Abstract
High-throughput procedures are an important requirement for future large-scale genetic studies such as genotyping of single nucleotide polymorphisms (SNPs). Matrix-assisted laser desorption/ ionisation mass spectrometry (MALDI-MS) has revolutionised the analysis of biomolecules and, in particular, provides a very attractive solution for the rapid typing of DNA. The analysis of DNA by MALDI can be significantly facilitated by a procedure termed ‘charge-tagging’. We show here a novel approach for the generation of charge-tagged DNA using a photocleavable linker and its implementation in a molecular biological procedure for SNP genotyping consisting of PCR, primer extension, photocleavage and a chemical reaction prior to MALDI target preparation and analysis. The reaction sequence is amenable to liquid handling automation and requires no stringent purification procedures. We demonstrate this new method on SNPs in two genes involved in complex traits.
INTRODUCTION
After the completion of the Human Genome Project, analysis of DNA variation is one of the most challenging tasks in genome research. Multiple applications include genotyping of single nucleotide polymorphisms (SNPs) for the dissection of complex diseases, pharmacogenetics, marker-assisted plant or animal breeding, and traceability (1,2). High-throughput procedures are necessary for these large-scale studies and require that SNP typing technologies become affordable (3). Matrix-assisted laser desorption/ionisation mass spectrometry (MALDI-MS) is an efficient technology for large-scale genome variation analysis as it can be used to obtain direct and rapid measurement of nucleic acids (4,5). Many procedures for genotyping SNPs by MALDI-MS have been published during the last few years (6,7). In principle, these methods follow a similar concept. After preparation of genomic DNA, a specific fragment including a polymorphic site of interest is amplified by PCR. The resulting product is purified and subsequently used to generate an allele-specific product. The most popular procedure for the generation of allele-specific products is the primer extension reaction because of its robustness and simplicity (8). A primer is either extended by using a DNA polymerase and a specific combination of dNTPs and ddNTPs or by simply elongating with a single ddNTP at the polymorphic site. The resulting DNA molecules are purified and prepared for MALDI analysis. MALDI mass spectrometers allow automatic detection and the use of standard microtitre plate formats, typically 384 wells. The SNP analysis from the MALDI spectra can be performed automatically by appropriate programs such as GenoTools (http://www.bruker-daltonik.de/genotools.html), which allows high-throughput allele calling without any user interaction (9).
An inherent problem in analysing DNA by MALDI-MS is its negatively charged sugar-phosphate backbone (10). DNA as a polyanion forms salt adducts with cations, in particular with alkali ions such as sodium and potassium thereby severely limiting the signal intensity and resolution. The tendency of cations to interfere with DNA increases significantly with DNA length and is one main reason for the limitations in DNA analysis. Furthermore, components of reaction buffers like detergents are not compatible with MALDI and have to be removed prior to analysis.
The type of purification determines sample quality and the robotic solution that can be applied to sample preparation (7). MALDI analysis of DNA generally requires stringent purification procedures, such as magnetic bead separation or reversed-phase binding (11). However, most of these procedures are cumbersome in high-throughput processes and significantly contribute to the costs of the assays.
A further limitation of MALDI is that reliable and reproducible DNA analysis can only be performed on DNA stretches less than ∼25 nt (6,7). Several approaches focused on generating short DNA products by site-specific cleavage, as smaller molecules in a mass range of 1000–2000 Da lead to increased sensitivity and resolution (7). Recently, a commercially available procedure for genotyping SNPs termed GenoSNIP assay (part of the GenoLink package of Bruker Saxonia) was introduced (http://www.bsax.de). The principle of the GenoSNIP assay is that DNA products are small compared to alternative procedures like the MassArray and the PinPoint assay (12,13). The GenoSNIP assay consists of PCR, shrimp alkaline phosphatase (SAP) digestion, primer extension, photocleavage and stringent purification by streptavidin-coated microtitre plates and MALDI analysis (http://www.bsax.de). Extension primers are converted by nucleotide extension with ddNTPs. These primers contain an o-nitrobenzyl moiety replacing a nucleotide and allow for selective photocleavage (http://www.inmerge.com/aspfolder/ASMSSchedule2.asp). This photocleavable linker substitutes for one nucleotide and thus creates an abasic site in the oligonucleotide but does not prevent annealing to the target sequence. By exposure to UV irradiation, the genotyping products are separated from their surface and, most importantly, the major part of the non-informative primer is removed. Therefore the resulting products are shifted in a mass range, which is much more accessible for MALDI detection (size range of 1000–2000 Da instead of 5000– 7000 Da). After desalting, the DNA oligomers can be detected by MALDI-MS. The stringent purification is done by usage of streptavidin-coated 96-well microtitre plates.
The only MALDI-based procedures for SNP genotyping that completely circumvent stringent purification procedures are the GOOD assays (14–16). The GOOD assays consist of a PCR optionally followed by SAP digestion, a primer extension with primers containing phosphorothioate linkages (and additionally for the GOOD assay in the positive ion mode nucleobases carrying aminopropargyl side chains at which a positive charge tag is attached by using, for example, 6-trimethylammoniumhexyryl-N-hydroxy-succinimidyl ester), a phosphodiesterase II digestion and an alkylation reaction. The principle of the GOOD assays is that the allele-specific DNA products are charge-tagged (17). Charge-tagging means that DNA is carrying either a single excess positive or a single excess negative charge. Thus, the allele-specific DNA products are modified in such a way that the detection sensitivity is increased 100-fold and the sensitivity to impurities such as the preparation buffers in MALDI is decreased significantly.
An inherent problem of the GOOD assays can be the usage of sequence-specific phosphodiesterase II (from calf spleen). Phosphodiesterase II is an exonuclease that hydrolyses 3′-phosphomononucleotides from oligonucleotides containing a 5′-hydroxyl terminus (18). Therefore, the use of phosphodiesterase precluded the performance of the GOOD assay on a surface. Furthermore, as for many enzymes, the phosphodiesterase II contributes to a significant part of the cost of the GOOD assays.
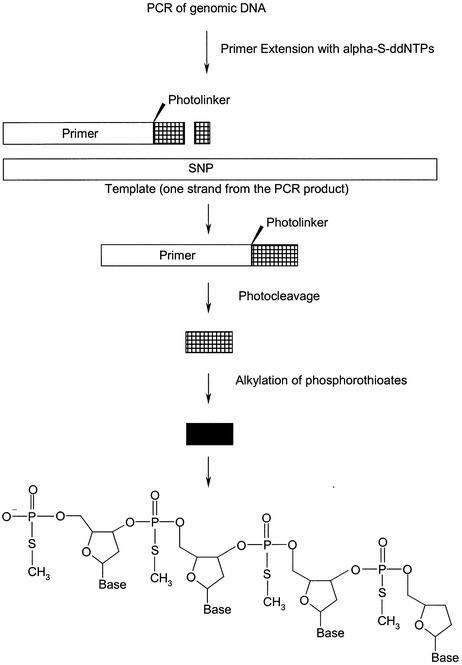
In this article we present a novel procedure for efficient analysis of SNPs that does not require stringent purification. The key to this procedure is the implementation of extension primers containing phosphorothioates and a photocleavable linker (abbreviated with L) at the 3′ end. The primers are placed immediately next to the position of the SNP and extended with α-S-ddNTPs. As shown in Figure 1, after photocleavage and alkylation, small DNA oligomers with a single negative fixed charge are generated. The negative fixed charge derives from the 5′-phosphate of the 3′-cleavage product of the primer. In this way, charge-tagged molecules are obtained without phosphodiesterase digestion. Thus, the phosphodiesterase digestion of all GOOD assays and the stringent purification of the GenoSNIP assay have become redundant. In a nutshell, the reaction sequences of both procedures are merged resulting in a significantly shortened and simplified procedure for efficiently genotyping SNPs.
Figure 1.
The principle of the procedure starting with a PCR followed by allele-specific primer extension reaction with a primer containing a photolinker and phosphorothioates at the 3′ end. The unmodified part of the primer is cut off by exposure to UV light and the modified part is then alkylated. The resulting product contains a DNA backbone with one negative charge deriving from the 5′-end. Thus, the product is negatively charge-tagged. Products are diluted and transferred onto a MALDI target for analysis.
MATERIALS AND METHODS
Oligonucleotides for PCR were obtained from MWG (Ebersberg, Germany). These primers were purified by MWG using their HPSF® technology. Oligonucleotides for primer extension containing phosphorothioates and the photocleavable linker were synthesised by Biotez (Berlin, Germany) using conventional phosphoramidite chemistry (G. Herrmann, personal communication). For the introduction of the photocleavable linker the PC-o-nitrophenyl-CE-phosphoramidite [o-nitrophenyl-1-O-(2-cyanoethyl-N,N-diisopropylphosphoramidyl)-3-O-(4,4′-dimethoxy-triphenylmethyl)-1,3-propandiol], which is commercially available from Bruker Saxonia (Leipzig, Germany), was used. dNTPs were purchased from Roche Diagnostics (Mannheim, Germany) and α-S-ddNTPs were synthesised by Biolog (Bremen, Germany). Taq polymerase was produced by Roman Pawlik (Max-Planck Institute for Molecular Genetics, Berlin, Germany). Thermosequenase and SAP were obtained from Amersham Buchler (Braunschweig, Germany). Chemical reagents were purchased from Aldrich (Steinheim, Germany). The MALDI matrix (α-cyano-4-hydroxy-cinnamic acid methyl ester) and the GenoTools software are available from Bruker Saxonia (Leipzig, Germany). The thermocycling procedures were carried out in a MJ PTC 200 Thermocycler obtained from Biozym (Hess. Oldendorf, Germany). Plastic ware was obtained from Abgene (Hamburg, Germany) and Eppendorf (Hamburg, Germany).
DNA isolation
Anonymous DNAs were prepared from leucocyte filters that were obtained from a blood donation centre in Berlin. Filters were flushed with 80 ml of PBS, and the cells were subsequently collected by centrifugation for 15 min at 3000 r.p.m. in a D-50 Sorvall centrifuge. The erythrocytes were lysed by resuspending the cells in erythrocyte lysis buffer (155 mM NH4Cl, 5 mM NH4HCO3, 0.1 mM EDTA, 40 µM KCl, pH 7.4) and incubation on ice for 15 min. The remaining leucocytes were collected again by centrifugation. The cells were resuspended in approximately the same volume of PBS as their packed cell volume (p.c.v.), mixed with an equal volume of 1.5% low melting point agarose and filled in block moulds of ∼100 µl. After solidifying, the blocks were pushed into a solution containing 500 mM EDTA, 1% N-lauroyl sarcosinat and 10 mM Tris–HCl, pH 9.0 mixed with 1 mg/ml proteinase K and incubated overnight at 50°C.
On the next day the blocks were washed five times for 30 min in 1× TE buffer (pH 8.0), incubated for 2 h in 1× TE buffer (pH 8.0) with 1 mM PMSF followed by four to five further washes in TE.
PCR
The PCRs were performed in a 3 µl volume and covered with 6 µl of oil. The primers used are listed in Table 1.
Table 1. Primers used in this study.
| SNP | PCR primers | Extension primers |
|---|---|---|
| PECAM1 gene, G58+A | CATTTTGCATTTCTCTCCACCGCAGGGCAGGTTCATAAATAAG | ATGTTCCGAGAAGAACLptGptAptT |
| OPRM1, IVS+691 | GAGTGATGTTACCAGCCTGAAAGGAAAAATTAGCAGCAACA | GGTGTTAATACTGCCATTTTGCTLptAptTptT |
| OPRM1, rs510769 | TGTGATGGGTGCTCTAGACAAAGAAACCTGCAATACTTGCTGAA | GATATATGGCATTTCACATTCACALptGptTptA |
| OPRM1, rs607759 | AAGCTCTAAAACATGGAAAGGAAATCATGCAATGAAGGGGTCTTAT | GCCTAAAATTGAATGGCTCTALptGptAptC |
| OPRM1, rs476685 | TTTTCTGCATGAAGGACACATTCCAATTTCAGTGGCCTTGGATC | GCAGAGAATGACAACCTGGAATLptTptTptA |
| OPRM1, rs3778148 | TAGCTGAAATGTTCCACTACACTGTCTCCCATTTGGATTCCTTGCG | CTGAGAGCTAATGTTTCAAAGAALptCptTptT |
| OPRM1, rs3778150 | ATTCCAAGTCAGAAGACAACTCCTGATGACTGTTTTCCATCCAAGA | TCCTTACAAATGCTATTGLptGptTptT |
| OPRM1, rs3778152 | GTCCCCAAGCTCCAGAACACAAAGAGGTCACCAGTGGTTCAAGC | AATGTGATATTTTAAAGGGLptCptCptT |
PECAM1 gene, SNP G58+A (SNP information provided by F. Cambien (Paris, France). One microlitre of genomic DNA (5 ng) was used as template for PCR. Five picomoles of the forward and 5 pmol of the reverse primer were used in 40 mM Tris base, 32 mM (NH4)2SO4, 50 mM KCl and 2 mM MgCl2 at pH 8.8 with 50 µM dNTPs and 0.2 U Taq DNA polymerase in a 3 µl volume. The reaction was denatured for 4 min at 94°C, then thermocycled for 30 s at 94°C, for 45 s at 65°C and 30 s at 72°C, repeating the cycle 30 times. The PCR product size was 218 bp.
OPRM1, SNP IVS+691(19). One microlitre of genomic DNA (10 ng) was used as template for PCR. Five picomoles of the forward and 5 pmol of the reverse primer were used in 40 mM Tris base, 32 mM (NH4)2SO4, 50 mM KCl and 2 mM MgCl2 at pH 8.8 with 50 µM dNTPs and 0.2 U Taq DNA polymerase in a 3 µl volume. The reaction was denatured at 95°C for 2 min, then thermocycled for 20 s at 95°C, 30 s at 68°C and 30 s at 72°C, repeating the cycle 30 times. The PCR product size was 753 bp. According to our experience the PCR product size seems to be less important. However, we recommend to generate PCR products with a size <1000 bp. For the amplification of a stretch of genomic DNA containing SNPs rs510769, rs607759, rs476685, rs3778148, rs3778150 and rs3778152 the following protocol was applied: 1 µl of genomic DNA (5 ng), 5 pmol of the forward and 5 pmol of the reverse primer were mixed in 40 mM Tris base, 32 mM (NH4)2SO4, 50 mM KCl and 2 mM MgCl2 at pH 8.8 with 50 µM dNTPs and 0.2 U Taq DNA polymerase in a 3 µl volume. The reaction was denatured at 95°C for 2 min, then thermocycled for 15 s at 95°C, 30 s at 56°C and 60 s at 72°C, repeating the cycle 40 times. The PCR product sizes were between 310 and 401 bp. The SNPs were extracted from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp).
Shrimp alkaline phosphatase (SAP) digestion. A 0.25 µl (1 U/µl) aliquot of SAP and 1.75 µl of 50 mM Tris base (pH 8.0) were added to the PCR and incubated for 1 h at 37°C. The SAP was denatured for 10 min at 90°C.
Primer extension
The primers used are listed in Table 1.
In the case of the SNP G58+A in the PECAM1 gene, 5 pmol of primer, 1 U Thermosequenase, 40 mM Tris base (pH 8.8), 2 mM MgCl2, 100 µM α-S-ddNTPs were added to the PCR in a final volume of 7 µl. An initial denaturing step of 1 min at 95°C was used followed by 35 cycles of 10 s at 95°C, 30 s at 50°C and 15 s at 72°C.
For SNPs of the OPRM1 gene, 5 pmol of primer, 1 U Thermosequenase, 40 mM Tris base (pH 8.8), 2 mM MgCl2, 100 µM α-S-ddNTPs were added to the PCR in a final volume of 7 µl. An initial denaturing step of 2 min at 94°C was used followed by 35 cycles of 10 s at 94°C, 30 s at 50°C and 15 s at 72°C.
Photocleavage. The 384-plate or the PCR tubes were put on a UV transilluminator for 1 min. The maximum wavelength was 312 nm.
Alkylation reaction. Twelve microlitres of acetonitrile, 3 µl of bidistilled water and 6 µl of methyliodide were added. The reaction was incubated at 40°C for 35 min. Upon cooling, a biphasic system was obtained. Ten microlitres of bidistilled water were added. As methyliodide and acetonitrile are used for these assays, the procedures are potentially toxic. Therefore, we highly recommend to perform liquid handling of alkylation reagents in a fume hood. After the incubation, the volatile methyliodide is in the lower phase of the reaction mix. Thus, evaporation is inhibited.
Preparation for MALDI analysis. For the preparation of the MALDI target, 1 µl of a 1.5% solution of α-cyano-4-hydroxy-cinnamic acid methyl ester matrix in acetone was spotted on an aluminium target. A 5 µl aliquot of the upper layer of the alkylation reaction was sampled and diluted in 10 µl of 40% acetonitrile. From this solution, 0.5 µl was transferred onto the dried MALDI matrix. Alternatively, the sample preparation procedure developed by Gobom et al. using ‘anchor’ targets was applied (20).
Mass spectrometric analysis. Spectra were recorded on a Bruker Reflex III time-of-flight mass spectrometer. This mass spectrometer is equipped with a Scout MTP™ ion source with delayed extraction. Spectra were recorded in negative ion linear time-of-flight mode. Typical acceleration potentials were 18 kV. For delayed extraction, the extraction delay was 200 ns. On average, 50 laser shots per spectrum were accumulated.
GenoTools analysis. The settings of the software were similar to the default parameters of the parameter set termed ‘soft’. We decreased the relative intensity threshold from 0.2 to a value of 0.08, the calibration tolerance from 12 to ±3 Da and searched for signals in the mass range of 1370–1410 Da.
RESULTS
In this study, a novel method for the generation of charge-tagged DNA by photocleavage is shown. The principle of this reaction is applied to genotyping of SNPs by MALDI-MS. The entire procedure requires only a single tube or well into which reagents are dispensed in successive reaction steps. In contrast to most SNP genotyping methods using MALDI-MS, the final charge-tagged DNA product is transferred onto the MALDI target without stringent purification.
To exemplify the novel procedure, the analysis of three SNPs, SNP G58+A in the PECAM1 gene and SNPs IVS2+69 and rs3778150 in the opioid receptor, mu1 (OPRM1) gene, is shown. The PECAM1 gene is a candidate gene for cardiovascular disease (21), while DNA variants in the OPRM1gene were associated with substance dependence (19).
A stretch of DNA containing SNP G58+A in the PECAM1 gene and SNPs IVS2+691 and rs3778150 in the OPRM1 were amplified by PCR in a 3 µl volume. Extension primers containing a photocleavable linker and phosphorothioates at the 3′ end were used for primer extension with α-S-ddNTPs in a total volume of 7 µl. The primer for SNP G58+A in the PECAM1 gene was extended with α-S-ddGTPs and α-S-ddATPs. The primer for SNP IVS2+691 in the OPRM1 was elongated with α-S-ddGTPs and α-S-ddCTPs and the primer for SNP rs3778150 was extended with α-S-ddTTPs and α-S-ddCTPs. In the primer for SNP G58+A, an adenine was replaced by the photocleavable linker, while for SNP IVS+691, a cytidine, and for SNP rs3778150, an adenine was replaced.
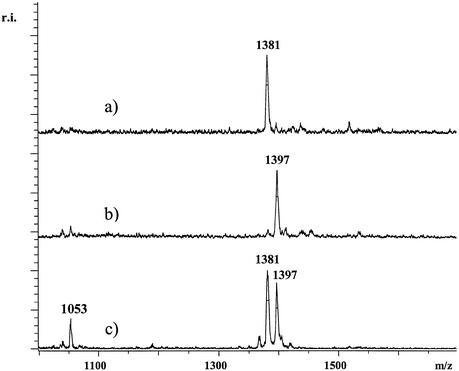
In Figure 2, a genotyping experiment for SNP G58+A in the PECAM1 gene is shown. In Figures 3 and 4, SNP genotyping experiments were performed for SNPs IVS2+691 and rs3778150 in the OPRM1 gene. The small cleavage products of all three SNPs were measured with high sensitivity, accuracy and resolution. High reproducibility in measurements was observed, accumulating on average only 50 laser shots per analysis. In most spectra signal-to-noise ratios higher than 7:1 and near baseline isotopic resolutions were obtained in the linear detection mode. Thirty DNAs were used for this study. As a control, the DNAs were genotyped by the standard GOOD assay in negative ion mode as was described elsewhere (14). The genotypes obtained by the method presented here and the standard GOOD assay matched completely. To get an idea of the robustness of the procedure, for all three SNPs 10 genotyping experiments were performed with one of these 30 DNAs in parallel and analysed by MALDI-MS. The experiments confirmed high reliability meaning that in each case spectra with a similar quality to those shown here were obtained. Furthermore, we took 12 out of the 30 DNAs and genotyped SNP G58+A in the PECAM1 gene for each of the 12 DNA samples eight times. The result of this experiment is displayed in Figure 5 showing 96 data points and a quality assessment according to GenoTools analysis. The quality of the spectra were similar. As shown in Figure 6, the automatic analysis using the GenoTools software consistently resulted in the correct genotypes.
Figure 2.
For the SNP G58+A of the PECAM1 gene primer 5′-ATGTTCCGAGAAGAACLptGptAptT was used. Respective products of the assay were pGptAptTpt[G/A] with masses 1397 and 1381 Da. Trace (a) shows the analysis of a DNA homozygous for A while in trace (b) the analysis of DNA homozygous for G is shown. In (c) the analysis of heterozygous DNA is displayed. At 1053 Da, residual primer of the primer extension reaction is observed. The spectra were not smoothed or otherwise manipulated.
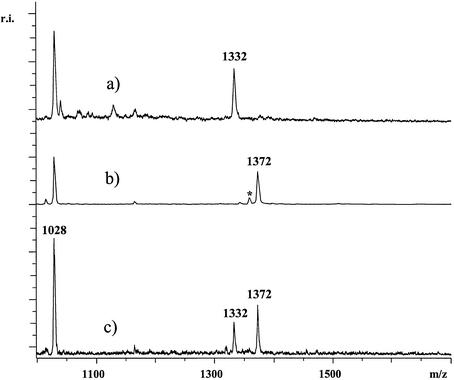
Figure 3.
For the SNP IVS2+691 of the OPRM1 gene primer 5′-GGTGTTAATACTGCCATTTTGCTLpt AptTptT was used. Respective products were pAptTptTpt[G/C] with masses 1372 and 1332 Da. Trace (a) shows the analysis of a DNA homozygous for A while in trace (b) the analysis of DNA homozygous for G is shown. In (c) the analysis of heterozygous DNA is displayed. At 1028 Da, residual primer of the extension reaction is observed. In trace (b) at 1358 Da a very small signal deriving from under-alkylated products was observed (signal indicated with an asterisk). The spectra were not smoothed or otherwise manipulated.
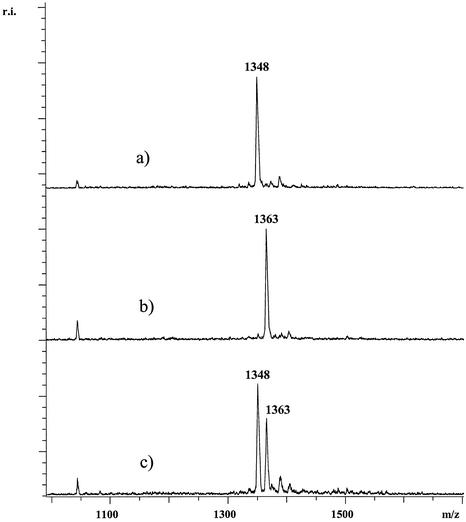
Figure 4.
For the SNP rs3778150 of the OPRM1 gene primer 5′-TCC TTA CAA ATG CTA TTG LptGptTpt T was used. Respective products were pGptTptTpt[T/C] with masses 1363 and 1348 Da. Trace (a) shows the analysis of a DNA homozygous for C while in trace (b) the analysis of DNA homozygous for T is shown. In (c) the analysis of heterozygous DNA is displayed. The spectra were not smoothed or otherwise manipulated.
Figure 5.
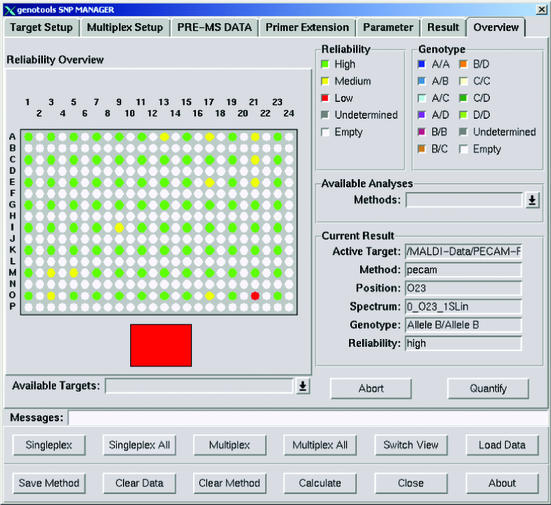
For automatic analysis the GenoTools software was used. An overview of the genotype analysis of the described experiment for SNP G+58A in the PECAM1 gene, using 12 different DNAs (in columns) and performing eight experiments in parallel (in rows), is shown on a 384-well plate target. Green points correspond to spectra with high, yellow points with medium and red points with low reliability. White points are empty positions, grey points would indicate undetermined results due to very low spectra quality.
Figure 6.
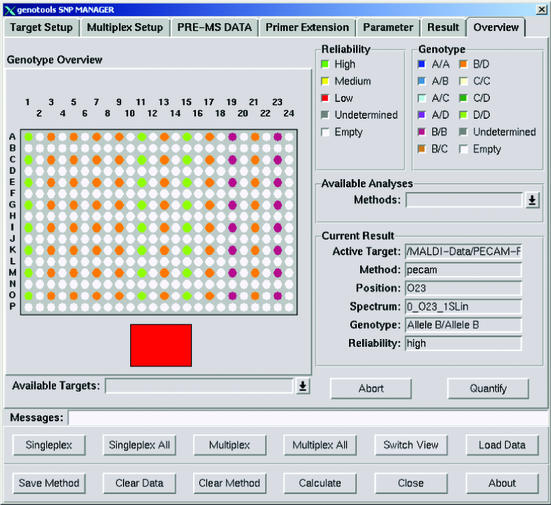
The corresponding biological alleles of the analysis shown in Figure 5 are coded A, B, C and D. Green points stand for homozygote D/D (G/G), orange points for heterozygotes B/D (A/G) and violet points for homozygous B/B (A/A) DNA.
In addition to the results of experiments shown in this article, we successfully established protocols for SNPs rs510769 (G/A polymorphism), rs607759 (C/T polymorphism), rs476685 (C/T polymorphism), rs3778148 (G/T polymorphism) and rs3778152 (G/A polymorphism) in the OPRM1. The respective modified extension primers are listed in Table 1 using the same primer extension protocols as for the two examples shown in Figures 3 and 4.
The allele-specific, charge-tagged DNA products generated by the method presented here are exclusively detectable under the chosen MALDI conditions. In particular, a MALDI matrix is used that has a pKa ∼8. Thus, the matrix does not significantly protonate DNA in solution, but its absorption maximum matches the emission wavelength of a nitrogen laser, which is the laser of choice in most MALDI mass spectrometers. As was shown recently, use can be made of the remarkable discriminative behaviour of this matrix. It was found empirically that its selectivity is towards single-charged DNA compounds (17). As was shown for the GOOD assays, due to the charge-tagged DNA products, no stringent purification was required. Assuming ∼4 pmol of primer extension products in the upper aqueous phase after alkylation (∼20 µl) and the subsequent 2-fold dilution, 0.5 µl of a solution containing 100 fmol of products is dispersed onto a single spot. Thus, ∼50 fmol of charge-tagged DNA products can be analysed easily without stringent purification, which would be very difficult using native, multiple-charged DNA diluted in buffers (17).
As was shown previously, the efficiency of the photocleavage reaction is roughly quantitative (22). This was verified by conventional MALDI DNA analysis of the primers using standard dried droplet preparation with 3-hydroxypicolinic acid.
In the GenoSNIP assay, the photocleavage reaction is performed for 20 min using a special lamp with a wavelengh maximum at 366 nm (M. Kostrzewa, personal communication). To accelerate this step we used UV light at 312 nm. After 20 s, ∼50% of the oligonucleotides were converted. One minute was sufficient to efficiently cut off the unmodified part of the primers. By usage of commercially available UV transilluminators (an integral part of each molecular biological laboratory) the photocleavage reaction can be applied easily without a special UV lamp. The photocleavage can be performed before or after the alkylation reaction used to neutralise the phosphorothioate linkages of the primer. Thus, the DNA products are negatively charge-tagged and, in contrast to the GenoSNIP assay, can be analysed by MALDI-MS without further stringent purification.
DISCUSSION
Clearly, the exposure of DNA to UV light potentially can involve side reactions such as crosslinking oligonucleotides or thymine dimer formation and therefore reduce the efficiency of the procedure. However, no corresponding products were detected either in the mentioned experiment analysing the photocleavage reaction as a function of exposure time or in the genotyping experiments using a matrix that is discriminative for charge-tagged DNA. Due to the usage of a α-cyano-4-hydroxy-cinnamic acid methyl ester matrix in the genotyping experiments, only charge-tagged DNA molecules could be detected efficiently. The probability of observing side products containing presumably more than one negative charge is low. However, in some genotyping experiments (in particular if the UV exposure time was >2 min) we have observed side products with a mass 136 Da higher than the main products. Thus, after the photocleavage reaction the corresponding side products contained the o-nitrophenyl group. These side products were also negatively charge-tagged. However, these signals did not disturb the interpretation of the experiments as they were in a completely different mass range and their signal intensity was significantly lower than the signals of the main products. If the reaction conditions were adhered to strictly, no side products were observed, as can be seen from the data shown here.
As the photocleavable linker creates an abasic site, the optimisation of the primer extension reaction is crucial. The UV-cleavable building block decreases the hybridisation temperature of the oligonucleotides since it does not form hydrogen bonds. As was shown for the GenoSNIP assay, this could be compensated for by extending the 5′ site of the primers and decreasing the annealing temperature during the primer extension reaction (http://www.inmerge.com/ aspfolder/ASMSSchedule2.asp). Utilising this strategy, we performed genotyping of eight different SNPs, which were performed on 30 DNAs, using the same protocol for each SNP without investing time in optimisation of primer extension reaction conditions. The photocleavable linker is preferably located at the fourth nucleotide from the 3′ end of the primers. Typically, primers are placed immediately next to the position of the SNP and extended only with α-S-ddNTPs. If primers with more than three phosphorothioates were used, the primer extension is not feasible. By synthesising phosphorothioates into oligonucleotides, stereocentres were introduced. Thus, the more phosphorothioate linkages were introduced, the more the hybridisation efficiency was reduced.
Another limitation of the use of phosphorothioates is the isotopic distribution of sulphur. 32S has a relative abundance of 100 and 34S has a striking relative abundance of 4.442 (23). Therefore, the more sulphur atoms that are integrated into DNA, the more complex in terms of isotopic distribution the resulting mass spectrum is, resulting in peak broadening and decreased sensitivity of the products.
The method presented here is in particular tailored to genotyping of SNPs. To apply this method to typing inserts or deletions more factors must be considered, for example, due to the fact that only small products can be detected reasonably. The analysis of such polymorphisms cannot be performed directly in each case. However, in many cases deletions or insertions can be analysed indirectly by making clever use of sequence information.
Residual dNTPs of the PCR had to be removed because the subsequent primer extension using common DNA polymerases such as Thermosequenase was otherwise disturbed. As was shown recently, by usage of a novel DNA polymerase for the primer extension reaction, termed Tma 31 FS, the enzymatic purification of the PCR can be avoided (16). In contrast to other commercially available DNA polymerases (for example Thermosequenase and DeepVent), this polymerase preferably incorporates (α-S-)ddNTPs over (α-S-)dNTPs. Unfortunately, this polymerase is not (yet) commercially available and the price is not assessable. Therefore, as is done in most standard MALDI-based genotyping assays, users are still constrained to perform enzymatic purification of PCRs by SAP digestion. However, SAP is by far the cheapest enzyme of the GOOD assays. It would be interesting to test novel engineered DNA polymerases with regard to their efficiency to extend ddNTPs in the presence of dNTPs.
As was seen in the spectra of Figures 2, 3 and 4, tiny signals deriving from under-alkylated products with a mass 14 Da lower than the main products were observed. The mass difference between a thymine and a cytosine is 15 Da. Thus, one could imagine that a (T/T) homozygote might falsely be interpreted as a (C/T) heterozygote. Due to the fact that the peak intensities of under-alkylated side products in worst cases were only about a fifth of the main products, such signals can be correctly interpreted by appropriate software, as is shown in Figure 6. The same alkylation chemistry used in our procedure is applied for the GOOD assay, which was recently validated in large-scale using automated GenoTools analysis (3), where the alkylation reaction did not affect the genotype quality.
Recently, a procedure termed the simplified GOOD assay was presented (16). This method consists of a PCR, a primer extension with oligonucleotides containing methylphosphonates and a phosphodiesterase digestion. The three enzymes required for these reactions contribute to the major part of the costs of the assay, and the primer extension reaction with primers containing methylphosphonates is only feasible with Tma 31 FS DNA polymerase. In the procedure presented here the phosphodiesterase digestion is replaced by photocleavage, which accelerates the process significantly and reduces the costs. The principle of the simplified GOOD assay was the replacement of phosphorothioates by methylphosphonates rendering the alkylation reaction unnecessary. A further advantage of the procedure presented here using an alkylation reaction is that the reaction mixtures can be covered with oil. Oil significantly interferes with MALDI preparation. However, during the alkylation reaction, a biphasic system is obtained. Thus, the oil is concentrated in the lower phase, while the DNA products remain in the upper phase. No sealing is required due to the alkylation reaction. Sealing is essential for the simplified GOOD assay but cumbersome for automation. An alternative to circumvent this would be an additional extraction procedure like the alkylation reaction.
The substitution of phosphorothioates by methylphosphonates in the extension primers containing a photocleavable linker is currently not possible for several reasons. Most importantly, primer extension with modified dNTPs or ddNTPs that would efficiently incorporate a charge-neutral linkage in an extension product is presently not feasible as, to our knowledge, such compounds are not commercially available. Unmodified dNTPs or ddNTPs would incorporate a second negative charge in the final product. Such products are not charge-tagged. Therefore, these products are detectable with a significantly lower sensitivity in the mass spectrometer without stringent purification. Another obstacle is that there is currently no phosphoramidite of the photocleavable linker available to synthesise oligonucleotides with a methylphosphonate linkage or another charge-neutral linkage.
The procedure shown here provides a novel SNP genotyping methodology that can be miniaturised in order to provide a high-throughput technology platform with low consumption of DNA and reagents and highly accurate results. After the establishment of such a platform including liquid handling in 384- or 1536-well plate formats, the evaluation of the procedure presented in this article can be performed with hundreds of SNPs and DNAs; for example, the genotype frequencies can be tested for agreement with Hardy–Weinberg expectations. For efficient, large-scale genotyping a computer software facilitating the assay design would be beneficial for users, but is presently not available.
Many factors contribute to the costs of SNP genotyping projects. Several considerations can be taken into account, for example, the initial set-up costs of detection instruments and robots. Although automation is a large up-front investment, for high-throughput it results in overall cost reduction and increased reliability. A second point is the consumption and cost of reagents. In this regard it is important to consider the price of the primers containing phosphorothioates and a photocleavable linker. Phosphorothioates do not significantly contribute to the final price of an oligonucleotide. The price for the photocleavable linker is presently ∼20 euros and therefore contributes to ∼50% of the primers obtained from Biotez. Considering 2000 experiments with such oligonucleotides (0.01 µmol scale) the primers would presently contribute ∼2 cent to a single experiment.
As for any other mass spectrometric procedure consisting of PCR and an allele-specific reaction, the entire procedure can be extended for physical haplotyping of SNPs by using allele-specific PCR (24).
The time-consuming steps of the presented procedure are the PCR and the primer extension reaction, both requiring a thermocycler usually equipped with peltier elements. Novel developments in thermocycler technology like 20K thermal cyclers that are four times faster than existing technology could complete 30 cycles in <20 min (http://www.sequenom. com). Thus, by using these novel technical devices it should be possible to perform the whole SNP typing procedure including thermocycling and liquid handling in ∼3 h.
The ability to multiplex reactions is of considerable importance for reducing the cost per SNP genotype and analysis time. However, in most approaches using mass spectrometry, multiplexing brings about reduced robustness and reliability, so that in high-throughput applications single or duplex reactions are performed. We are presently investigating the limits of multiplexing reactions using our approach. The long-term goal is the development of a methodology making use of the multichannel detection capacity of mass spectrometers. Therefore, further development will focus on the production of arrays containing thousands of different primers for querying SNPs or other DNA variations such as inserts and deletions. The implementation of the photocleavable linker has made feasible the generation of allele-specific and charge-tagged DNA products on a surface.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Carola Burgtorf for the DNA samples, Anett Smyra for technical assistance and Anahid Powell for critical reading of this manuscript. This work was supported by the German National Genome Research Network (NGFN), grant 01GR0155, and the Max-Planck Society.
REFERENCES
- 1.Weeks D.E. and Lathrop,G.M. (1995) Polygenic disease: methods for mapping complex disease traits. Trends Genet., 11, 513–519. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy J.J. and Hilfiker,R. (2000) The use of single-nucleotide polymorphism maps in pharmacogenomics. Nat. Biotechnol., 18, 505–508. [DOI] [PubMed] [Google Scholar]
- 3.Lechner D., Lathrop,G.M. and Gut,I.G. (2002) Large-scale genotyping by mass spectrometry: experience, advances and obstacles. Curr. Opin. Chem. Biol., 6, 31–38. [DOI] [PubMed] [Google Scholar]
- 4.Hillenkamp F., Karas,M., Beavis,R.C. and Chait,B.T. (1991) Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem., 63, 1193A–1203A. [DOI] [PubMed] [Google Scholar]
- 5.Berkenkamp S., Kirpekar,F. and Hillenkamp,F. (1998) Infrared MALDI mass spectrometry of large nucleic acids. Science, 281, 260–262. [DOI] [PubMed] [Google Scholar]
- 6.Griffin T.J. and Smith,L.M. (2000) Single-nucleotide polymorphism analysis by MALDI-TOF mass spectrometry. Trends Biotechnol., 18, 77–84. [DOI] [PubMed] [Google Scholar]
- 7.Pusch W., Wurmbach,J.H., Thiele,H. and Kostrzewa,M. (2002) MALDI-TOF mass spectrometry-based SNP genotyping. Pharmacogenomics, 3, 537–548. [DOI] [PubMed] [Google Scholar]
- 8.Chicurel M. (2001) Faster, better, cheaper genotyping. Nature, 412, 580–582. [DOI] [PubMed] [Google Scholar]
- 9.Pusch W., Kraeuter,K.O., Froehlich,T., Stalgies,Y. and Kostrzewa,M. (2001) Genotools SNP manager: a new software for automated high-throughput MALDI-TOF mass spectrometry SNP genotyping. Biotechniques, 30, 210–215. [DOI] [PubMed] [Google Scholar]
- 10.Nordhoff E., Ingendoh,A., Cramer,R., Overberg,A., Stahl,B., Karas,M., Hillenkamp,F. and Crain,P.F. (1992) Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun. Mass. Spectrom., 6, 771–776. [DOI] [PubMed] [Google Scholar]
- 11.Sauer S. and Gut,I.G. (2002) Genotyping single nucleotide polymorphisms by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci., 782, 73–87. [DOI] [PubMed] [Google Scholar]
- 12.Tang K., Fu,D.-J., Julien,D., Braun,A., Cantor,C.R. and Koester,H. (1999) Chip-based genotyping by mass spectrometry. Proc. Natl Acad. Sci. USA, 96, 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haff L.A. and Smirnov,I.P. (1997) Multiplex genotyping of PCR products with MassTag-labeled primers. Nucleic Acids Res., 25, 3749–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer S., Lechner,D., Berlin,K., Lehrach,H., Escary,J.L., Fox,N. and Gut,I.G. (2000) A novel procedure for efficient genotyping of single nucleotide polymorphisms. Nucleic Acids Res., 28, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauer S., Lechner,D., Berlin,K., Plancon,C., Heuermann,A., Lehrach,H. and Gut,I.G. (2000) Full flexibility genotyping of single nucleotide polymorphisms by the GOOD assay. Nucleic Acids Res., 28, e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer S., Gelfand,D.H., Boussicault,F., Bauer,K., Reichert,F. and Gut,I.G. (2002) Facile method for automated genotyping of single nucleotide polymorphisms by mass spectrometry. Nucleic Acids Res., 30, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gut I.G., Jeffery,W.A., Pappin,D.J.C. and Beck,S. (1997) Analysis of DNA by ‘charge tagging’ and matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom., 11, 43–50. [Google Scholar]
- 18.Bernardi A. and Bernardi,G. (1968) Studies on acid hydrolases. IV. Isolation and characterization of spleen exonuclease. Biochim. Biophys. Acta, 155, 360–370. [PubMed] [Google Scholar]
- 19.Hoehe M.R., Kopke,K., Wendel,B., Rohde,K., Flachmeier,C., Kidd,K.K., Berrettini,W.H. and Church,G.M. (2000) Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum. Mol. Genet., 9, 2895–2908. [DOI] [PubMed] [Google Scholar]
- 20.Gobom J., Schuerenberg,M., Mueller,M., Theiss,D., Lehrach,H. and Nordhoff,E. (2001) Alpha-cyano-4-hydroxycinnamic acid affinity sample preparation. A protocol for MALDI-MS peptide analysis in proteomics. Anal. Chem., 73, 434–438. [DOI] [PubMed] [Google Scholar]
- 21.Newman P.J. (1997) The biology of PECAM-1. J. Clin. Invest., 100, S25–S29. [PubMed] [Google Scholar]
- 22.Hausch F. and Jaschke,A. (2000) Multifunctional DNA conjugates for the in vitro selection of new catalysts. Nucleic Acids Res., 28, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pretsch E., Clerc,T., Seibl,J. and Simon,W. (1981) Tabellen zur Strukturaufklärung organischer Verbindungen mit spektroskopischen Methoden. Springer-Verlag, Berlin, Heidelberg, New York.
- 24.Tost J., Brandt,O., Boussicault,F., Derbala,D., Caloustian,C., Lechner,D. and Gut,I.G. (2002) Molecular haplotyping at high throughput. Nucleic Acids Res., 30, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]