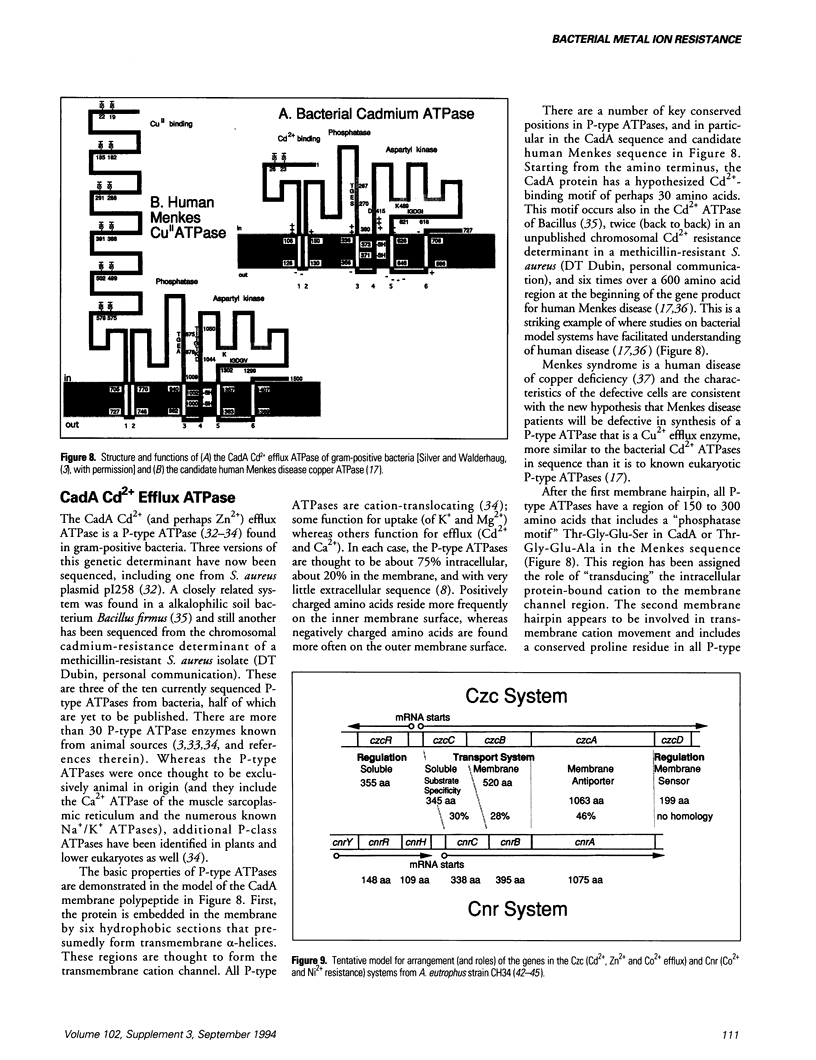
Bacterial plasmids contain specific genes for resistances to toxic heavy metal ions including Ag+, AsO2-, AsO4(3-), Cd2+, Co2+, CrO4(2-), Cu2+, Hg2+, Ni2+, Pb2+, Sb3+, and Zn2+. Recent progress with plasmid copper-resistance systems in Escherichia coli and Pseudomonas syringae show a system of four gene products, an inner membrane protein (PcoD), an outer membrane protein (PcoB), and two periplasmic Cu(2+)-binding proteins (PcoA and PcoC). Synthesis of this system is governed by two regulatory proteins (the membrane sensor PcoS and the soluble responder PcoR, probably a DNA-binding protein), homologous to other bacterial two-component regulatory systems. Chromosomally encoded Cu2+ P-type ATPases have recently been recognized in Enterococcus hirae and these are closely homologous to the bacterial cadmium efflux ATPase and the human copper-deficiency disease Menkes gene product. The Cd(2+)-efflux ATPase of gram-positive bacteria is a large P-type ATPase, homologous to the muscle Ca2+ ATPase and the Na+/K+ ATPases of animals. The arsenic-resistance system of gram-negative bacteria functions as an oxyanion efflux ATPase for arsenite and presumably antimonite. However, the structure of the arsenic ATPase is fundamentally different from that of P-type ATPases. The absence of the arsA gene (for the ATPase subunit) in gram-positive bacteria raises questions of energy-coupling for arsenite efflux. The ArsC protein product of the arsenic-resistance operons of both gram-positive and gram-negative bacteria is an intracellular enzyme that reduces arsenate [As(V)] to arsenite [As(III)], the substrate for the transport pump. Newly studied cation efflux systems for Cd2+, Zn2+, and Co2+ (Czc) or Co2+ and Ni2+ resistance (Cnr) lack ATPase motifs in their predicted polypeptide sequences. Therefore, not all plasmid-resistance systems that function through toxic ion efflux are ATPases. The first well-defined bacterial metallothionein was found in the cyanobacterium Synechococcus. Bacterial metallothionein is encoded by the smtA gene and contains 56 amino acids, including nine cysteine residues (fewer than animal metallothioneins). The synthesis of Synechococcus metallothionein is regulated by a repressor protein, the product of the adjacent but separately transcribed smtB gene. Regulation of metallothionein synthesis occurs at different levels; quickly by derepression of repressor activity, or over a longer time by deletion of the repressor gene at fixed positions and by amplification of the metallothionein DNA region leading to multiple copies of the gene.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown N. L., Rouch D. A., Lee B. T. Copper resistance determinants in bacteria. Plasmid. 1992 Jan;27(1):41–51. doi: 10.1016/0147-619x(92)90005-u. [DOI] [PubMed] [Google Scholar]
- Cha J. S., Cooksey D. A. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelly J., Tümer Z., Tønnesen T., Petterson A., Ishikawa-Brush Y., Tommerup N., Horn N., Monaco A. P. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nat Genet. 1993 Jan;3(1):14–19. doi: 10.1038/ng0193-14. [DOI] [PubMed] [Google Scholar]
- Collard J. M., Provoost A., Taghavi S., Mergeay M. A new type of Alcaligenes eutrophus CH34 zinc resistance generated by mutations affecting regulation of the cnr cobalt-nickel resistance system. J Bacteriol. 1993 Feb;175(3):779–784. doi: 10.1128/jb.175.3.779-784.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey D. A., Azad H. R., Cha J. S., Lim C. K. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl Environ Microbiol. 1990 Feb;56(2):431–435. doi: 10.1128/aem.56.2.431-435.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Morby A. P., Turner J. S., Whitton B. A., Robinson N. J. Deletion within the metallothionein locus of cadmium-tolerant Synechococcus PCC 6301 involving a highly iterated palindrome (HIP1). Mol Microbiol. 1993 Jan;7(2):189–195. doi: 10.1111/j.1365-2958.1993.tb01110.x. [DOI] [PubMed] [Google Scholar]
- Huckle J. W., Morby A. P., Turner J. S., Robinson N. J. Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol Microbiol. 1993 Jan;7(2):177–187. doi: 10.1111/j.1365-2958.1993.tb01109.x. [DOI] [PubMed] [Google Scholar]
- Ivey D. M., Guffanti A. A., Shen Z., Kudyan N., Krulwich T. A. The cadC gene product of alkaliphilic Bacillus firmus OF4 partially restores Na+ resistance to an Escherichia coli strain lacking an Na+/H+ antiporter (NhaA). J Bacteriol. 1992 Aug;174(15):4878–4884. doi: 10.1128/jb.174.15.4878-4884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Silver S. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Silver S. Regulation and expression of the arsenic resistance operon from Staphylococcus aureus plasmid pI258. J Bacteriol. 1992 Jun;174(11):3684–3694. doi: 10.1128/jb.174.11.3684-3694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur P., Rosen B. P. Plasmid-encoded resistance to arsenic and antimony. Plasmid. 1992 Jan;27(1):29–40. doi: 10.1016/0147-619x(92)90004-t. [DOI] [PubMed] [Google Scholar]
- Levy S. B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992 Apr;36(4):695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesegang H., Lemke K., Siddiqui R. A., Schlegel H. G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol. 1993 Feb;175(3):767–778. doi: 10.1128/jb.175.3.767-778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney P. C. A consensus structure for membrane transport. Res Microbiol. 1990 Mar-Apr;141(3):374–383. doi: 10.1016/0923-2508(90)90015-i. [DOI] [PubMed] [Google Scholar]
- Mellano M. A., Cooksey D. A. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J Bacteriol. 1988 Jun;170(6):2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. D., Jasalavich C. A., Cooksey D. A. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J Bacteriol. 1993 Mar;175(6):1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra T. K. Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid. 1992 Jan;27(1):4–16. doi: 10.1016/0147-619x(92)90002-r. [DOI] [PubMed] [Google Scholar]
- Nies D. H. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol. 1992 Dec;174(24):8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H., Nies A., Chu L., Silver S. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7351–7355. doi: 10.1073/pnas.86.19.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid. 1992 Jan;27(1):17–28. doi: 10.1016/0147-619x(92)90003-s. [DOI] [PubMed] [Google Scholar]
- Nucifora G., Chu L., Misra T. K., Silver S. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc Natl Acad Sci U S A. 1989 May;86(10):3544–3548. doi: 10.1073/pnas.86.10.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt A., Suter H., Krapf R., Solioz M. An ATPase operon involved in copper resistance by Enterococcus hirae. Ann N Y Acad Sci. 1992 Nov 30;671:484–486. doi: 10.1111/j.1749-6632.1992.tb43836.x. [DOI] [PubMed] [Google Scholar]
- Olafson R. W., McCubbin W. D., Kay C. M. Primary- and secondary-structural analysis of a unique prokaryotic metallothionein from a Synechococcus sp. cyanobacterium. Biochem J. 1988 May 1;251(3):691–699. doi: 10.1042/bj2510691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Rogers S. D., Bhave M. R., Mercer J. F., Camakaris J., Lee B. T. Cloning and characterization of cutE, a gene involved in copper transport in Escherichia coli. J Bacteriol. 1991 Nov;173(21):6742–6748. doi: 10.1128/jb.173.21.6742-6748.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen B. P., Dey S., Dou D., Ji G., Kaur P., Ksenzenko MYu, Silver S., Wu J. Evolution of an ion-translocating ATPase. Ann N Y Acad Sci. 1992 Nov 30;671:257–272. doi: 10.1111/j.1749-6632.1992.tb43801.x. [DOI] [PubMed] [Google Scholar]
- Rosenstein R., Peschel A., Wieland B., Götz F. Expression and regulation of the antimonite, arsenite, and arsenate resistance operon of Staphylococcus xylosus plasmid pSX267. J Bacteriol. 1992 Jun;174(11):3676–3683. doi: 10.1128/jb.174.11.3676-3683.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S., Ji G., Bröer S., Dey S., Dou D., Rosen B. P. Orphan enzyme or patriarch of a new tribe: the arsenic resistance ATPase of bacterial plasmids. Mol Microbiol. 1993 May;8(4):637–642. doi: 10.1111/j.1365-2958.1993.tb01607.x. [DOI] [PubMed] [Google Scholar]
- Silver S., Nucifora G., Chu L., Misra T. K. Bacterial resistance ATPases: primary pumps for exporting toxic cations and anions. Trends Biochem Sci. 1989 Feb;14(2):76–80. doi: 10.1016/0968-0004(89)90048-0. [DOI] [PubMed] [Google Scholar]
- Silver S., Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992 Mar;56(1):195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K. J., Yoon K. P., Lynn A. R. ATP-dependent cadmium transport by the cadA cadmium resistance determinant in everted membrane vesicles of Bacillus subtilis. J Bacteriol. 1992 Jan;174(1):116–121. doi: 10.1128/jb.174.1.116-121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulpe C., Levinson B., Whitney S., Packman S., Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper-transporting ATPase. Nat Genet. 1993 Jan;3(1):7–13. doi: 10.1038/ng0193-7. [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Distefano M. D., Moore M. J., Shewchuk L. M., Verdine G. L. Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts. FASEB J. 1988 Feb;2(2):124–130. doi: 10.1096/fasebj.2.2.3277886. [DOI] [PubMed] [Google Scholar]
- Wu J., Rosen B. P. The ArsR protein is a trans-acting regulatory protein. Mol Microbiol. 1991 Jun;5(6):1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x. [DOI] [PubMed] [Google Scholar]
- Yoon K. P., Misra T. K., Silver S. Regulation of the cadA cadmium resistance determinant of Staphylococcus aureus plasmid pI258. J Bacteriol. 1991 Dec;173(23):7643–7649. doi: 10.1128/jb.173.23.7643-7649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K. P., Silver S. A second gene in the Staphylococcus aureus cadA cadmium resistance determinant of plasmid pI258. J Bacteriol. 1991 Dec;173(23):7636–7642. doi: 10.1128/jb.173.23.7636-7642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


