Abstract
Although many leukaemia-associated nuclear oncogenes are well characterized, little is known about the molecular details of how they alter gene expression. Here we examined transcription factor complexes and chromatin structure of the human c-FMS gene in normal and leukaemic cells. We demonstrate by in vivo footprinting and chromatin immunoprecipitation assays that this gene is bound by the transcription factor AML1 (RUNX1). In t(8;21) leukaemic cells expressing the aberrant fusion protein AML1–ETO, we demonstrate that this protein is part of a transcription factor complex binding to extended sequences of the c-FMS intronic regulatory region rather than the promoter. The AML1–ETO complex does not disrupt binding of other transcription factors, indicating that c-FMS is not irreversibly epigenetically silenced. However, AML1–ETO binding correlates with changes in the histone modification pattern and increased association of histone deacetylases. Our experiments provide for the first time a direct insight into the chromatin structure of an AML1–ETO-bound target gene.
Keywords: AML1–ETO/c-FMS/chromatin/HDAC-1/t(8;21) leukaemia
Introduction
In recent years, considerable evidence has accumulated confirming the critical importance of the AML1 (acute myeloid leukaemia or RUNX1) transcription factor in normal haematopoiesis. In embryogenesis, AML1 is expressed at high levels in haematopoietic stem cells (HSCs) and cells committed to all haematopoietic lineages including erythroid precursors (Speck, 2001). High levels of expression are found in developing HSCs within the aorta–gonad–mesonephros region where wild-type levels of expression are essential for normal HSC development (Cai et al., 2000; North et al., 2002). Analysis of mice deficient for the AML1 transcription factor demonstrated that these intra-aortic haematopoietic clusters are absent (North et al., 1999) and fetal liver haematopoiesis is deficient (Okuda et al., 1996).
In its wild-type form, AML1 is part of the core binding factor (CBF) complex that exists as a heterodimer together with the CBFβ subunit. It binds to the consensus DNA sequence PyGPyGGT and is involved in the regulation of transcription from a range of haematopoietic-specific genes (reviewed in Scandura et al., 2002). However, the AML1 gene is also an important proto-oncogene. Chromosomal translocations affecting the AML1 gene are a recurring feature in acute leukaemia, with t(8;21), t(16;21) and t(12;21) being the most frequently observed mutations (reviewed in Lutterbach and Hiebert, 2000). The (8;21) translocation occurs in ∼12% of all cases of acute myeloid leukaemia. The result of this mutation is a fusion of almost the entire ETO gene to the N-terminus of the AML1 gene, which encodes the DNA-binding domain of the AML1 transcription factor. This creates a 752 amino acid aberrant fusion protein. A significant amount of evidence from biochemical studies and transient transfection assays has firmly established that this fusion dramatically changes the properties of the CBF complex with regard to transcriptional regulation. Like many other transcription factors, AML1 has been shown to be able to either activate or repress transcription by collaborating with a number of different transcription factors on AML1-responsive target genes (Petrovick et al., 1998; Lutterbach et al., 2000). This is the result of the recruitment of chromatin modification activities such as histone acetylases (HATs) that promote transcriptional activation, or repressive histone deacetylase (HDAC) complexes (Kitabayashi et al., 1998). In contrast, a wealth of biochemical evidence shows that AML1–ETO possesses potent transcriptional repressor activity and within the cell is associated with large multiprotein assemblies containing HDACs (mostly HDAC-1, -2 and -3) and co-repressor complexes such as N-Cor/Sin3A (Lutterbach et al., 1998; Wang et al., 1998; Amann et al., 2001). Transient transfection experiments demonstrated that AML1–ETO is a dominant-negative inhibitor of AML1 function (Frank et al., 1995; Meyers et al., 1995; Westendorf et al., 1998; Melnick et al., 2000), providing a molecular explanation for the finding that mice carrying only one knocked-in AML1–ETO allele display a phenotype almost identical to AML1-deficient mice (Yergeau et al., 1997; Okuda et al, 1998). A recent report has also described the direct interaction of AML1–ETO with an AML1 target gene and correlated this interaction with the downregulation of the expression of this gene in leukaemic cells (Linggi et al., 2002). However, until now, it has not been known how the binding of AML1–ETO affects the chromatin structure of its target genes and thus causes deregulated gene expression.
One of the target genes of AML1 that is affected by AML1–ETO expression is the human c-FMS gene (Zhang et al., 1994). The c-FMS gene locus encodes the receptor for colony-stimulating factor-1 (CSF-1R), which is essential for macrophage development in vivo (Dai et al., 2002). AML1–ETO overexpression profoundly inhibits macrophage differentiation and causes growth arrest and apoptosis (Burel et al., 2001). The c-FMS promoter contains a functional binding site for AML1, which is also recognized by AML1–ETO (Rhoades et al., 1996; Zhang et al., 1996). However, the sequence containing this binding site is not conserved between mouse and human. We recently have characterized the regulatory elements within the second intron of the murine c-fms locus (Himes et al., 2001; Tagoh et al., 2002), which are highly conserved between mouse and human (Himes et al., 2001). One of these elements (c-FMS intronic regulatory element or FIRE) is absolutely required for c-fms expression in macrophage cell lines and transgenic mice (Himes et al., 2001; Sasmono et al., 2003) and contains several functional AML1-binding sites (Tagoh et al., 2002). We have shown that the occupancy of all transcription factor-binding sites in FIRE is acutely regulated during macrophage differentiation, which contrasts with the promoter which is fully assembled in early macrophage progenitors (Tagoh et al., 2002).
In this study, we have applied this knowledge to study the regulation of the human c-FMS gene in myeloid cells. Using human cell lines and cells from normal as well as leukaemic donors, we studied chromatin fine structure, transcription factor occupancy and histone modification patterns along the c-FMS regulatory region during myelopoiesis, and asked whether the chromatin structure and expression of this gene are altered in leukaemic cells. We have shown that the conserved intronic elements also function in human cells, and characterized transcription factors binding to the c-FMS promoter and to intronic regulatory elements by in vivo footprinting. We have employed chromatin immunoprecipitation (ChIP) assays to confirm that FIRE is a bona fide target region of AML1. We studied the recruitment of activating and repressing chromatin-modifying activities as a function of the developmental stage and correlated this with alterations in the histone modification states. Most importantly, we contrasted these findings with chromatin structure studies of leukaemic cells with and without the t(8;21). We can clearly demonstrate that although FIRE is occupied by transcription factors, expression of c-FMS in t(8;21) cells is consistently low. This correlates with significant alterations in the chromatin structure at c-FMS intronic elements.
Results
Human c-FMS mRNA expression levels in normal and leukaemic cells
In contrast to the mouse gene, very little was known about the regulation of the human c-FMS gene. To analyse the effect of the overexpression of a specific oncogene on c-FMS regulation, we initially had to establish the expression pattern, transcription factor occupancy of cis-regulatory elements and chromatin structure in both human cell culture models and primary cells. We therefore examined c-FMS chromatin structure and mRNA expression levels in cells from normal donors, leukaemic patients and myeloid cell lines with and without t(8;21), and correlated these results with the cell type and the differentiation stage of the cells. We used HeLa cells and normal fibroblasts as c-FMS-non-expressing controls. Positively sorted CD34-expressing cells from normal bone marrow (NBM CD34+) served as an immature precursor cell population, and cultured monocytes/macrophages from peripheral blood represented mature myeloid cells. We also analysed samples from the bone marrow of leukaemia patients with or without a t(8;21). However, as these leukaemic cell populations are significantly heterogeneous, we analysed immature leukaemic blast cells purified by cell sorting as described in Materials and methods. In addition, we used the CD34-positive Kasumi-1 cell line, which was originally derived from an AML patient with t(8;21) (Asou et al., 1991). As a leukaemic cell line without t(8;21), we used the promyelocytic leukaemia HL60 cell line, which could be differentiated further into monocytes/macrophages by culturing them with phorbol 12-myristate 13-acetate (PMA) as described in Materials and methods.
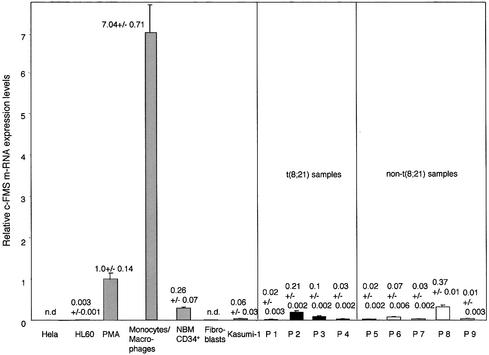
We employed real-time PCR to compare c-FMS expression in the different cell types (Figure 1). Of all expressing cell types, the highest level of c-FMS mRNA was found in cultured monocytes/macrophages, followed by PMA-differentiated HL60 cells, which upregulate c-FMS expression 340-fold compared with undifferentiated HL60 cells. The CD34-positive Kasumi-1 cells express significantly less c-FMS mRNA than NBM CD34+ cells. This was also the case with sorted cells from patients suffering from a leukaemia with or without a t(8;21), indicating that the presence of AML1–ETO does not lead to a significant upregulation of c-FMS expression.
Fig. 1. Kasumi-1 cells and leukaemic blast cells from leukaemia patients express low levels of c-FMS mRNA compared with induced HL60 cells and primary cells (monocytes/macrophages and NBM CD34+ cells). Relative mRNA expression levels in the indicated cell types were measured by real-time PCR as described in Materials and methods. n.d., not detectable. The numbers indicate average relative mRNA levels from at least two independent RNA preparations and/or three independent real-time PCR experiments.
c-FMS intronic regulatory elements display a strong DNase I-hypersensitive site in t(8;21) cells
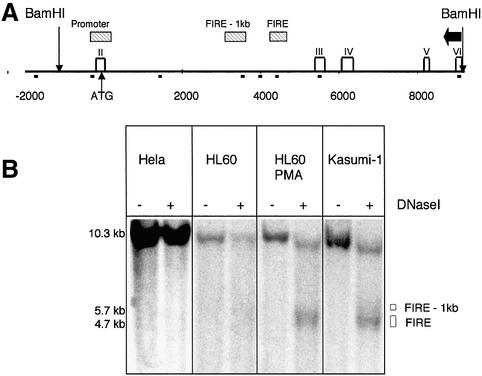
Our previous analysis of the murine c-fms locus identified several regions of DNase I hypersensitivity, all of which coincided with regions of high sequence conservation between mouse and human genes and served as an indicator for the position and the activity of c-fms regulatory elements (Himes et al., 2001) (Figure 2A). Three major DNase I-hypersensitive sites (DHSs) were identified, corresponding to the promoter, and intronic elements, one of which is FIRE. In order to examine whether these elements were also active in human cells, we mapped the DHSs in c-FMS-expressing and non-expressing cells as indicated in Figure 2. Whilst at the promoter only a weak DHS was detected in all tested cell types except HeLa cells (data not shown), we observed significant and highly reproducible differences in DHS intensity at the intronic regulatory elements. Transcription factor occupancy at FIRE in the murine gene is dependent on the differentiation stage of the cells and correlates well with alterations of mRNA expression levels observed during the maturation of primary myeloid precursor cells in vitro (Tagoh et al., 2002). In accordance with these results, a low c-FMS expression level in undifferentiated HL60 cells correlated with the presence of a weak DHS at FIRE, which gained intensity after PMA-induced differentiation. Surprisingly, despite their low mRNA expression levels, we observed the formation of a DHS over the c-FMS intronic regulatory region in Kasumi-1 cells that was as strong as that of PMA-treated HL60 cells. This phenomenon was observed at all DNase I concentrations (data not shown).
Fig. 2. The c-FMS intronic regulatory element displays a strong DHS in Kasumi-1 cells despite a low level of c-fms mRNA expression. (A) A schematic map of the first 12 kb of the human c-FMS gene showing non-coding regions with significant sequence homology between mouse and human as hatched horizontal bars. Exons are indicated as white rectangles and exon numbers are depicted on top. Note that exon I is a non-coding exon and is far upstream of the transcriptional start site close to the PDGF receptor gene (Visvader and Verma, 1989). BamHI sites are indicated. Small black squares below the line indicate primer positions for ChIP experiments presented in Figures 5–8. The probe used for indirect end labelling is displayed as a horizontal black arrow, and open squares represent exons. (B) A strong DHS was found over the intronic regulatory elements in Kasumi-1 cells. This region was only weakly hypersensitive in HL60 cells, which increased significantly with PMA treatment.
Transcription factors binding to c-FMS regulatory elements in normal and leukaemic cells with and without t(8;21): c-FMS promoter and intronic elements are direct targets of AML1
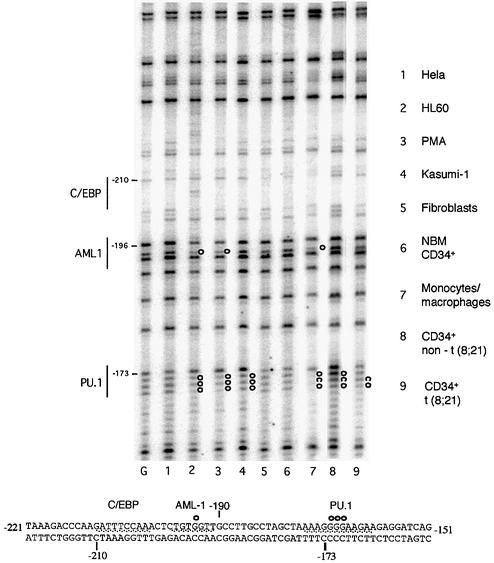
DHS mapping gives important information about DNase I accessibility, but does not indicate the extent of transcription factor-binding site occupancy in the different cell types. It also restricted our analysis to cell lines, because this type of assay required more cells than we could purify from primary tissues. We therefore performed in vivo dimethylsulfate (DMS) footprinting experiments, which are highly sensitive and highlight specific DNA–protein contacts mostly at G(N7). Figure 3 shows an in vivo footprint of the c-FMS promoter. The PU.1 site at –173 bp, that is also present at a similar position in the mouse gene, displays an altered DMS reactivity in both undifferentiated and differentiated HL60 cells, as well as primary macrophages. This also holds true for the AML1 site at –196 bp, which is absent in the mouse gene, thus validating previously obtained in vitro results (Zhang et al., 1994). The extent of protection is identical between the cells, confirming our results from the murine c-fms gene that saw no difference in promoter occupancy during myeloid precursor maturation (Tagoh et al., 2002). NBM CD34+ showed very little change in DMS reactivity as compared with naked DNA, confirming that the c-FMS gene may only be transcribed in a subset of this primitive precursor population, as described previously (Miyamoto et al., 2002). Interestingly, Kasumi-1 cells show clear protection at the PU.1 site that is very similar to that of mature cells, but little protection of the AML1 site. The AML1 site is also not protected in cells from leukaemia patients (Figure 3, lanes 8 and 9).
Fig. 3. In vivo DMS footprinting of the human c-FMS promoter reveals significant protections over the PU.1 and AML-1 sites previously identified with in vitro assays (Zhang et al., 1994). c-FMS non-expressing cells have similar DMS reactivity compared with naked DNA. Both undifferentiated and PMA-differentiated HL60 cells show similar occupancy to primary macrophages, with Kasumi-1 cells and CD34+ blast cells from leukaemia patients (P2 and P9) showing weaker footprints. The lower panel shows the promoter sequence, with protections from DMS reactivity indicated as white circles. G, G reaction with naked DNA.
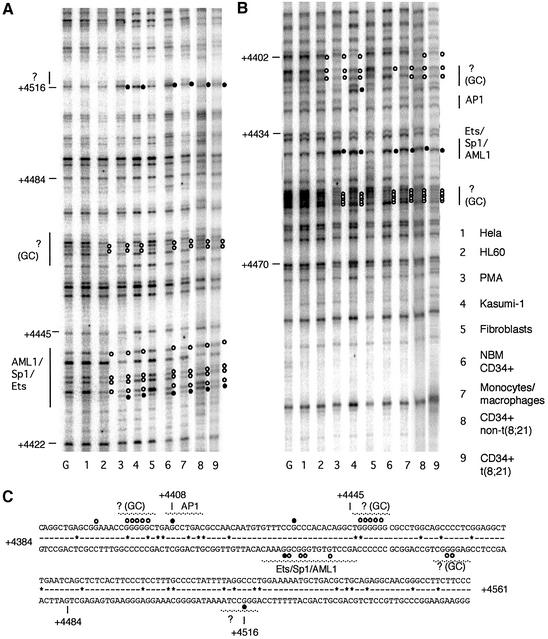
We performed a similar DMS in vivo footprinting experiment to obtain first insights into the transcription factors bound by human FIRE and to examine whether the high DNase I accessibility at this region in Kasumi-1 cells could be explained by a recruitment of additional DNA-binding proteins to FIRE. The results are depicted in Figure 4. FIRE contains a central 100% conserved cluster of 24 bp that contains overlapping functional Ets/AML1/Sp1 consensus binding sites (Tagoh et al., 2002). This element is footprinted to varying extents in all c-FMS-expressing cells tested, including undifferentiated HL60, Kasumi-1 cells, leukaemic blast cells and NBM CD34+ cells (Figure 4A). We also observed protection at GC-rich elements flanking the Ets/AML1/Sp1, which match consensus sequences for AP2 or MZF1 (Morris et al., 1994). Two results are noteworthy: induced and uninduced HL60 cells, primary macrophages and NBM CD34+ all display the same general in vivo footprinting pattern, indicating no differences in protein–DNA interactions at the level of G(N7) contacts. The degree of protection varies with the c-FMS expression level, as was observed with the murine gene (Tagoh et al., 2002). Most importantly, the same pattern of G(N7) contacts is also observed in Kasumi-1 cells and cells from leukaemia patients. However, the degree of DMS reactivity for all other FIRE factor-binding sites as compared with HL60 cells is, if anything, weaker. We conclude from these experiments: (i) that the expression of AML1–ETO does not disrupt or alter the contacts of an enhancer complex at its base (the DNA); and (ii) that the high DNase I accessibility seen in Kasumi-1 cells is not caused by an additional recruitment of transcription factors to DNA.
Fig. 4. In vivo DMS footprinting of the human c-FMS intronic regulatory element (FIRE) reveals extensive protections and enhancements of DMS reactivity on both DNA strands. (A) The lower DNA strand shows extensive protections particularly over the AML1/SP-1/Ets-binding site which are present with varying penetration in all c-FMS-expressing cells. There are further protections in these cells over a GC-rich region at +4450. (B) The upper DNA strand has protections of GC-rich regions flanking the AML1/Sp1/Ets site, which has a prominent enhancement of G +4434 in normal bone marrow CD34+ cells, CD34+ blast cells from leukaemia patients (P2 and P9), Kasumi-1, PMA-treated HL60 and primary macrophages. (C) The FIRE sequence, with protections and enhancements indicated as white and black filled circles, respectively. Sequences conserved between mouse and man are indicated by a line; sequence deviations are displayed as (*). G, G reaction with naked DNA.
AML1–ETO is part of an extended transcription factor complex binding to the c-FMS regulatory region
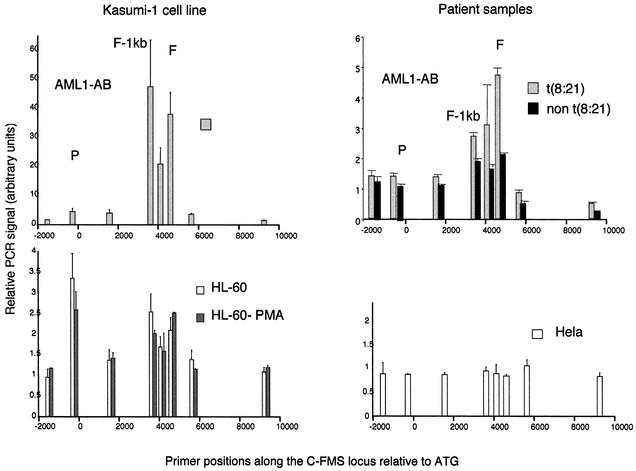
In vivo footprinting experiments indicate the occupancy of specific transcription factor-binding sites, but do not provide direct evidence of the nature of the factors binding to a specific consensus sequence, as different transcription factors from one factor family can recognize the same DNA sequence. In addition, when binding sites that are bound by different proteins in vitro overlap, as seen with FIRE, only ChIP assays can confirm whether a specific transcription factor is bound in vivo. Figure 5 depicts a ChIP assay performed with undifferentiated and PMA-treated HL60 cells, HeLa cells, total bone marrow cells from leukaemia patients and Kasumi-1 cells using an antibody against the RUNT domain of AML1. This domain is present in both wild-type AML1 and AML1–ETO. Specifically precipitated DNA fragments were quantified using real-time PCR and SyBr green as previously described (Göttgens et al., 2002), using primers across the c-FMS regulatory region as indicated in the figure. In order to control for precipitation efficiency, specific signals were calculated relative to a non-specific precipitation with rabbit IgG and then normalized against signals obtained with primers specific for GAPDH exon 6 as detailed in Materials and methods.
Fig. 5. Chromatin immunoprecipitation of AML1 in Kasumi-1, HL60, HL60-PMA and HeLa cells as well as cells from leukaemia patients with or without a t(8;21) with quantification of recovered DNA at eight points along the c-FMS locus. The locations of the promoter, FIRE –1 kb and FIRE regions are indicated (P, F-1kb and F), and DNA was quantified as in Materials and methods. With Kasumi-1 chromatin and chromatin from primary t(8;21) cells, the AML1 antibody significantly enriches the DNA from the intronic regulatory elements with very little promoter enrichment. Precipitation of HL60 and HL60-PMA chromatin with an AML1 antibody enriched both promoter and intronic DNA although amounts precipitated were significantly less than seen with Kasumi-1 chromatin. No specific DNA enrichment was seen with HeLa chromatin.
No enrichment of any c-FMS sequences was obtained with HeLa cells. With induced and uninduced HL60 cells, we saw a 2- to 3-fold enrichment of c-FMS promoter sequences and FIRE. Relative PCR signals were similar for the intronic elements and the promoter. From our ChIP assays, it appeared that the second conserved region in the c-FMS intron harbours another functional AML1-binding site. Sequence analysis indeed revealed AML1 consensus sequences in this element, which are presently being characterized further. A similar level of AML1 immunoprecipitation was seen with cells from a patient with an M2 leukaemia with no t(8;21).
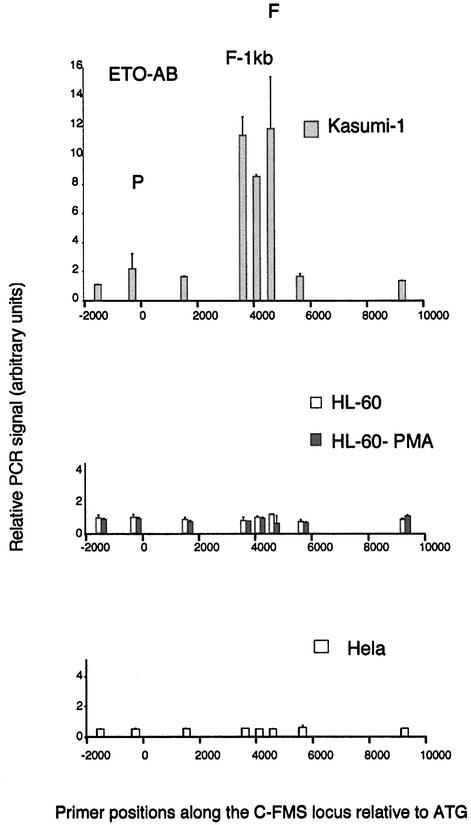
The pattern of specific AML1 target region enrichment with both Kasumi-1 cells and t(8;21) leukaemia patients turned out to be markedly different. Although we found a similar pattern of G(N7) contacts to that in HL60 cells, and sequence analysis revealed no other AML1 consensus sequences in this area, we obtained up to 50-fold enrichment of intronic sequences from Kasumi-1 cells. In contrast, the enrichment at the promoter resembled that of induced HL60 cells. A similar general pattern was observed with a t(8;21) leukaemia patient. The patient experiments were performed with 10 times fewer cells than used for the cell line experiments, yet the degree of specific enrichment still remained significantly higher than in non-t(8;21) control cells. As t(8;21) cells express the AML1–ETO protein as well as the AML1 protein, we repeated the experiment with an antibody specific for ETO (Figure 6). The result was the same, indicating that the c-FMS intronic elements are strongly bound by the AML1–ETO oncoprotein. Because the ETO antibody did not give the same high enrichment as the AML1 antibody, we have been unable to repeat the experiments convincingly with t(8;21) patient samples. In order to exclude that the increased enrichment of FIRE sequences observed with the t(8;21) cells is not due to the developmental stage of the leukaemic samples, we repeated the ChIP experiments using the AML1 antibody with an M2 non-t(8;21) sample where we were able to use more material. Although the cells were blocked at the same developmental stage as the t(8;21) samples, we observed the same low AML1-specific enrichment as PMA-treated HL60 cells (data not shown). We have not been able to find a ChIP-capable antibody against the AML1 C-terminus, and we are therefore unable currently to say whether wild-type AML1 is also bound to c-FMS sequences in Kasumi-1 cells. With the ETO antibody, no c-FMS sequences were precipitated in HL60 cells, HeLa cells (Figure 6) and non-t(8;21) cells from patients (data not shown), indicating that c-FMS regulatory elements are not a natural target for ETO. Taken together, we conclude that the c-FMS intronic elements and not the promoter are high affinity targets for AML1–ETO, which forms extended complexes in Kasumi-1 cells and primary cells from patients. We infer from this result that the formation of these extended complexes is the most likely reason for the high DNase I accessibility observed at c-FMS intronic elements in Kasumi-1 cells.
Fig. 6. Chromatin immunoprecipitation of ETO in Kasumi-1, HL60, HL60-PMA and HeLa cells with quantification of recovered DNA at eight points along the c-FMS locus. The locations of the promoter, FIRE –1 kb and FIRE regions are indicated (P, F-1kb and F), and DNA was quantified as in Materials and methods. With Kasumi-1 chromatin, the ETO antibody significantly enriches the DNA from the intronic regulatory elements with very little promoter enrichment. No specific DNA enrichment was seen with the ETO antibody and chromatin other than Kasumi-1.
Histone modification at AML1–ETO target regions
As outlined in the Introduction, AML1–ETO is a potent repressor of AML1 transactivation (Frank et al., 1995; Meyers et al., 1995; Westendorf et al., 1998; Melnick et al., 2000). This is also true for the c-FMS regulatory elements. Repression of the c-FMS promoter by AML1–ETO in myeloid cells has been demonstrated before (Vangala et al., 2003). We extended these studies by performing transient transfection experiments in macrophage cell lines and measuring the effect of AML1/AML1–ETO expression on luciferase constructs carrying the SV40 basic promoter and the c-FMS promoter alone or together with FIRE inserted in its downstream position. We could clearly demonstrate an inhibitory effect of AML1–ETO overexpression on AML1 transactivation of the c-FMS elements, but not of the SV40 basic promoter (data not shown).
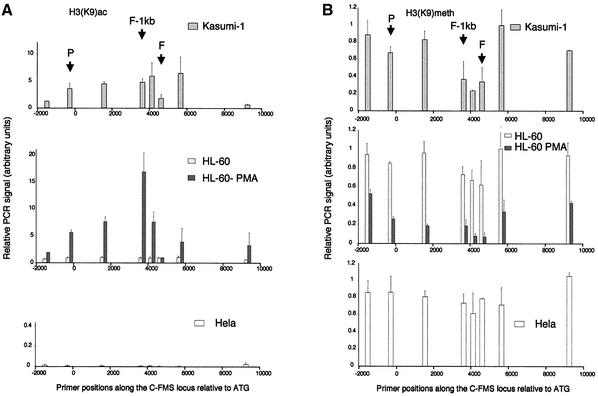
One basis for the difference in the transactivation potential of AML1 and AML1–ETO is that they recruit different types of histone-modifying complexes. To gain first insights into the in vivo effects of AML1–ETO binding to c-FMS on the differential modification of histone tails, we assayed c-FMS chromatin in the different cell lines by ChIP with antibodies against acetylated and methylated Lys9 of histone H3 (Figure 7A and B, respectively). These modifications are mutually exclusive, whereby Lys9 methylation is a hallmark of inactive chromatin and acetylation is found at active loci (for recent reviews see Eberharter and Becker, 2002; Kouzarides, 2002). H3 acetylation levels in HeLa cells were uniformly low across the c-FMS locus. Interestingly, although already bound by transcription factors, this also held true for uninduced HL60 cells. In contrast, PMA-treated HL60 cells displayed a significant increase in H3 Lys9 acetylation upstream and downstream of FIRE, but surprisingly not over FIRE itself. The highest level of H3 acetylation was seen at the promoter-proximal conserved region (FIRE –1 kb). Although strongly DNase I sensitive, the acetylation level of the intronic regulatory region in Kasumi-1 cells was consistently 2- to 3-fold lower than that of induced HL60 cells. Figure 7A shows results with an antiserum against mono-acetylated H3 Lys9. We observed similar results with antibodies against di-acetylated forms of H3 (Lys9 and Lys14), as well as with antibodies against fully acetylated H4 (data not shown).
Fig. 7. Chromatin immunoprecipitation of (A) acetylated Lys9 of histone H3 and (B) methylated Lys9 of histone H3 in Kasumi-1, HL60, HL60-PMA and HeLa cells, with quantification of recovered DNA at eight points along the c-FMS locus. The locations of the promoter, FIRE –1 kb and FIRE regions are indicated (P, F-1kb and F), and DNA was quantified as in Materials and methods. Differentiation of HL60 cells with PMA induces a marked increase in acetylation up- and downstream of FIRE, while FIRE remains relatively deacetylated. Kasumi-1 H3 is acetylated either side of FIRE although relatively less than HL60-PMA chromatin. HeLa H3 is not acetylated along this locus. Both HL60 and HeLa H3 are methylated at K9 across the locus, while PMA differentiation of HL60 cells induces general demethylation, most markedly over FIRE. Kasumi-1 histone H3 (K9) appears relatively methylated, with some reduction over the intronic regulatory region.
Figure 7B shows the results of ChIP assays examining H3 Lys9 methylation. In this case, we normalized real-time PCR signals to signals obtained with primers specific for the α-fetoprotein gene exon 3, which is only active in fetal hepatocytes (Chen et al., 1997). The c-FMS locus in HeLa cells displays the same degree of H3 (Lys9) methylation as the control gene. To our surprise, this was also true for uninduced HL60 cells. In contrast, differentiation of HL60 cells resulted in a significant demethylation of H3 tails at Lys9, with the lowest levels in the intronic regulatory region. The higher level of histone methylation in Kasumi-1 cells paralleled a significantly lower level of H3 Lys9 acetylation. H3 Lys9 at the promoter appeared to be almost completely methylated, and in the intronic regulatory region only a slight reduction of methylation levels could be seen.
Altered equilibrium of CBP and HDAC-1 recruitment to AML1–ETO-bound c-FMS intronic elements
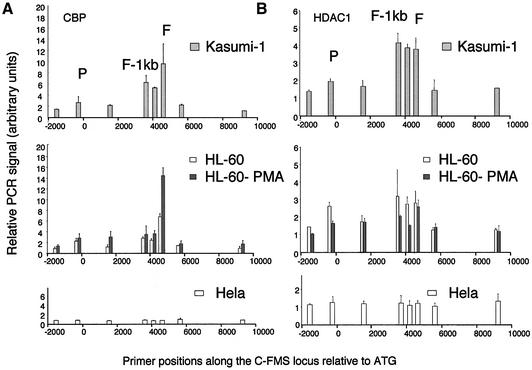
To examine whether the differences in histone modification between AML1- and AML1–ETO-expressing cells were due to differences in the recruitment of histone modification activities in vivo, we performed ChIP assays with antibodies against CBP (Figure 8A) and HDAC-1 (Figure 8B). It is clear that FIRE and not the promoter is the main element recruiting CBP in both undifferentiated and induced HL60 cells. At first sight, this result was puzzling because, even in undifferentiated HL60 cells where histone acetylation levels across the locus were low, a significant amount of CBP was recruited. Even more intriguing was that while PMA treatment increased CBP recruitment 2-fold, histone acetylation levels at FIRE itself remained low. In Kasumi-1 cells, in spite of the strong DHS, low amounts of CBP similar to those in unstimulated HL60 cell were bound to FIRE.
Fig. 8. Chromatin immunoprecipitation of (A) CBP and (B) HDAC-1 in Kasumi-1, HL60, HL60-PMA and HeLa cells, with quantification of recovered DNA at eight points along the c-fms locus. The locations of the promoter, FIRE –1 kb and FIRE regions are indicated (P, F-1kb and F), and DNA was quantified as in Materials and methods. CBP clearly binds to FIRE in both HL60 and PMA-treated HL60 cells, with little binding at other points across the locus. CBP is also bound at FIRE and FIRE –1 kb in Kasumi-1 cells, with no binding in HeLa cells. With Kasumi-1 chromatin, anti-HDAC-1 enriched DNA from the intronic elements with a pattern similar to the precipitation seen with AML1 and ETO antibodies. Low level HDAC-1 binding was also found in HL60 and PMA-treated HL60 cells, with no HDAC-1 binding observed with HeLa cells.
Our analysis of the recruitment of HDAC-1 to c-FMS chromatin revealed an interesting result (Figure 8B). HL60 cells showed a 2-fold enrichment over the promoter and FIRE, which may explain the low level of acetylation over the locus despite the presence of CBP and provides a molecular explanation for the dynamic regulation of FIRE activity that was also seen with the murine locus (Tagoh et al., 2002). The fact that HDAC-1 is recruited at FIRE even in PMA-treated HL60 cells points to a dynamic equilibrium of activators and repressors bound at this regulatory element. The HDAC-1 distribution across the c-FMS locus in Kasumi-1 cells was different. The level of enrichment over the intronic regulatory region was higher and, most importantly, the pattern of HDAC-1 association followed precisely the pattern of AML1–ETO association (Figure 6). No HDAC-1-containing complex was recruited to the promoter, suggesting that the c-FMS intronic region is the main target for the repressive complex recruited by AML1–ETO in t(8;21) leukaemic cells. No targeted recruitment of HDAC-1 could be observed in HeLa cells.
Discussion
Both the c-FMS promoter and the c-FMS intronic regulatory region are targets for AML1, but the intronic regulatory regions are the main target for AML1–ETO
Previous experiments have demonstrated that the promoter of the human c-FMS gene is regulated by AML1 and is also a target for AML1–ETO (Zhang et al., 1994; Rhoades et al., 1996). Our in vivo footprinting results confirm the occupancy of the promoter AML1 site in committed myeloid cells. However, our in vivo footprinting assays, transfection data and ChIP experiments demonstrate that c-FMS intronic elements appear to be additional targets for AML1 action. In contrast to the promoter site, FIRE is 100% conserved between man and mouse and has been shown by in vitro DNA binding assays to bind AML1 (Tagoh et al., 2002). Although this binding site deviates slightly from the ideal AML1 consensus sequence (PyGPyGGT), it is most likely that a juxtaposed Ets-binding site converts it into a high affinity AML1 site. AML1 and Ets proteins are known to interact and form high affinity binding complexes on AML1/Ets composite elements (Kim et al., 1999; Gu et al., 2000). Murine c-fms intronic elements are absolutely required for the correct expression of c-fms constructs in stably transfected cells and transgenic mice (Himes et al., 2001; Sasmono et al., 2003), and FIRE activity is acutely regulated during macrophage differentiation (Tagoh et al., 2002). The most convincing result to confirm this notion is our finding that AML1–ETO forms extended complexes over the intronic regulatory regions, but not the promoter in Kasumi-1 cells and primary t(8;21) cells. This finding stresses the importance of assaying AML1–ETO function in the proper sequence context.
Our finding of low c-FMS expression in sorted leukaemic blast cells contrasts with one previous study. Rhoades et al. (1996) compared c-FMS RNA from unsorted bone marrow samples from patients with and without t(8;21) and unsorted normal bone marrow. They found that t(8;21) correlated with increased c-FMS expression. The reason for this discrepancy is uncertain but may be due to differences in the highly variable cell composition of unsorted leukaemic and normal bone marrow, or may reflect the low patient numbers studied. Indeed, c-FMS expression increases when AML1–ETO is removed from Kasumi cells with inhibitory RNA (Heidenreich et al., 2003), and expression microarray studies from leukaemia patients have found no increased c-FMS expression in t(8;21) cells (Debernardi et al., 2003).
AML1–ETO binding correlates with the establishment of a repressive chromatin structure but does not lead to irreversible epigenetic silencing
It was shown recently that AML1–ETO binding to the p14 ARF gene promoter correlates with a repression of this gene in AML patients carrying t(8;21) (Linggi et al., 2002). Our experiments suggest a molecular explanation for the repressive activity of AML1–ETO at the level of chromatin structure and correlate well with a wealth of biochemical evidence that demonstrated that AML1–ETO is a dominant-negative regulator of AML1 transactivation. We demonstrate that (i) c-FMS expression in purified CD34+ t(8;21) cells is generally lower than in NBM-CD34+ cells and monocyte/macrophage cells; (ii) AML1–ETO overexpression downregulates c-FMS expression and inhibits AML1 induced transactivation; (iii) AML1–ETO forms extended complexes on c-FMS intronic regulatory elements that at the same time are strongly DNase I hypersensitive; (iv) these complexes co-localize with co-repressor complexes containing HDAC-1; and last, but not least, (v) the recruitment of these co-repressors to c-FMS chromatin in t(8;21) cells correlates with a lower level of H3 and H4 acetylation and a higher level of H3 (Lys9) methylation. Our data are therefore consistent with a model by which the binding of AML1–ETO leads to alterations in the chromatin structure of its target genes.
An interesting aspect that warrants further discussion is our finding of a strong DHS in the c-FMS intronic regulatory region in t(8;21) Kasumi-1 cells. As the in vivo footprinting shows that transcription factor complexes are clearly bound at the promoter and FIRE, it indicates that in Kasumi-1 cells, c-FMS is not irreversibly epigenetically silenced and packaged into DNase I-resistant heterochromatin. We do not find a change in transcription factor occupancy on an AML1–ETO target region in cell lines and patient cells or, in other words, AML1–ETO interacts with factors recognizing the same binding sites as AML1. It does not recruit additional factors to the DNA or inhibit binding of factors normally present. This finding is interesting in light of a recent study of chromatin structure observed with PML-RAR leukaemias. Here, PML-RAR binding leads to DNA hypermethylation and epigenetic silencing of the RARβ2 gene over time (Di Croce et al., 2002). With AML1–ETO, however, our data imply that repression requires the continuous presence of the aberrant transcription factor, a finding that may be of therapeutic relevance. That this is indeed true was demonstrated elegantly by experiments in Kasumi-1 cells in which expression of AML1–ETO was blocked by expression of an AML1–ETO-specific RNAi molecule (Heidenreich et al., 2003). After downregulation of AML1–ETO, these cells immediately started to upregulate CSF-1 receptor expression on their surface. This indicates that it is the expression of AML1–ETO itself and not an additional mutation that is responsible for the block in differentiation and the low level of c-FMS expression.
Implications of the mechanism of AML1–ETO action for the development of t(8;21) leukaemias
Our experiments show that the alterations in chromatin structure and expression in c-FMS correlating with the binding of the AML1–ETO complex are quite subtle. Our data show clearly that in their natural state, c-FMS regulatory elements such as FIRE dynamically recruit repressive and activating co-factors. We therefore suggest that AML1–ETO acts by shifting the equilibrium of activating and repressing transcription factor complexes rather than by disrupting their formation altogether. In AML1–ETO-expressing cells, transcription factors are bound to c-FMS regulatory regions and a certain amount of CBP is recruited even in the presence of the AML1–ETO complex. This correlates with low but not absent histone acetylation. We speculate that such subtle deregulation events may happen with a large number of AML1 target genes. Expression of AML1–ETO during embryogenesis is lethal (Yergeau et al., 1997; Okuda et al., 1998). However, recent studies using a conditional AML1–ETO allele demonstrated that AML1–ETO expression on its own does not cause leukaemia, but requires at least one additional mutation for the manifestation of a clinical phenotype (Yuan et al., 2001). Mice expressing AML1–ETO exclusively in the haematopoietic system only show some evidence of haematopoietic deregulation, such as hyper-proliferation of blast cells and developmental abnormalities in myelopoiesis (de Guzman et al., 2002). These experiments clearly show that the expression of AML1–ETO is compatible with haematopoietic cell differentiation once an epigenetic imprint that defines a haematopoietic stem cell has been established. From this point onwards, development is quite stable and can only be disrupted by further mutations that eventually lead to the massive deregulation of global gene expression observed in recent expression microarray studies of AMLs (Schoch et al., 2002). It will be interesting to compare the chromatin structure of different AML1–ETO target genes in order to understand better the molecular details of the deregulation of gene expression by this oncoprotein.
Materials and methods
Cell culture
HL60 cells (ATCC CCL-240) were grown in Iscove’s medium containing 10% fetal calf serum (FCS), 100 U/ml penicillin and 100 µg/ml streptomycin (P/S). When indicated, HL60 cells were cultured with 50 ng/ml PMA (Sigma) for 72 h prior to harvesting. HeLa cells and human fibroblasts (ATCC CCL-110) were grown in Dulbecco’s modified Eagle’s medium with 10% FCS and P/S. Kasumi-1 cells (Deutsche Sammlung von Mikroorganismen und Zellkulturen ACC220) were grown in RPMI with 10% FCS and P/S. CD34+ cells were isolated from bone marrow aspirates from fully consented normal donors and using a CD34+ isolation kit with MACS columns (Miltenyi Biotec) followed by cell sorting using a FACS Vantage cell sorter (Beckton Dickinson) to obtain fractions of >95% CD34+ cells (data not shown). Leukaemic blasts were sorted on the basis of forward/sideward scatter and CD34+/CD33+. The presence of the t(8;21) was confirmed with standard cytogenetic and RT–PCR techniques (data not shown). Normal monocyte/macrophages were obtained from peripheral blood by culturing mononuclear cells in Iscove’s medium containing 10% FCS, P/S and 50 ng/ml M-CSF (R+D Systems). Adherent monocyte/macrophages were harvested after 24 h in culture.
RT–PCR
RNA was extracted and reverse transcribed using standard techniques. PCRs were performed using real-time quantitative PCR (ABI Prism 7700 sequence detection system, Perkin Elmer) with SYBR green. Relative expression was calculated as a c-FMS/GAPDH ratio expressed relative to HL60 PMA. Primers were designed using Primer Express™1.5 software. The following primers were used (F, forward primer; R, reverse primer): c-FMS F, AGCACGAGAACATCGTCAACC; R, TTCGCAGAAAGTT GAGCAGGT; GAPDH F, AACAGCGACACCCACTCCTC; R, CATA CCAGGAAATGAGCTTGACAA.
DNase I-hypersensitive site mapping
DHS mapping was performed as described (Cockerill, 2000). Essentially, nuclei were prepared from cell cultures and incubated with increasing concentrations of DNase I (Roche) for 15 min at 4°C. Following proteinase K treatment and DNA extraction, 20 µg of DNA was digested per sample with BamHI then electrophoresed and subjected to Southern blotting. An c-FMS-specific probe was made with the following primers: c-FMS 8647, AGCCCAGGAATATAACAGCTA; and c-FMS 9121, CGGAAGAACATGGAGGTG.
In vivo footprinting analysis
In vivo DMS footprinting was performed exactly as described (Tagoh et al., 2002) with 1 µg of purified and piperidine-cleaved genomic DNA (Maxam and Gilbert, 1980) from DMS-treated cells used as starting material for LM-PCR amplification. These were HeLa, HL60, PMA-treated HL60, Kasumi-1 cell lines and CD34+ normal and leukaemic bone marrow, normal fibroblasts and monocyte/macrophage cells. DMS-treated and piperidine-cleaved naked genomic DNA was used as a control. Alterations in G(N7) DMS reactivity at c-FMS cis-regulatory elements were visualized by LM-PCR employing the following primers. Promoter: P1, biotin-CTACTAGCTCCGCAGGGATCG; P2, ACACG TTCCTCTCCTCTGCACTG; P3, CTCTCCTCTGCACTGGCTGTTTG TCTTG. FIRE (upper strand): FU1, biotin-AGAGTTAGTCACTATG TTTATTA; FU2, ACCCCCACACATCTGTGTTTACTG; FU3, ACC CCCACACATCTGTGTTTACTG. FIRE (lower strand): FL1, biotin-GTCAGCGTCAGAGCCCAGCCT; FL2, TAGGAGGCTGGGGAAGC AGAA; FL3, TGGGGAAGCAGAAGTGAGAACATCC. LM-PCR-generated bands were visualized and quantified with a phosphoimager screen on a Molecular Imager™ FX (Bio-Rad), using Quantity One software.
Chromatin immunoprecipitation assay and real-time PCR analysis
ChIP assays were performed as follows: 108 cells (or 107 cells from patients) were incubated at room temperature with 1/10 cross-linking solution (11% formaldehyde, 50 mM HEPES pH 8, 0.1 M NaC1, 1 mM EDTA, 0.5 mM EGTA) for 30 min and quenched for 5 min with 125 mM glycine (final concentration). After two washes with ice-cold phosphate-buffered saline (PBS), cell pellets were re-suspended in 40 ml of buffer A (10 mM HEPES pH 8, 10 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.25% Triton X-100) and incubated at 4°C for 10 min with gentle shaking. Adherent cells (HL60-PMA and HeLa) were scraped into ice-cold PBS freshly supplemented with 2.5 mM phenylmethylsulfonyl fluoride (PMSF) and harvested by centrifugation at 500 g at 4°C for 5 min before re-suspension in buffer A. After centrifugation at 500 g at 4°C for 5 min, cells were re-suspended into 40 ml of buffer B (10 mM HEPES pH 8, 200 mM NaC1, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 0.01% Triton X-100), incubated for 10 min and centrifuged as before. Nuclei were sonicated on ice (12 × 30 s at 1 min intervals) in 5 ml of IP buffer (25 mM Tris–HCl pH 8, 2 mM EDTA, 150 mM NaC1, 1% Triton X-100, 0.1% SDS, 2.5 mM PMSF) containing 5 µl of protease cocktail inhibitor (Sigma, P-8340) with 1 ml of glass beads (212–300 µm, acid-washed, Sigma G-1277). After centrifugation at 14 000 g for 10 min at 4°C, chromatin preparations were stored at –80°C in 5% glycerol. Sonicated chromatin from 107 cells was used for each immunoprecipitation. The volume was first adjusted to 500 µl using fresh IP buffer and pre-cleared with 10 µl of protein A–agarose solution [0.5 µl of 5 mg/ml salmon sperm DNA and 1 µl of 10 mg/ml bovine serum albumin (BSA) per 10 µl of washed 50% bead suspension, Sigma P-2545] for 1 h at 4°C using a rotating wheel. The protein A–agarose beads were then pelleted for 20 s at 1500 g and the supernatant was incubated overnight at 4°C on a rotating wheel with 5 µl of normal rabbit IgG (Upstate Biotechnology, 12-370), 25 µl of anti-CBP, 50 µl of anti-ETO antibodies (Santa-Cruz) or 5 µl of anti-dimethyl-histone H3 (Lys9), anti-acetyl-histone H3 (Lys9), anti-diacetyl-histone H3 (Upstate Biotechnology), anti-acetyl histone H4 (gift from C.Crane-Robinson), anti-HDAC-1 (Upstate Biotechnology), 50 µl of anti-AMLrunt or 50 µl of anti-AML1 antibodies (CalBiochem). Then, 10 µl of protein A–agarose solution were added for an additional 2 h. The protein A–agarose beads were pelleted with the immunoprecipitate for 20 s at 1500 g and washed four times with 800 µl of RIPA buffer (10 mM Tris–HCl pH 8, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaC1, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycholate, 1 mM PMSF), once with 800 µl of LiCl buffer (0.25 M LiC1, 0.5% NP-40, 0.5% Na deoxycholate, 1 mM EDTA, 10 mM Tris–HCl pH 8) and twice with 800 µl of TE (10 mM Tris–HCl pH 8.0, 1 mM EDTA). The immune complexes were eluted by adding 500 µl of elution buffer (1% SDS, 0.1 M NaHCO3, 200 mM NaCl). The cross-link was reversed at 65°C overnight. After 1 h incubation at 45°C with a PK solution (10 µl of 0.5 M EDTA, 20 µl of 1 M Tris–HCl pH 6.5 and 20 µg of proteinase K), DNA was extracted by phenol/CHC13, ethanol precipitated, resuspended into 40 µl of TE and stored at 4°C.
PCRs were performed using real-time quantitative PCR (ABI Prism 7700 sequence detection system, Perkin Elmer) with SYBR green. Primers were designed using Primer Express™1.5 software. The following primers were used. c-FMS upstream promoter (–1.5 kb) F, TGTTATATCAGGATTTTTCAATTTCAAGA; R, AGCCACTATAC CCAGCCTAAAGTTC; c-FMS promoter (–0.4 kb) F, TCACTCT GAAGACCCTCATTTGTG; R, CAAGAGCAGCCAGACCGTTAG; c-FMS downstream promoter (+1.4 kb) F, GCCTGGACTTCTTA TTTTGCAATG; R, CCTGTGCTTTCTACTGCATGTAAGTT; c-FMS FIRE –1 kb (+3.5 kb) F, GTGTAAAAAAGGAACAAATGGAATCA; R, ATTGCTACATTTCTAATGTCCCTTCTG; c-FMS FIRE –0.5 kb (+4.0 kb) F, CTGAAAGGACTTGAGAGAGGAAAC; R, CCCACCAT AGAGCCCAGAAT; c-FMS FIRE (+4.5 kb) F, AGAAGGCCC GTTGCCTCT; R, GCTCTCACTTCCCTCCTTTGC; c-FMS exon 3 (+5.5 kb) F, TCCCCGTGTTTTGGAAGGT; R, CTGATGGCTCCAG CAGCAT; c-FMS c-FMS exon 6 (+9.5 kb) F, CTTCCGGGTGG TAGGTAAGCA; R, ACATCAGCTTCCTCAACAAAACC; GAPDH exon 6 F, CAAGGCTGTGGGCAAGGT; R, GGCCATGCCAGTGA GCTT; α-fetoprotein exon 3 F, GCATCGATCCCACTTTTCCA; R, TCTTCATATGCTTCACAGCTTGTG. The distance in kb from the ATG site of c-FMSlocus-specific primers is indicated in parentheses. The amount of DNA precipitated in each experiments was quantified by comparison with standard curve values obtained from amplification reactions carried out with serial dilutions of human genomic DNA. To control the specificity of the amplification reaction and to make sure that only one product was generated, products were examined by using a dissociation curve program (Dissociation Curves 1.0) and were analysed by gel electrophoresis. Relative PCR signals for all primers were first calculated as a signal ratio obtained with the specific antibody versus signals observed with IgG control (non-specific background). In order to correct for the efficiency of precipitation in different experiments with different cell types, signals were then normalized to those obtained with the GAPDH exon 6 primers, except for H3 methylation mapping experiments where signals were normalized to α-fetoprotein exon 3. Data presented in Figures 5–8 represents the mean ± SD of quantifications from between two and four separate immunoprecipitations on at least two separate chromatin preparations for each cell type.
Acknowledgments
Acknowledgements
We thank Dan Tenen for AML1 and AML1–ETO overexpression vectors, Drs D.Swirsky and S.Richards for help with the patient samples, H.Dickinson for cytogenetic analysis, and Charlotte Stephenson for technical support. Work in C.B.’s laboratory is supported by grants from the Leukaemia Research Fund, the City of Hope Medical Center, the Wellcome Trust, the MRC, the AICR and Yorkshire Cancer Research. G.A.F. is a recipient of an MRC Clinical Training Fellowship.
References
- Amann J.M., Nip,J., Strom,D.K., Lutterbach,B., Harada,H., Lenny,N., Downing,J,R., Meyers,S. and Hiebert,S.W. (2001) ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol., 21, 6470–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asou H., Tashiro,S., Hamamoto,K., Otsuji,A., Kita,K. and Kamada,N. (1991) Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood, 77, 2031–2036. [PubMed] [Google Scholar]
- Burel S.A., Harakawa,N., Zhou,L., Pabst,T., Tenen,D.G. and Zhang,D.E. (2001) Dichotomy of AML1–ETO functions: growth arrest versus block of differentiation. Mol. Cell. Biol., 21, 5577–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., de Bruijn,M., Ma,X., Dortland,B., Luteijn,T., Downing,R.J. and Dzierzak,E. (2000) Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity, 13, 423–431. [DOI] [PubMed] [Google Scholar]
- Chen H., Egan,J.O. and Chiu,J.F. (1997) Regulation and activities of α-fetoprotein. Crit. Rev. Eukaryot. Gene Expr., 7, 11–41. [DOI] [PubMed] [Google Scholar]
- Cockerill P.N. (2000) Identification of DNase I hypersensitive sites within nuclei. Methods Mol. Biol., 130, 29–46. [DOI] [PubMed] [Google Scholar]
- Dai X.M., Ryan,G.R., Hapel,A.J., Dominguez,M.G., Russell,R.G., Kapp,S., Sylvestre,V. and Stanley,E.R. (2002) Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies and reproductive defects. Blood, 99, 111–120. [DOI] [PubMed] [Google Scholar]
- Debernardi S., Lillington,D.M., Chaplin,T., Tomlinson,S., Amess,J., Rohatiner,A., Lister,T.A. and Young,B.D. (2003) Prognostically important chromosomal events in acute myeloid leukaemia can be deduced from gene expression analysis. Genes Chromosomes Cancer, in press. [DOI] [PubMed] [Google Scholar]
- de Guzman C.G., Warren,A.J., Zhang,Z., Gartland,L., Erickson,P., Drabkin,H., Hiebert,S.W. and Klug,C.A. (2002) Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1–ETO translocation. Mol. Cell. Biol., 22, 5506–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L. et al. (2002) Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science, 295, 1079–1082. [DOI] [PubMed] [Google Scholar]
- Eberharter A. and Becker,P.B. (2002) Histone acetylation: a switch between repressive and permissive chromatin: second in review series on chromatin dynamics EMBO Rep., 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R., Zhang,J., Uchida,H., Meyers,S., Hiebert,S.W. and Nimer,S.D. (1995) The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene, 11, 2667–2674. [PubMed] [Google Scholar]
- Göttgens B. et al. (2002) Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J., 21, 3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T.L., Goetz,T.L., Graves,B.J. and Speck,N.A. (2000) Auto-inhibition and partner proteins, core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1). Mol. Cell. Biol., 20, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich O., Krauter,J., Riehle,H., Hadwiger,P., John,M., Heil,G., Vornlocher,H.P. and Nordheim,A. (2003) AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood, 101, 3157–3163. [DOI] [PubMed] [Google Scholar]
- Himes S.R., Tagoh,H., Goonetilleke,N., Sasmono,T., Oceandy,D., Clark,R., Bonifer,C. and Hume,D.A. (2001) A highly conserved c-fms gene intronic element controls macrophage-specific and regulated expression. J. Leukocyte Biol., 70, 812–820. [PubMed] [Google Scholar]
- Kim W.Y., Sieweke,M., Ogawa,E., Wee,H.J., Englmeier,U., Graf,T. and Ito,Y. (1999) Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J., 18, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I., Yokoyama,A., Shimizu,K. and Ohki,M. (1998) Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J., 17, 2994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2002) Histone methylation in transcriptional control. Curr. Opin. Genet. Dev., 12, 198–209. [DOI] [PubMed] [Google Scholar]
- Linggi B. et al. (2002) The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat. Med., 8, 743–750. [DOI] [PubMed] [Google Scholar]
- Lutterbach B. and Hiebert,S.W. (2000) Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene, 245, 223–235. [DOI] [PubMed] [Google Scholar]
- Lutterbach B. et al. (1998) ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol., 18, 7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B., Westendorf,J.J., Linggi,B., Isaac,S., Seto,E. and Hiebert,S.W. (2000) A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem., 275, 651–656. [DOI] [PubMed] [Google Scholar]
- Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- Melnick A., Carlile,G.W., McConnell,M.J., Polinger,A., Hiebert,S.W. and Licht,J.D. (2000) AML-1/ETO fusion protein is a dominant negative inhibitor of transcriptional repression by the promyelocytic leukemia zinc finger protein. Blood, 96, 3939–3947. [PubMed] [Google Scholar]
- Meyers S., Lenny,N., Hiebert,S.W. (1995) The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol., 15, 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T., Iwasaki,H., Reizis,B., Ye,M., Graf,T., Weissman,I.L. and Akashi,K. (2002) Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell, 3, 137–147. [DOI] [PubMed] [Google Scholar]
- Morris J.F., Hromas,R. and Rauscher,F.J.3rd (1994) Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol. Cell. Biol., 14, 1786–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T., Gu,T.L., Stacy,T., Wang,Q., Howard,L., Binder,M., Marin-Padilla,M., Speck,N.A. (1999) Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development, 126, 2563–2575. [DOI] [PubMed] [Google Scholar]
- North T.E., de Bruijn,M.F., Stacy,T., Talebian,L., Lind,E., Robin,C., Binder,M., Dzierzak,E. and Speck,N.A. (2002) Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity, 16, 661–672. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen,J., Hiebert,S.W., Grosveld,G. and Downing,J.R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell, 84, 321–330. [DOI] [PubMed] [Google Scholar]
- Okuda T., Cai,Z., Yang,S., Lenny,N., Lyu,C.J., van Deursen,J.M., Harada,H. and Downing,J.R. (1998) Expression of a knocked-in AML1–ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood, 91, 3134–3143. [PubMed] [Google Scholar]
- Petrovick M.S, Hiebert,S.W., Friedman,A.D., Hetherington,C.J., Tenen,D.G. and Zhang,D.E. (1998) Multiple functional domains of AML1: PU.1 and C/EBPα synergize with different regions of AML1. Mol. Cell. Biol., 18, 3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades K.L., Hetherington,C.J., Rowley,J.D., Hiebert,S.W., Nucifora,G., Tenen,D.G. and Zhang,D.E. (1996) Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc. Natl Acad. Sci. USA, 93, 11895–11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmono R.T., Oceandy,D., Pollard,J.W., Tong,W., Pavli,P., Wainwright,B.J., Ostrowski,M.C., Himes,S.R. and Hume,D.A. (2003) A macrophage-colony-stimulating factor receptor (CSF-1R) green fluoresecent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood, 101, 1155–1163. [DOI] [PubMed] [Google Scholar]
- Scandura J.M., Boccuni,P., Cammenga,J., Nimer,S.D. (2002) Transcription factor fusions in acute leukemia: variations on a theme. Oncogene, 21, 3422–3444. [DOI] [PubMed] [Google Scholar]
- Schoch C. et al. (2002) Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc. Natl Acad. Sci. USA, 99, 10008–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N.A. (2001) Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol., 8, 192–196. [DOI] [PubMed] [Google Scholar]
- Tagoh H., Himes,R., Clarke,D., Leenen,P.J., Riggs,A.D., Hume,D. and Bonifer,C. (2002) Transcription factor complex formation and chromatin fine structure alterations at the murine c-fms (CSF-1 receptor) locus during maturation of myeloid precursor cells. Genes Dev., 16, 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangala R.K., Heiss-Neumann,M.S., Rangatia,J.S., Singh,S.M., Schoch,C., Tenen,D.G., Hiddemann,W. and Behre,G. (2003) The myeloid master regulator transcription factor PU.1 is inactivated by AML1–ETO in t(8;21) myeloid leukemia. Blood, 101, 270–277. [DOI] [PubMed] [Google Scholar]
- Visvader J. and Verma,I.M. (1989) Differential transcription of exon 1 of the human c-fms gene in placental trophoblasts and monocytes. Mol. Cell. Biol., 9, 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hoshino,T., Redner,R.L., Kajigaya,S. and Liu,J.M. (1998) ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC complex. Proc. Natl Acad. Sci. USA, 95, 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf J.J., Yamamoto,C.M., Lenny,N., Downing,J.R., Selsted,M.E. and Hiebert,S.W. (1998) The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-α, inhibits C/EBP-α-dependent transcription and blocks granulocytic differentiation. Mol. Cell. Biol., 18, 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau D.A. et al. (1997) Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1–ETO fusion gene. Nat. Genet., 15, 303–306. [DOI] [PubMed] [Google Scholar]
- Yuan Y. et al. (2001) AML1–ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc. Natl Acad. Sci. USA, 98, 10398–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.E., Fujioka,K., Hetherington,C.J., Shapiro,L.H., Chen,H.M., Look,A.T. and Tenen,D.G. (1994) Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol. Cell. Biol., 14, 8085–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.E., Hetherington,C.J., Meyers,S., Rhoades,K.L., Larson,C.J., Chen,H.M., Hiebert,S.W. and Tenen,D.G. (1996) CCAAT enhancer-binding protein (C/EBP) and AML1 (CBFα2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol. Cell. Biol., 16, 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]