Abstract
The post-translational modifications of histones are key to the modulation of chromatin structure. Distinct patterns of modifications established by histone-modifying enzymes control diverse chromosomal processes. Here, we report the purification and molecular characterization of the fission yeast Clr6 histone deacetyl ase involved in higher order chromatin assembly. We show that a chromodomain protein Alp13, which belongs to the conserved MRG protein family linked to cellular senescence in humans, is associated with Clr6. In addition, Clr6 interacts with homologs of the mammalian transcriptional co-repressors Sin3, Pst1 and Pst2, and a WD40 repeat-containing protein, Prw1. Alp13, Pst2 and Prw1 form a stable complex with Clr6 in the nucleus. Deletion of any of these factors causes progressive loss of viability and sensitivity to DNA-damaging agents, and impairs condensation/resolution of chromosomes during mitosis. This is accompanied by hyperacetylation of histones and a reduction in histone H3 Ser10 phosphorylation, which correlates with chromosome condensation during mitosis. These results link the MRG family protein Alp13 to histone deacetylation, and suggest that Clr6 and its associated factors are essential for fundamental chromosomal events.
Keywords: chromosome segregation/Clr6/fission yeast/histone deacetylase/mitosis/S.pombe
Introduction
In eukaryotic cells, organization of the genome into higher order structures plays an important role in diverse chromosomal processes including gene regulation, recombination, replication, chromosomal condensation and segregation. Higher order chromatin assembly has been linked to the post-translational modifications of N-terminal histone tails, including acetylation, phosphorylation, ubiquitylation, ADP-ribosylation and methyl ation (reviewed in Jenuwein and Allis, 2001; Zhang and Reinberg, 2001). Among these covalent modifications, the reversible acetyl modification at the lysine residues of histone tails has long been known to correlate with transcriptional regulation. The findings that many transcriptional coactivators including GCN5, PCAF and CBP/p300 possess intrinsic histone acetyltransferase (HAT) activity (Struhl and Moqtaderi, 1998), and that transcriptional co-repressors associate with histone deacetylases (HDACs) (Knoepfler and Eisenman, 1999), provide direct molecular links between histone acetylation states and transcriptional regulation.
In mammalian cells, HDAC1 and HDAC2 are tightly associated with two closely related WD40 repeat-containing proteins, RbAp48 and RbAp46, to form a core repressor complex. This core complex is a constitutive component of two distinct multiprotein complexes referred to as Sin3 and NuRD/Mi2 (Knoepfler and Eisenman, 1999). Mammalian Sin3 was identified originally as a co-repressor that interacts with the DNA-binding heterodimeric transcriptional repressor Mad-Max, and was shown to be structurally homologous to budding yeast transcriptional repressor Sin3 (Ayer et al., 1995; Schreiber-Agus et al., 1995). The mSin3–HDAC complex also contains two other proteins, SAP30 and SAP18 (Laherty et al., 1998; Zhang et al., 1998). The NuRD complex-containing Mi2/CHD family proteins have ATP-dependent nucleosome-remodeling and HDAC activity (Knoepfler and Eisenman, 1999). Besides their role in promoter-specific transcriptional repression, HDACs also play a role in the global deacetylation of histones throughout large chromosomal domains (Vogelauer et al., 2000; Robyr et al., 2002), which might have crucial functions in higher order chromatin assembly and chromosome architecture.
The fission yeast Schizosaccharomyces pombe contains three classical HDACs, named Clr3, Clr6 and Hda1 (Grewal et al., 1998; Bjerling et al., 2002). Our previous studies have shown that Clr3 is a histone H3 Lys14-specific deacetylase that acts in concert with the H3 Lys9-specific methyltransferase Clr4 to establish the histone code for recruitment of Swi6 protein, which is essential for heterochromatin formation at the silent mating-type region and centromeres (Nakayama et al., 2001; Bjerling et al., 2002; Grewal and Elgin, 2002). The Clr6 protein, unlike its budding yeast homolog Rpd3, is essential for viability and it acts in a redundant manner with Clr3 to silence heterochromatic loci. A conditional mutation in clr6 also causes mis-segregation of chromosomes and UV light sensitivity (Grewal et al., 1998). It has been hypothesized that Clr6 participates in global deacetylation of histones, affecting chromatin maturation throughout the genome (Grewal, 2000). However, a detailed role for Clr6 in chromatin assembly and genome organization remains to be fully explored.
Here we describe the genetic and biochemical characterization of Clr6, and investigate its role in the dynamics of chromatin organization. We find that Clr6 interacts with the Sin3 homologs Pst1 and Pst2, the RbAp48-related histone-binding protein named Prw1 and a novel factor Alp13 that belongs to the highly conserved MORF4 (mortality factor on chromosome 4)-related gene (MRG) family, in addition to several as yet unidentified factors. We show that three major polypeptides, Pst2, Alp13 and Prw1, are present together in the same complex with Clr6, and are important for the condensation/resolution of chromosomes during mitosis. These analyses provide novel insight into the role of HDACs in general chromatin assembly and chromosome dynamics.
Results
Immunoaffinity purification of Clr6 HDAC
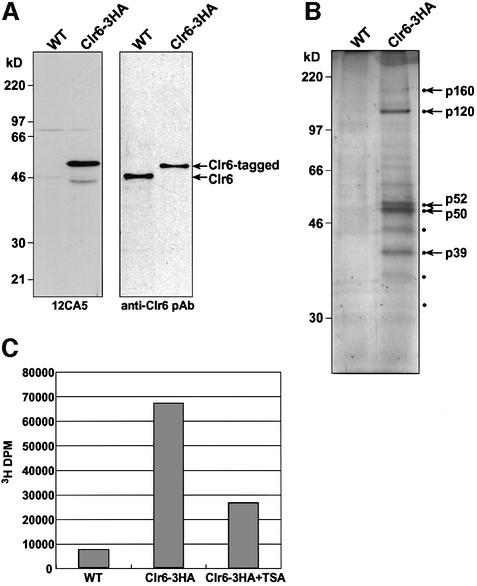
To characterize Clr6 and identify its interacting partners, we utilized an affinity purification procedure. A strain expressing the Clr6 protein fused with a triple hemagglutinin (HA) epitope tag at the C-terminus (Clr6-HA) was constructed by a PCR-based method (Bahler et al., 1998). The expression of tagged clr6+ is under the control of its native regulatory elements so as to achieve wild-type levels of expression. Western analysis with affinity-purified antibodies against recombinant Clr6 revealed the expected size protein band of molecular mass 50 kDa representing Clr6-HA in the tagged strain, and a slightly smaller size band of 46 kDa in control wild-type extracts (Figure 1A). A protein band corresponding to Clr6-HA was also detected with anti-HA monoclonal (12CA5) antibody (Figure 1A). The strain expressing Clr6-HA does not show any obvious phenotypes associated with mutant clr6, suggesting that Clr6-HA can functionally replace the wild-type protein.
Fig. 1. Immunoaffinity purification of Clr6. (A) Whole-cell extracts from wild-type cells (WT) or cells expressing the Clr6-HA fusion protein were resolved on a 10% SDS–polyacrylamide gel and subjected to western blot analyses using anti-HA monoclonal (12CA5) antibody and anti-Clr6 polyclonal antibody. (B) Silver staining of an SDS–polyacrylamide gel resolving the immunoprecipitated fractions using 12CA5-coupled beads. Protein bands present in the purified fractions from Clr6-HA but not wild-type cell extracts are indicated by dots. Major protein bands pointed out by arrows were excised from the gel and subjected to peptide sequencing analyses. (C) Immuno precipitated fractions from the wild-type or Clr6-HA strain were tested for intrinsic HDAC activity. The HDAC activity of Clr6 is inhibited by TSA.
To purify the Clr6 HDAC from cellular extracts, we employed an immunoaffinity purification scheme using anti-HA monoclonal antibody. Whole-cell extracts from either Clr6-HA or wild-type cells were partially purified by ammonium sulfate precipitation and incubated with 12CA5-coupled beads. Bound proteins were eluted from the beads by adding an excess of free HA peptide and analyzed by SDS–PAGE. Several polypeptides present in the purified fraction from Clr6-tagged extracts, but absent in wild-type fractions, were recognized (Figure 1B). Among many bands observed in the Clr6-purified fraction, four major polypeptides, tentatively designated p120, p52, p50 and p39, were found to be present in a nearly equal ratio (1:1:1.2:1). In addition, we also detected several minor polypeptides including p160, p43, p35 and p31 that were present only in the Clr6-HA lane. The four major protein bands were also observed in purified fractions from wild-type cells using anti-Clr6 polyclonal antibodies (data not shown).
To confirm whether the affinity-purified fraction contained a functional HDAC, we performed in vitro HDAC activity assays. Affinity-purified Clr6-HA and control wild-type fractions were incubated with [3H]acetyl-labelled histones. Quantitation of released [3H]acetyl groups revealed that the Clr6-HA fraction possesses deacetylase activity and that this activity is sensitive to trichostatin A (TSA), a specific inhibitor of HDACs (Figure 1C). Based on these results, Clr6 is a functional HDAC that seems to be associated with multiple proteins.
Identification of Clr6-associated proteins
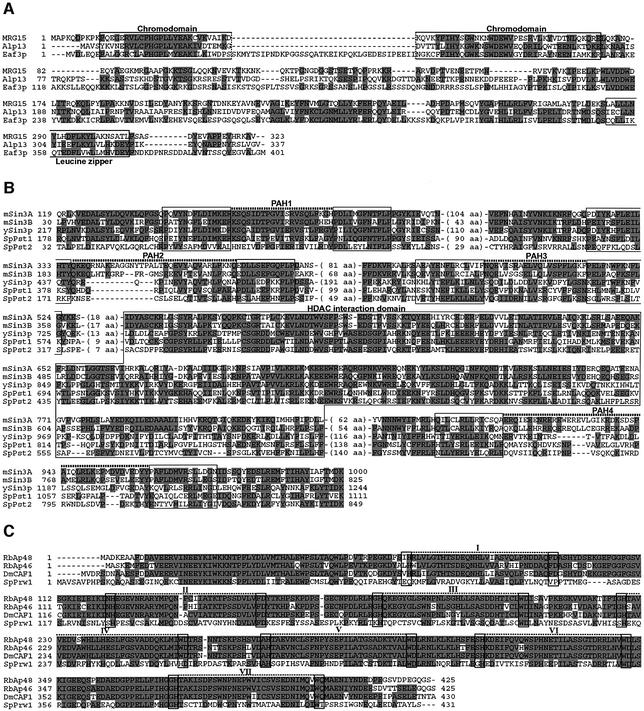
To determine the identities of Clr6-associated polypeptides, we performed a large-scale purification. Protein bands for p160, p120, p52, p50 and p39 were excised from the polyacrylamide gel and subjected to peptide sequencing analyses. Partial sequencing of p50 showed a perfect match to the Clr6 protein (Table I), consistent with its migration (Figure 1B). Microsequencing analysis of p160 and p120 revealed several peptides matching the Pst1 and Pst2 proteins, respectively (Table I; Figure 2B). The calculated molecular weight of both Pst1 (171.4 kDa) and Pst2 (124.8 kDa) is in good agreement with their predicted molecular mass (p160 or p120). Pst1 and Pst2 share high homology with the Sin3A and Sin3B proteins from higher eukaryotes, known for their role as transcriptional co-repressors (Figure 2B) (Dang et al., 1999; Knoepfler and Eisenman, 1999). Sin3 family proteins share conserved paired amphipathic helix (PAH) domains by which Sin3 associates with various repressor proteins. It has also been shown that mammalian Sin3A exists in a large complex including HDAC1 and HDAC2 (Knoepfler and Eisenman, 1999).
Table I. Peptide sequences of p160, p120, p52, p50 and p39.
| Protein | Determined sequences | Positions |
|---|---|---|
| p160 | 1. AIEQQPTILSSTDSNIPRPGTVK | 133–155 |
| 2. QLNVTDALSYLDLVK | 179–193 | |
| 3. TATSSVEETTPR | 489–500 | |
| 4. LEEERYEYDR | 655–664 | |
| 5. SLDHQGVSFK | 772–781 | |
| 6. ASSSPIHHANNNGLR | 901–915 | |
| 7. SSVKEDYVSESTER | 933–946 | |
| 8. NLLNESGNGK | 1364–1373 | |
| 9. STAPDFETSSHRPER | 1454–1468 | |
| p120 | 1. DYPDLLEYLNIFLPSSYK | 87–104 |
| 2. LLNLYVQGIIDR | 280–291 | |
| 3. KTPYEEAMTK | 386–395 | |
| 4. INELPEEERETYTLEEGLGLPSK | 424–446 | |
| 5. DKFSIFLDQVFR | 608–619 | |
| 6. GPDYDVNAPNIVGNKPVR | 620–637 | |
| 7. SQSNSQNSLSDESGNVTPVSK | 678–698 | |
| 8. SQSNSQNSLSDESGNVTPVSKK | 678–699 | |
| 9. KQLSQPAAAIK | 699–709 | |
| 10. QLSQPAAAIK | 700–709 | |
| 11. IIPNPVSQK | 780–788 | |
| 12. LIYGIVDQSAFEDYLR | 819–834 | |
| 13. FVTSLVEQNSSASPK | 869–883 | |
| 14. ANLIVDQSLDTQR | 925–937 | |
| 15. GTSDNTAEVNFDADINALFIP | 1055–1075 | |
| p52 | 1. QAQASEEGINQEK | 12–24 |
| 2. NSPFLYDLIITR | 36–47 | |
| 3. LHTLEGHEDIVTK | 318–330 | |
| 4. ISFSPHEEPILASTSADR | 331–348 | |
| 5. ISFSPHEEPILASTSADRR | 331–349 | |
| p50 | 1. LNVLSNNMENHNTR | 340–353 |
| 2. QYLDSITSEIIENLR | 354–368 | |
| 3. TPGDFTFENAEK | 381–392 | |
| 4. QNIAKEEIMDER | 393–404 | |
| p39 | 1. SRESSTVTVDGDSHELPSR | 105–123 |
| 2. ESSTVTVDGDSHELPSR | 107–123 | |
| 3. SESPIPQQVK | 129–138 | |
| 4. NEETTKPENNEKDDFEEEPPLPK | 147–169 | |
| 5. YLVLHKDEYFIK | 311–322 | |
| 6. EYQNAPPNYR | 323–332 |
Fig. 2. Sequence alignment of the Clr6-interacting factors with similar proteins from other organisms. (A) The predicted amino acid sequences of human MRG15, S.cerevisiae Eaf3p and S.pombe Alp13 were aligned using Clustal W. Identical amino acids are indicated by shaded boxes. Open boxes indicate the conserved chromo domain and leucine zipper motif, respectively. (B) Amino acid sequence alignment of murine Sin3A (mSin3A), murine Sin3B (mSin3B), S.cerevisiae Sin3p (ySin3), S.pombe Pst1 (SpPst1) and S.pombe Pst2 (SpPst2). Conserved paired amphipathic helix (PAH) motifs and HDAC-interacting domains are indicated by open boxes, respectively. (C) Amino acid sequence alignment of human RbAp48 (RbAp48), human RbAp46 (RbAp46), Drosophila melanogaster p55 subunit of CAF-1 (DmCAF1) and S.pombe Prw1 (SpPrw1). Conserved WD40 repeats (I–VII) are indicated by open boxes and hatched lines.
Several peptides from p52 matched a hypothetical protein (accession No. O14021) with a predicted molecular mass of 48.5 kDa (Table I). This protein contains evolutionarily conserved domains known as WD40 repeats and is highly homologous to the RbAp48 protein of mammals (Figure 2C). The gene coding for p52 is thus named prw1+ (S.pombe RbAp48-related WD40 protein 1). RbAp48 was first described as a protein associated with Rb (Qian et al., 1993) and later demonstrated to exist in an HDAC complex (Taunton et al., 1996). RbAp48 and its related proteins were also found to be components of the human chromatin assembly factor CAF1 and a yeast B-type histone H4 acetyltransferase (Tyler et al., 1996; Verreault et al., 1998).
p39 is a member of the highly conserved MRG family of chromodomain proteins
The peptide sequences obtained from p39 matched the predicted amino acid sequence of the Alp13 protein (Table I; Figure 2A). The predicted molecular weight of Alp13 protein is 39.1 kDa, consistent with the apparent molecular mass of p39. alp13+ was identified originally in a screen for mutant cells showing altered polarity (Radcliffe et al., 1998). Sequence analysis revealed that Alp13 is a member of the MRG protein family present in most eukaryotes (Figure 2A; Bertram and Pereira-Smith, 2001). Members of the MRG protein family contain an N-terminal chromodomain, a motif found in proteins associated with higher order chromatin packaging. In addition, they also contain a conserved leucine zipper at their C-terminus. Although the actual functions of MRG family proteins are not known, the human MORF4 was identified based on its ability to induce replicative senescence in immortal human cell lines (Bertram et al., 1999). Another protein MRG15 has been shown to associate with the Rb protein (Leung et al., 2001). The Saccharomyces cerevisiae MRG family member Eaf3p was found in an NuA4 HAT complex (Eisen et al., 2001).
Alp13, Prw1 and Pst2 are stably associated with Clr6 in vivo
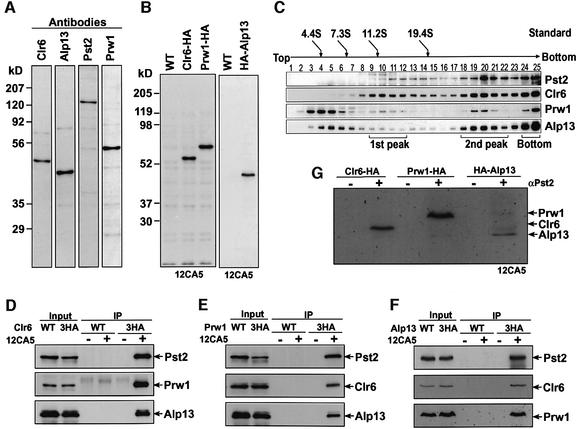
To provide evidence that the major polypeptides present in purified fractions such as Alp13, Prw1 and Pst2 form a stable complex with Clr6, and to characterize these proteins further, we prepared polyclonal antibodies against each of these factors. The respective affinity-purified antibodies specifically recognized Clr6, Alp13, Pst2 or Prw1 (Figure 3A). We also constructed strains expressing Prw1 or Alp13 tagged with triple HA tag (Figure 3B). To determine whether Clr6 is present in multiple protein complexes, we fractionated partially purified cellular extract from the Clr6-HA strain by ultracentrifugation on a 5–40% linear sucrose gradient. Clr6 was detected in at least three peaks by western analysis. The first peak with a sedimentation coefficient of ∼11S contained Clr6 and Prw1 but not Pst2 or Alp13 proteins. All four proteins (Clr6, Alp13, Pst2 and Prw1) were present in the second ∼30S peak, suggesting that they might form a complex. These proteins are also detected in the unresolved bottom fraction.
Fig. 3. Alp13, Pst2 and Prw1 are stably associated with Clr6. (A) Western blot analyses using affinity-purified polyclonal antibodies against each factor. Whole-cell extracts from the wild-type strain were resolved by 10% SDS–PAGE and examined by immunoblotting using antibodies against Clr6, Alp13, Pst2 or Prw1. (B) Whole-cell extracts prepared from wild-type (WT) or strains expressing either Clr6-HA, Prw1-HA or HA-Alp13 were examined by immunoblotting using anti-HA monoclonal antibody (12CA5). (C) Partially purified cell extract from the Clr6-HA strain was fractionated on a 5–40% sucrose gradient and analyzed by western blot using the antibodies shown in (A). Bovine serum albumin (4.4S), aldolase (7.3S), catalase (11.2S) and thyroglobulin (19.4S) were used for sedimentation standards. Peak fractions of Clr6 are indicated underneath the blots. (D–F) Extracts prepared from wild-type cells (WT) or strains expressing Clr6-HA, Prw1-HA or HA-Alp13 were treated with (+) or without (–) 12CA5 antibody, and immunoprecipitated (IP) fractions were analyzed by western blotting using the polyclonal antibodies shown in (A). Lanes labeled input show the equivalent of 20% of the input extracts used in IP lanes. (G) Extracts prepared from Clr6-HA, Prw1-HA or HA-Alp13 were used to perform immunoprecipitation with (+) or without (–) Pst2 antibody, and immunoprecipitated fractions were analyzed by western blotting analysis with anti-HA (12CA5) antibody.
To examine further whether Alp13, Prw1 and Pst2 are present in the same complex with Clr6 in vivo, we carried out a series of immunoprecipitation experiments. First, we performed immunoprecipitation with anti-HA antibody by using extracts prepared from either a wild-type strain or a Clr6-HA-strain. Western analysis revealed that Pst2, Prw1 and Alp13 co-immunoprecipitated with Clr6-HA, as expected, whereas no band was detected in the control wild-type strain (Figure 3D). When we immunoprecipitated the Prw1-HA or HA-Alp13 protein using anti-HA antibody, each of the three other proteins was detected in the precipitated fraction (Figure 3E and F). Finally, we also observed that Alp13, Prw1 and Clr6 co-immunoprecipitate with Pst2 protein (Figure 3G). Based on these results, we suggest that Pst2, Prw1 and Alp13, the three major polypeptides detected in Clr6 purified fractions (see Figure 1A), form a stable protein complex. Interestingly, Pst1 did not co-immunoprecipitate with Alp13 or Pst2 proteins, suggesting that Pst1 might exist in a distinct Clr6-containing complex (our unpublished data).
Alp13, Prw1 and Pst2 are present in the nucleus and co-localized with Clr6
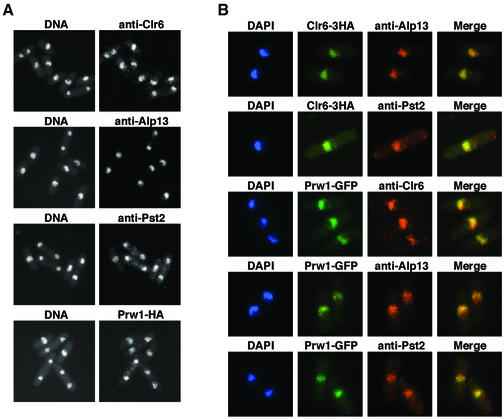
To investigate the subcellular localization of Clr6-associated proteins, we performed indirect immunofluorescence analyses using the polyclonal antibodies described above, or the monoclonal anti-HA (12CA5) antibody with tagged strains. Our analyses showed that all four proteins were localized predominantly in the nucleus on chromatin but excluded from the nucleolus (Figure 4A and B). Interestingly, all four proteins localized to several bright foci exclusively on the 4′,6-diamino-2-phenylindole (DAPI)-stained chromosomes, in addition to uniform distribution throughout the nucleus. We observed no obvious changes in the localization of these proteins at different stages of the cell cycle, as determined by tubulin staining used to visualize microtubules (data not shown).
Fig. 4. Nuclear localization of Clr6 and its associated factors. (A) Fixed wild-type cells were stained with antibodies to Clr6, Alp13 or Pst2. Cells expressing Prw1-HA were also fixed and stained using 12CA5 antibody. DNA was visualized by DAPI. (B) Clr6-HA cells were fixed and doubly stained using 12CA5 and anti-Alp13 or Pst2 antibodies. Cells expressing the Prw1–GFP fusion protein were fixed and stained with antibodies against Clr6, Alp13 or Pst2. DNA was visualized by DAPI. Merged images of two different antibodies, or GFP signals are also shown.
We also performed double immunofluorescence experiments to examine the co-localization of Alp13, Pst2, Prw1 and Clr6. First, to determine whether Alp13 or Pst2 co-localizes with the Clr6 protein, cells expressing Clr6-HA were doubly stained with anti-HA antibody and a polyclonal antibody against Alp13 or Pst2. The data suggest that Alp13 and Pst2 predominantly co-localize with Clr6-HA (Figure 4B), except that a small fraction of Alp13 was present in the Clr6-free, DAPI-stained areas of the nucleus. To analyze co-localization of these proteins with Prw1 further, we made a strain in which the prw1+ gene was tagged with green fluorescent protein (GFP) at the C-terminus. The Prw1–GFP protein showed an identical localization pattern to the Prw1-HA detected using anti-HA antibody (data not shown). We also found that most of the fluorescent Prw1–GFP co-localized with Alp13, Pst2 and Clr6 (Figure 4B). These data, along with results presented above, suggest that Alp13, Pst2 and Prw1 are associated with Clr6 and might be essential for the assembly of a functional HDAC complex.
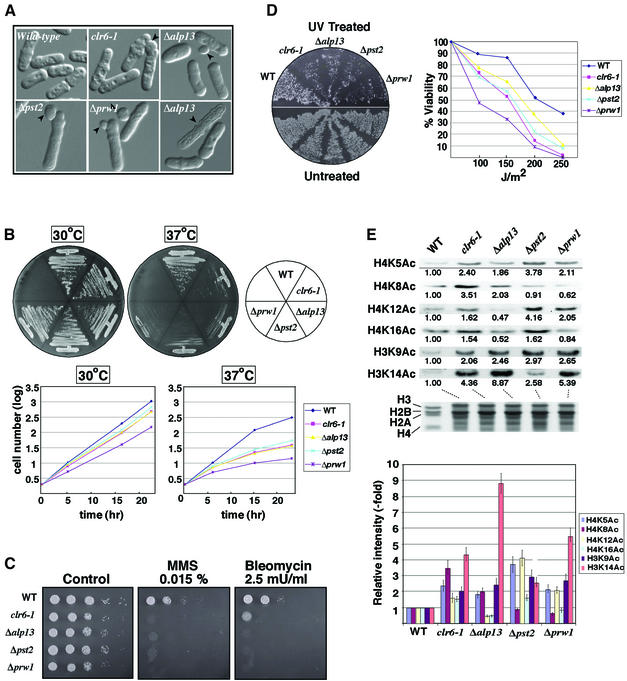
Deletion of alp13+, prw1+ or pst2+ causes aberrant cell morphology, temperature-sensitive growth defects and sensitivity to DNA-damaging agents
We next sought to investigate the biological effects of mutations in Clr6-associated factors by constructing their deletion alleles. Disruptions were verified by PCR and genomic Southern hybridization of heterozygous diploid cells. Tetrad analysis of the sporulated diploids revealed that the G418-resistant segregants carrying deletion alleles grew significantly more slowly than their wild-type counterparts, suggesting that all three genes are important for normal growth (data not shown). Microscopic examination revealed that cells carrying alp13, pst2 or prw1 deletion alleles showed phenotypes similar to clr6 mutant cells. For example, a significant population of cells containing deletion alleles possessed a bud-like protrusion similar to clr6 mutant cells (Figure 5A). In addition, deletion strains often contained dead cells typically in the dividing stage of the cell cycle (Figure 5A; data not shown).
Fig. 5. Deletion of alp13, pst2 or prw1 causes increased histone acetylation, sensitivity to DNA-damaging agents, temperature sensitivity and abnormal morphology. (A) Wild-type and mutant cells were observed under the light microscope. Arrowheads indicate abnormal protrusions or dead cells. (B) Temperature sensitivity (Ts–) of the gene-disrupted strains. The strains previously grown under permissive growth condition (30°C) were replicated onto yeast extract adenine (YEA) medium plates and incubated for 2 days at 30 or 37°C. Since mutant strains displayed a gradual Ts– growth defect, the cells grown overnight were replicated further onto YEA and incubated at the respective temperatures for 2 days (top panel). The strains were cultured in liquid YEA medium at 30 or 37°C. Cell numbers in samples collected at different time points were calculated based on the OD595 (bottom panel). (C) Bleomycin and MMS sensitivity. Serial dilutions of the indicated cultures were plated on the indicated medium and incubated at 30°C for 3 days. (D) UV sensitivity. Patches of the indicated strains were replicated onto YEA plates in duplicate, and one of the plates was UV irradiated (200 J/m2). Plates were incubated in the dark at 30°C for 3 days (top panel). Wild-type and mutant strains were exposed to the indicated doses of UV light, and viable colonies were counted after growth for 3 days (bottom panel). (E) Acetylation levels of bulk histones in wild-type (WT) and mutant strains. Histones were extracted from each strain and examined by western blotting using site-specific histone acetylation antibodies (provided by C.D.Allis, B.Turner or Upstate Biotechnology). Signal intensities for each band were measured using NIH image. Relative band intensities were calculated by comparison with wild-type cells, and means of values from three independent experiments are shown underneath each lane (left panel) and depicted by bars (right panel).
We also found that all three deletions cause sensitivity to DNA-damaging agents [such as UV, methyl methanesulfonate (MMS) and bleomycin] and irreversible temperature-sensitive (Ts–) growth defects (Figure 5B–D). Interestingly, Δalp13, Δpst2 and Δprw1 cells grown at the restrictive temperature became unusually elongated with abnormal cell shapes and progressively lost viability ∼10 cell divisions after shifting to 37°C. Microscopic examination of cells grown at the restrictive temperature revealed that mutant cells did not stop dividing uniformly but lost viability heterogeneously, as suggested by the irregular shapes of the micro-colonies. To confirm these results further, we compared the growth rate of wild-type and mutant cultures in a liquid medium. In comparison with normal exponential growth at 30°C, deletion strains displayed a gradual decrease in their growth rate at 37°C (Figure 5B, bottom panel). Although the basis for the sensitivity to DNA-damaging agents and irreversible growth defects exhibited by mutant strains remains unknown, these results underscore the importance of the association of Alp13, Prw1 and Pst2 with Clr6, and their possible role in chromatin assembly.
Alp13, Prw1, Pst2 and Clr6 are required for deacetylation of histones in vivo
As shown above, purified Clr6 possesses HDAC activity. It was therefore possible that mutations in alp13, pst2, prw1 or clr6 might cause an increase in histone acetylation levels. To examine this possibility, we compared the levels of histone acetylation in wild-type and mutant cells. Bulk histones prepared from wild-type and from Δalp13, Δpst2, Δprw1 or clr6-1 mutants were analyzed by western analysis with antibodies for specific acetylated residues (such as acetylated Lys5, Lys8, Lys12 or Lys16 of histone H4, and Lys9 or Lys14 of histone H3). The results presented in Figure 5E demonstrate that mutant strains show a significant increase in histone acetylation levels compared with the wild-type control. Mutation in clr6 results in elevated acetylation levels at all residues tested on the histone H3 and H4 tails. However, deletion of alp13, pst2 or prw1 had differential effects on the acetylation status of histone tails. Interestingly, we noticed that Δprw1 causes an increase in the acetylation of H4 Lys5 and Lys12, a pattern of acetylation known to be associated with newly synthesized histones (Sobel et al., 1995). Moreover, Δprw1 cells showed an increase in acetylation of H3 Lys9 and Lys14. Cells carrying Δalp13 were specifically defective in the removal of acetyl groups present on H3 Lys9 and Lys14, as well as H4 Lys5 and Lys8. The Δpst2 cells showed a significant increase in acetylation of most lysine residues on H3 and H4 tails, except H4 Lys8. These data suggest that Clr6 and its associated factors are involved in the deacetylation of histones in vivo. The biological significance of the differential effects on histone acetylation observed in different deletion backgrounds is not clear. However, it can be imagined that Alp13, Pst2 and Prw1 help in recruiting Clr6 HDAC activity to perform diverse functions, or that these factors also have a role in other aspects of histone metabolism.
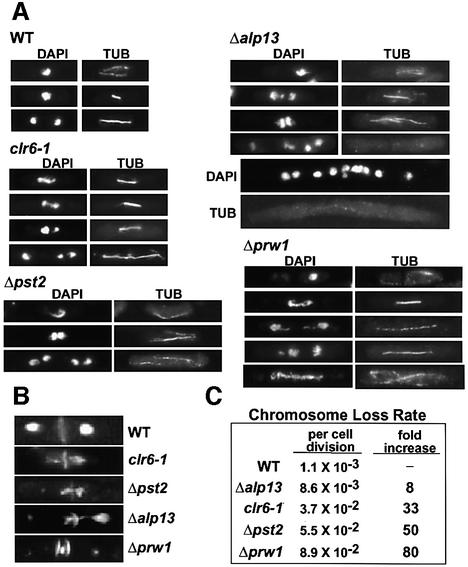
Alp13, Prw1, Pst2 and Clr6 are essential for maintenance of genomic integrity
We previously showed that a mutation in the evolutionarily conserved domain of clr6 severely affects the fidelity of chromosome segregation (Grewal et al., 1998); however, the exact cause of this phenotype is not known. We investigated the nature of segregation defects in the clr6 mutant cells and studied whether deletion of alp13, pst2 or prw1 also affects chromosome segregation. Cells were stained with DAPI to visualize DNA, and with anti-tubulin (TAT1) antibody to indicate their position in the cell cycle. Our analysis revealed that mutant strains display defects in the segregation, resolution and/or condensation of chromosomes during mitosis (Figure 6A), and mis-segregated the mini-chromosome Ch16 (a 530 kb derivative of chromosome 3; Niwa et al., 1986) at a significantly higher rate than wild-type cells (Figure 6C). In wild-type late anaphase cells with fully elongated spindles, chromosomes had completed segregation and were located at the spindle poles in all cells (Figure 6A). We found that clr6, alp13 and prw1, but not pst2 mutant cells, showed a high incidence of lagging chromosomes. Cells with chromosomes stuck in the middle portion of the mitotic spindle were observed frequently (25% in clr6, 19% in alp13 and 35% in prw1). These cells have apparently normal-looking mitotic spindles but clearly fail to segregate chromosomes evenly to the opposite poles. We also observed a significant proportion of mutant cells with aberrant septum formation in the absence of chromosome segregation (Figure 6B). Chromatin in these cells is often fragmented. Cells carrying a deletion of prw1 also exhibit stretching of chromatin along an elongating mitotic spindle, indicating severe defects in condensation or resolution of chromosomes (Figure 6A).
Fig. 6. Cells carrying mutated alleles of clr6, alp13, pst2 and prw1 have defects in chromosome condensation/resolution during mitosis. (A) Wild-type and mutant cells grown at 30°C were fixed with formaldehyde and stained with DAPI to visualize DNA, and with the anti-tubulin antibody TAT1 (TUB) to reveal microtubules. Mutant cells show a high incidence of lagging chromosomes and stretching of chromatin along the spindle during mitosis. The appearance of unusually elongated cells with multiple nuclei was often observed in mutant backgrounds, in particular in Δalp13 cells. (B) Exponentially growing wild-type and mutant cells were stained with DAPI to visualize chromatin, and with calcofluor to visualize the septum. (C) Wild-type and mutant cultures carrying mini-chromosome Ch16 were plated onto adenine-limiting medium. The loss of Ch16 results in red colonies owing to adenine auxotrophy. The rate of mini-chromosome loss was assayed as described previously (Grewal et al., 1998).
Another prevalent phenotype observed was that mutant cultures often contained exceptionally elongated cells with multiple nuclei and/or cells with multiple septa. The multinucleate phenotype, presumably indicating cytokinesis defects, was more pronounced in Δalp13 and Δpst2 cells. These results strongly suggest that the HDAC Clr6 and its associated proteins are important for proper chromosome resolution and/or condensation during mitosis.
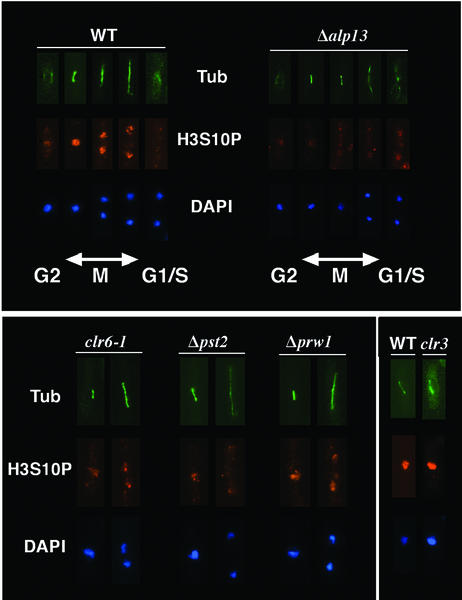
Mitotic defects in mutant cells are correlated with reduction in histone H3 Ser10 phosphorylation
The phosphorylation of H3 Ser10 has been correlated with chromosome condensation and mitosis in a broad range of species (Cheung et al., 2000). Interestingly, deletion of the Ark1/Aim1 aurora kinase responsible for H3 Ser10 phosphorylation in fission yeast causes mitotic defects virtually indistinguishable from the phenotypes described above (Petersen et al., 2001; Leverson et al., 2002; Rajagopalan and Balasubramanian, 2002). It was therefore formally possible that the chromosome segregation defects caused by mutations in Clr6 or its associated factors are functionally linked to changes in H3 Ser10 phosphorylation. Immunofluorescence analysis using site-specific antibodies to H3 phosphorylated at Ser10 revealed that in wild-type cells, H3 Ser10 phosphorylation was first visible at the G2/M phase of the cell cycle and was localized mainly to one or two discrete foci corresponding to heterochromatic loci, where histones are known to be hypoacetylated (Figure 7A; Petersen et al., 2001). H3 Ser10 phosphorylation staining was strongest in early M phase cells with short spindles, and became dispersed throughout the nucleus by mid-metaphase. However, the intensity of the signal diminished as cells entered anaphase, and almost all staining had disappeared in G1/S cells (Figure 7).
Fig. 7. The effects of mutations in clr6, alp13, pst2 or prw1 on histone H3 Ser10 phosphorylation. Exponentially growing wild-type and mutant cells were fixed with formaldehyde and stained with DAPI, with antibodies specific to phosphorylated H3 Ser10 (H3S10P), and with anti-tubulin TAT1 antibodies (TUB) to indicate the cell cycle stage. The top panel shows wild-type and Δalp13 cells during different stages of the cell cycle, while the bottom panel shows clr6, Δpst2 and Δprw1 cells during early and late mitosis. The effect of mutation in Clr3, an H3 Lys14-specific deacetylase, on H3 Ser10 phosphorylation was also examined.
We found that H3 Ser10 phosphorylation levels were considerably reduced in alp13, pst2 and clr6 mutant cells, except at 2–3 foci near the nuclear periphery, presumably representing heterochromatic loci (Figure 7). Deletion of prw1 had a relatively weak effect on H3 Ser10 phosphorylation. Mutation in the H3 Lys14-specific deacetylase Clr3, known to be dispensable for maintenance of genomic stability (Grewal et al., 1998; Bjerling et al., 2002), did not affect H3 Ser10 phosphorylation (Figure 7). Based on the results presented above, chromosome segregation defects in mutant cells are correlated with defects in H3 Ser10 phosphorylation.
Discussion
In this study, we report the biochemical and genetic characterization of the Clr6 HDAC required for higher order chromatin assembly and chromosome segregation in fission yeast (Grewal et al., 1998). The data presented demonstrate that Clr6 is associated with several highly conserved proteins, among which Alp13, Pst2, and Prw1 are present together in the same complex with Clr6, and are important for the maintenance of genomic integrity. Our analyses suggest that these Clr6-interacting proteins are essential for fundamental chromosomal events.
Clr6 is associated with RbAp48- and Sin3-related proteins
Clr6 interacts with several conserved proteins known to be present in HDAC complexes from diverse species, in addition to unique polypeptides such as Alp13. Two of these proteins, Pst1 and Pst2, belong to the Sin3 family of transcriptional co-repressors known to be part of HDAC complexes isolated from budding yeast and mammals (Knoepfler and Eisenman, 1999). Our analysis indicates that Pst1 and Pst2 are present in two distinct Clr6-containing complexes. Although the major proportion of Clr6 forms a stable complex with Pst2, along with Alp13 and Prw1 proteins, a small fraction of Clr6 is present in a distinct Pst1-containing complex that does not include Alp13 or Pst2 (Figure 3; our unpublished data). We suggest that the catalytic subunit Clr6 is a component of multiple protein complexes, presumably serving distinct cellular functions.
Prw1 protein shares strong sequence similarity to the histone-binding protein RbAp48. Mammalian RbAp48 and its related proteins are known to be associated with CAF-1, in addition to HDAC and HAT complexes, and these proteins are thought to serve as molecular bridges between histone-modifying enzymes and core histones (Verreault et al., 1998). Since sucrose gradient fractionation of S.pombe extracts separates Prw1 into multiple peaks (Figure 3C), it is tempting to speculate that Prw1 is also included in additional chromatin-modifying activities, similar to its mammalian counterparts. The presence of Prw1 in additional chromatin-modifying activities might explain the relatively severe chromosome condensation/segregation defects caused by deletion of prw1+.
Role of MRG family proteins in histone modifications
We found that the Clr6-interacting protein Alp13 is a member of the MRG protein family, of which the founding member, MORF4, is capable of inducing replicative senescence in immortal cell lines (Bertram et al., 1999). Members of the MRG family contain a highly conserved chromodomain motif at their N-terminus and are present in most eukaryotic species (Bertram and Pereira-Smith, 2001). Recent studies suggest that MRG proteins might have a conserved role in post-translational modification of histones. Similar to Alp13, the human MRG family proteins interact with the HDAC-containing Sin3 co-repressor, in addition to their association with retinoblastoma (Rb) tumor suppressor (Leung et al., 2001; Yochum and Ayer, 2002). Given that Rb also associates with RbAp48, and that transcriptional repression by Rb involves recruitment of HDAC activity (Brehm et al., 1998; Luo et al., 1998), MRG proteins probably play a regulatory role in the modulation of chromatin structure by directing recruitment of HDACs. Interestingly, members of the MRG family, such as human MRG15, are also present in HAT complexes (Eisen et al., 2001; Pardo et al., 2002). To this end, Alp13 may also interact with additional chromatin-modifying proteins (see also Figure 3C). We speculate that MRG family members, including Alp13, serve as regulatory transcriptional oscillators that direct recruitment of both HDAC and HAT complexes to specific chromosomal domains.
Histone deacetylation and global chromosome functions
We showed that clr6, alp13, pst2 and prw1 mutants are defective in condensation/resolution of chromosomes during mitosis. Although the precise cause of these chromosomal abnormalities remains to be explored, we hypothesize that Clr6-mediated removal of acetyl moieties from histone tails might facilitate the establishment of specific histone modification patterns that are essential for the chromatin association of factors involved in chromosome stability. For example, it is possible that deacetylation of histones by Clr6, in combination with other histone modifications, helps chromosomal binding of condensin and/or cohesin required for condensation of chromosomes during mitosis and sister chromatid cohesion, respectively (Sutani et al., 1999; Tomonaga et al., 2000). Although it remains to be tested whether Clr6 HDAC activity is required for chromosomal arm cohesin, we recently found that a mutation in clr6 does not affect preferential enrichment of cohesin at centromeres (our unpublished data), which is essential for proper kinetochore assembly and chromosome segregation (Bernard et al., 2001; Nonaka et al., 2002). Another possibility is that Clr6 has protein targets other than histones, and that acetylation of these proteins helps regulate proper progression through mitosis.
Histone H4 is acetylated at Lys5 and Lys12 prior to its packaging into chromatin (Sobel et al., 1995; Parthun et al., 1996; Verreault et al., 1998). It has been suggested that these transient modifications might facilitate the recognition of newly synthesized histones by chromatin assembly factors, but are erased shortly after the deposition of histones onto newly replicated DNA (Verreault, 2000). Considering that Δprw1 cells show increased acetylation of histone H4 specifically at Lys5 and Lys12, we hypothesize that Prw1 helps in targeting Clr6 to deacetylate histones soon after or during nucleosome assembly. The chromosome segregation defects displayed by mutant cells, therefore, might be a product of defects in chromatin maturation.
The clr6, alp13, pst2 and prw1 mutants are sensitive to DNA-damaging agents. In principle, these defects could arise from the general relaxation of chromatin. However, it is possible that these factors have a more direct role in maintenance of genomic integrity through their involvement in the re-establishment of chromatin structure following DNA repair. In this regard, histone acetylation is directly required for DNA repair, and components of the NuA4 HAT complex, which contains MRG family protein Eaf3, are recruited to DNA repair sites (Bird et al., 2002). Human HDAC1 recently was shown to interact with the Hus1 protein (Cai et al., 2000), whose S.pombe homolog is implicated in the G2/M checkpoint induced by DNA damage or replication block (Caspari et al., 2000). Although it remains to be tested whether Clr6 also interacts with Hus1, it can be imagined that the Clr6 HDAC plays a role in DNA damage checkpoint control.
Histone deacetylation and cellular senescence
The deacetylation of histones has several connections to the replicative capacity of cells and aging (Guarente, 2000). The treatment of human fibroblasts with HDAC inhibitors reduces their replicative life span, which correlates with aberrant mitosis resulting in cells containing multiple nuclei and fragmented chromatin (Ogryzko et al., 1996; Qiu et al., 2000). In this regard, it is intriguing to note that mutations in Clr6 and its associated factors cause a progressive loss of viability that correlates with severe chromosomal abnormalities. These similar phenotypes in evolutionarily distant species further emphasize an important role for histone deacetylation in cellular senescence. It is possible that chromosomal pathologies caused by defects in the mitotic machinery contribute significantly to the aging process. Considering that the MRG family members share homology with Alp13 (Bertram et al., 2001), it is likely that the senescence induced by human MORF4 is also linked to abnormal histone acetylation patterns. Further analysis of the MRG protein family, as well as other HDAC components, may reveal fundamental mechanisms underlying genome integrity, cellular senescence and aging.
Materials and methods
Strains and media
Strains expressing epitope-tagged Clr6 (HU12) and Prw1 (SPM88) were constructed by a PCR-based module method as described previously (Bahler et al., 1998). To construct a strain expressing Alp13 tagged at its N-terminus, a construct carrying a triple HA tag fused to the alp13+ open reading frame at its 5′ end was used to replace the KAMX6 marker inserted at the alp13+ locus. The resultant strain expresses HA-alp13+ under the control of native promoter. Deletions of alp13, pst2 and prw1 were constructed by transforming diploid cells. Heterozygous diploids were sporulated to obtain haploid strains carrying deletion alleles.
Immunoaffinity purification and peptide sequencing
Exponentially growing cells of wild-type or Clr6-HA strains were harvested by centrifugation and washed with ice-cold STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3 pH 8.0). The cell pellets (1 × 1010 cells) were resuspended in ice-cold HB buffer [25 mM MOPS pH 7.2, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 15 mM MgCl2, 15 mM EGTA, 1 mM dithiothreitol (DTT), 0.1 mM Na3VO4, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with proteinase inhibitor cocktail (Complete™, Roche), and disrupted by vigorous agitation at 4°C. The cell lysates were recovered by adding 10 ml of HB500 (HB buffer + 0.5 M NaCl), and centrifuged at 10 000 r.p.m. for 15 min at 4°C. Ammonium sulfate was added to the supernatant to bring it to 40% saturation. The mixture was incubated on ice for 30 min with gentle stirring, and centrifuged at 10 000 r.p.m. for 30 min at 4°C (Eppendorf microfuge). The precipitated proteins were resuspended in 5 ml of HB500 and centrifuged further at 10 000 r.p.m. for 30 min at 4°C. The supernatant was pre-cleared by incubating with 250 µl of protein A–agarose (Gibco-BRL) for 1 h at 4°C. A 100 µl aliquot of antibody (12CA5)-coupled beads was incubated with the pre-cleared cell lysate at room temperature for 2–3 h. The beads were washed with 150 column volume of HBII buffer (HB buffer with 0.1% Triton X-100 and 10% glycerol), 70 column volumes of HB500, 50 column volumes of HB and 50 column volumes of HBII buffer. For elution, 1 ml of HBII containing 1 mg/ml HA peptide was added and incubated for 1 h or overnight at 4°C. The eluted fractions were visualized by Coomassie staining and excised for in-gel digestion using modified trypsin (Roche). The digests were analyzed by liquid chromatography–mass spectrometry using a Vydac C4 column and analyzed by an electrospray ionization ion trap mass spectrometer (Thermo-Finnigan LCQdeca). The acquired MS data were analyzed by SEQUEST software (Thermo-Finnigan) against the database from the National Center for Biotechnology Information (NCBI).
Antibodies
To obtain antibodies, the coding sequences for the N-terminal region of Pst2 (amino acids 1–218), or for full-length Alp13 (amino acids 1–337), Prw1 (amino acids 1–431) and Clr6 (amino acids 1–405) were PCR amplified from the S.pombe cDNA library. The amplified DNA was cloned into pCRII-TOPO vector (Invitrogen) and subcloned into pRSET vector (Invitrogen). His6-tagged recombinant proteins were produced in BL21 (DE3) cells by inducing expression with 0.1–1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and purified using an immobilized metal affinity chromatography (TALON, Clontech). The purified proteins were used for immunization of rabbits. Crude antisera were affinity purified by using an NHS-activated HiTrap system (Amersham Pharmacia).
Immunoprecipitation and western blotting
Crude cell extracts from wild-type, Clr6-HA, Prw1-HA or HA-Alp13 were partially purified by adding ammonium sulfate at 40% saturation. The lysates (500 µl of 5 mg/ml) were pre-cleared by adding 20 µl of protein A–agarose and incubated for 1 h at 4°C. The clarified extracts were incubated with 12CA5 antibody for 1 h at 4°C, and bound proteins were recovered by adding 20 µl of protein A–agarose beads. The beads were washed twice with 1 ml of HB buffer, once with 1 ml of HB500 and once with 1 ml of HB buffer. Proteins precipitated with the beads were separated by SDS–PAGE and detected by immunoblotting.
Sucrose gradient analysis
Crude cell extract from the Clr6-HA strain was partially purified by adding ammonium sulfate at 40% saturation. The pelleted protein was resuspended in buffer C (20 mM Tris–HCl pH 7.5, 50 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF) and loaded on 5–40% sucrose gradients prepared in buffer C. Centrifugation was performed in a Bechman MLS50 at 36 000 r.p.m. for 14 h at 4°C. Fractions of 200 µl were collected from the top, and aliquots were analyzed by SDS–PAGE and western blot.
Acknowledgments
Acknowledgements
We thank C.David Allis and Bryan Turner for the gift of histone antibodies, Keith Gull for TAT1 antibody, Amar Klar for strains, Robert Krimper, Chikako Nishimoto and Guillaume Lettre for their help, Rebecca Silverstein for sharing the HU12 strain prior to publication, and Ira Hall for critical reading of the manuscript. This work was supported by a research grant from the National Institutes of Health (GM59772) to S.G. K.E.’s laboratory is supported by MFR K2000-31X-12562, NFR 0990-302 and CF 4284 grants. S.G. is an Ellison Medical Foundation scholar.
References
- Ayer D.E., Lawrence,Q.A. and Eisenman,R.N. (1995) Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell, 80, 767–776. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A., Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure,J.F., Partridge,J.F., Genier,S., Javerzat,J.P. and Allshire,R.C. (2001) Requirement of heterochromatin for cohesion at centromeres. Science, 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Bertram M.J. and Pereira-Smith,O.M. (2001) Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene, 266, 111–121. [DOI] [PubMed] [Google Scholar]
- Bertram M.J. et al. (1999) Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol. Cell. Biol., 19, 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A.W., Yu,D.Y., Pray-Grant,M.G., Qiu,Q., Harmon,K.E., Megee,P.C., Grant,P.A., Smith,M.M. and Christman,M.F. (2002) Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature, 419, 411–415. [DOI] [PubMed] [Google Scholar]
- Bjerling P., Silverstein,R.A., Thon,G., Caudy,A., Grewal,S. and Ekwall,K. (2002) Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol., 22, 2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- Cai R.L., Yan-Neale,Y., Cueto,M.A., Xu,H. and Cohen,D. (2000) HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem., 275, 27909–27916. [DOI] [PubMed] [Google Scholar]
- Caspari T., Dahlen,M., Kanter-Smoler,G., Lindsay,H.D., Hofmann,K., Papadimitriou,K., Sunnerhagen,P. and Carr,A.M. (2000) Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol., 20, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P., Allis,C.D. and Sassone-Corsi,P. (2000) Signaling to chromatin through histone modifications. Cell, 103, 263–271. [DOI] [PubMed] [Google Scholar]
- Dang V.D., Benedik,M.J., Ekwall,K., Choi,J., Allshire,R.C. and Levin,H.L. (1999) A new member of the Sin3 family of corepressors is essential for cell viability and required for retroelement propagation in fission yeast. Mol. Cell. Biol., 19, 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A., Utley,R.T., Nourani,A., Allard,S., Schmidt,P., Lane,W.S., Lucchesi,J.C. and Cote,J. (2001) The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem., 276, 3484–3491. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. (2000) Transcriptional silencing in fission yeast. J. Cell. Physiol., 184, 311–318. [DOI] [PubMed] [Google Scholar]
- Grewal S.I.S. and Elgin,S.C. (2002) Heterochromatin: new possibilities for inheritance of structure. Curr. Opin. Genet. Dev., 12, 178–187. [DOI] [PubMed] [Google Scholar]
- Grewal S.I., Bonaduce,M.J. and Klar,A.J. (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics, 150, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2000) Sir2 links chromatin silencing, metabolism and aging. Genes Dev., 14, 1021–1026. [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Knoepfler P.S. and Eisenman,R.N. (1999) Sin meets NuRD and other tails of repression. Cell, 99, 447–450. [DOI] [PubMed] [Google Scholar]
- Laherty C.D. et al. (1998) SAP30, a component of the mSin3 corepressor complex involved in N-CoR- mediated repression by specific transcription factors. Mol. Cell, 2, 33–42. [DOI] [PubMed] [Google Scholar]
- Leung J.K., Berube,N., Venable,S., Ahmed,S., Timchenko,N. and Pereira-Smith,O.M. (2001) MRG15 activates the B-myb promoter through formation of a nuclear complex with the retinoblastoma protein and the novel protein PAM14. J. Biol. Chem., 276, 39171–39178. [DOI] [PubMed] [Google Scholar]
- Leverson J.D., Huang,H.K., Forsburg,S.L. and Hunter,T. (2002) The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell, 13, 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I.S. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Niwa O., Matsumoto,T., Yanagida,M. (1986) Costruction of mini-chromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol. Gen. Genet., 203, 397–405. [Google Scholar]
- Nonaka N., Kitajima,T., Yokobayashi,S., Xiao,G., Yamamoto,M., Grewal,S.I. and Watanabe,Y. (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol., 4, 89–93. [DOI] [PubMed] [Google Scholar]
- Ogryzko V.V., Hirai,T.H., Russanova,V.R., Barbie,D.A. and Howard,B.H. (1996) Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol. Cell. Biol., 16, 5210–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo P.S., Leung,J.K., Lucchesi,J.C. and Pereira-Smith,O.M. (2002) MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J. Biol. Chem., 277, 50860–50866. [DOI] [PubMed] [Google Scholar]
- Parthun M.R., Widom,J. and Gottschling,D.E. (1996) The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell, 87, 85–94. [DOI] [PubMed] [Google Scholar]
- Petersen J., Paris,J., Willer,M., Philippe,M. and Hagan,I.M. (2001) The S.pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci., 114, 4371–4384. [DOI] [PubMed] [Google Scholar]
- Qian Y.W., Wang,Y.C., Hollingsworth,R.E.,Jr, Jones,D., Ling,N. and Lee,E.Y. (1993) A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature, 364, 648–652. [DOI] [PubMed] [Google Scholar]
- Qiu L., Burgess,A., Fairlie,D.P., Leonard,H., Parsons,P.G. and Gabrielli,B.G. (2000) Histone deacetylase inhibitors trigger a G2 checkpoint in normal cells that is defective in tumor cells. Mol. Biol. Cell, 11, 2069–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P., Hirata,D., Childs,D., Vardy,L. and Toda,T. (1998) Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell, 9, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S. and Balasubramanian,M.K. (2002) Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics, 160, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D., Suka,Y., Xenarios,I., Kurdistani,S.K., Wang,A., Suka,N. and Grunstein,M. (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell, 109, 437–446. [DOI] [PubMed] [Google Scholar]
- Schreiber-Agus N., Chin,L., Chen,K., Torres,R., Rao,G., Guida,P., Skoultchi,A.I. and DePinho,R.A. (1995) An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell, 80, 777–786. [DOI] [PubMed] [Google Scholar]
- Sobel R.E., Cook,R.G., Perry,C.A., Annunziato,A.T. and Allis,C.D. (1995) Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl Acad. Sci. USA, 92, 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. and Moqtaderi,Z. (1998) The TAFs in the HAT. Cell, 94, 1–4. [DOI] [PubMed] [Google Scholar]
- Sutani T., Yuasa,T., Tomonaga,T., Dohmae,N., Takio,K. and Yanagida,M. (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev., 13, 2271–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Tomonaga T. et al. (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev., 14, 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J.K., Bulger,M., Kamakaka,R.T., Kobayashi,R. and Kadonaga,J.T. (1996) The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol., 16, 6149–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A. (2000) De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev., 14, 1430–1438. [PubMed] [Google Scholar]
- Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol., 8, 96–108. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Yochum G.S. and Ayer,D.E. (2002) Role for the mortality factors MORF4, MRGX and MRG15 in transcriptional repression via associations with Pf1, mSin3A and transducin-like enhancer of split. Mol. Cell. Biol., 22, 7868–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. and Reinberg,D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev., 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]