Abstract
The soxRS regulon functions in protecting Escherichia coli cells against superoxide and nitric oxide. When SoxR is activated by oxidation of its [2Fe–2S] cluster, it increases the synthesis of SoxS, which then activates its target gene expression. How the oxidized SoxR returns to and is maintained in its reduced state has been under question. To identity genes that constitute the SoxR-reducing system, we screened an E.coli mutant library carrying a chromosomal soxSp::lacZ fusion, for constitutive mutants. Mutations mapped to two loci: the rsxABCDGE operon (named for reducer of SoxR) that is highly homologous to the rnfABCDGE operon in Rhodobacter capsulatus involved in transferring electrons to nitrogenase, and the rseC gene in the rpoE–rseABC operon. In-frame deletion of each open reading frame in the rsxABCDGE operon produced a similar constitutive phenotype. The double mutation of rsx and rseC suggested that rsxABCDGE and rseC gene products act together in the same pathway in reducing SoxR. Electron paramagnetic resonance analysis of SoxR and measurement of re-reduction kinetics support the proposal that rsx and rseC gene products constitute a reducing system for SoxR.
Keywords: ferredoxin motif/iron–sulfur cluster/redox regulation/reduction of SoxR/superoxide stress
Introduction
Cells are equipped with a defense system to cope with the harmful effect of reactive oxygen species encountered during aerobic growth or environmental stress conditions. Bacteria have evolved sophisticated molecular mechanisms to sense the oxidant levels and to activate protective systems. Much progress has been made in understanding these mechanisms, especially in Escherichia coli. Escherichia coli cells utilize two transcription factors OxyR and SoxR to sense the oxidants and then induce various genes against oxidative stress that are involved in removing oxidants, repairing damaged cell components and maintaining reducing conditions in the cell (Bauer et al., 1999; Storz and Imlay, 1999; Pomposiello and Demple, 2001). Whereas OxyR responds primarily to H2O2 and nitrosylating agents, SoxR is known to respond primarily to superoxide and nitric oxide (Nunoshiba et al., 1993; Hausladen et al., 1996; Storz and Imlay, 1999).
SoxR is a 17 kDa transcriptional regulator of the MerR family (Amabile-Cuevas and Demple, 1991; Wu and Weiss, 1991). It forms a dimer in solution, with each monomer containing a [2Fe–2S] cluster (Hidalgo et al., 1995; Wu et al., 1995). For SoxR, the [2Fe–2S] cluster is not required for initial folding, or for maintaining its structure or DNA-binding activity. Instead, the [2Fe–2S] cluster undergoes reversible one-electron oxidation and reduction and thereby modulates its activity (Hidalgo and Demple, 1994; Wu et al., 1995). When the [2Fe–2S] center of SoxR is in the fully oxidized state (Fe3+–Fe3+), SoxR can activate the transcription of its only known target gene soxS. When it is reduced by one electron (Fe2+–Fe3+), the ability to activate soxS transcription is lost (Ding et al., 1996; Gaudu and Weiss, 1996). Thus, the Fe–S cluster serves as an elaborate redox-sensitive switch for SoxR activation to modulate soxS gene transcription. The mechanism of target gene activation by oxidized SoxR involves promoter distortion, being similar to that by MerR in response to Hg2+ (Ansari et al., 1995; Hidalgo and Demple, 1997) and to that by another MerR family member ZntR in response to Zn2+ (Outten et al., 1999). The oxidative activation of SoxR is distinguished from another well-known Fe–S-containing transcription factor Fnr, which contains a [4Fe–4S] cluster. Fnr loses its DNA-binding activity upon oxidation, due to disassembly of its Fe–S cluster (Lazazzera et al., 1996; Popescu et al., 1998).
It has been estimated that the [2Fe–2S] clusters in SoxR are >90% reduced during aerobic growth, as monitored by electron paramagnetic resonance (EPR) analysis of E.coli cells overproducing SoxR protein (Ding and Demple, 1997; Gaudu et al., 1997). Upon exposure of E.coli cells to paraquat, the EPR signal from the reduced [2Fe–2S] cluster in SoxR disappears rapidly, but returns within a few minutes after the withdrawal of the oxidative stress (Ding and Demple, 1997). The in vivo kinetics of the activation and inactivation of SoxR monitored by the increase and decrease in soxS mRNA level parallel the change in the oxidation and reduction state of SoxR monitored by EPR (Ding and Demple, 1997).
A question to be resolved is how the oxidized SoxR is reduced rapidly upon removal of oxidative stress condition and how the reduced state of SoxR is maintained against auto-oxidation during aerobic growth. It has been hypothesized that activation of the SoxR system by redox cycling agents might be mediated via limiting reductase activity through depletion of NADPH, a possible electron donor for the reductase (Liochev and Fridovich, 1992). Recently, an NADPH-dependent SoxR-reducing activity was isolated in E.coli, but the protein has not been characterized further (Kobayashi and Tagawa, 1999). Flavodoxins (FldAB) and ferredoxin-NADPH-oxidoreductase (Fpr), both being the components of the soxRS regulon, have been examined for SoxR-reducing activity, and turned out not to affect the level of soxS expression (Gaudu and Weiss, 1996, 2000). In this study, we explored factors involved in reducing SoxR, taking advantage of random Tn10 insertional mutagenesis, and selected mutants that constitutively express the soxS gene, excluding mutations in the soxR gene itself. We report on finding two genetic loci (rsx and rseC) whose products are necessary for maintaining the reduced state of SoxR.
Results
Tn10 insertion in rsx and rseC loci caused constitutive expression of soxSp::lacZ
To screen for genes involved in reducing SoxR, random insertional mutations were created in the chromosome of the ΔsoxRS strain of E.coli (BW829, Table I) using mini-Tn10. About 200 000 independent mutants were pooled and their genes were transduced by P1 phage into the reporter strain MS1343 containing a chromosomal copy of the soxSp::lacZ fusion gene. Constitutive mutants expressing a red color on MacConkey plates in the absence of added oxidants were selected. To exclude the possibility of second site mutations, we confirmed the mutant phenotype by re-transducing the Tn10 marker into fresh MS1343 host. From the transductants, we isolated four constitutive mutants. While soxS promoter-driven β-galactosidase activity in the wild-type is 80–100 Miller units at exponential phase (A600 = 0.5), the isolated mutants exhibited an ∼6- to 8-fold elevated basal level of soxS expression in the absence of any oxidants (Figure 1).
Table I. Bacterial strains used in this study.
| Strains | Relevant genotype | Reference |
|---|---|---|
| GC4468 | (argF-lac) 169 rpsL sup(Am) | Laboratory collection |
| MS1343 | GC4468, soxSp::lacZ, Ampr | This study |
| MC1306 | MS1343, rseB::Tn10 | This study |
| MC1392 | MS1343, rsxB::Tn10 | This study |
| MC1323 | MS1343, rsxB::Tn10 | This study |
| MC1353 | MS1343, rsxB::Tn10 | This study |
| MS11 | GC4468, ΔrsxC::kan | Koo (2001) |
| MC1393 | MS1343, ΔrsxC::kan | This study |
| MC1394 | MS1343, ΔrsxBC::kan | This study |
| BW900 | GC4468, soxR9::cat | Wu and Weiss (1991) |
| BW829 | GC4468, Δsox-8::cat | Tsaneva and Weiss (1990) |
| BW847 | GC4468, soxR4::cat | Tsaneva and Weiss (1990) |
| BM900 | MS1343, soxR9::cat | This study |
| BMC9-23 | MS1343, soxR9::cat, rsxB::Tn10 | This study |
| BMC9-06 | MS1343, soxR9::cat, rseB::Tn10 | This study |
| BM847 | MS1343, soxR4::cat (soxR4c) | This study |
| BMC847-93 | MS1343, soxR4c, ΔrsxC::kan | This study |
| CP367 | Temperature-sensitive polA mutant | Laboratory collection |
| DY330 | W3110 lacU169 gal490 cI857 (cro-bioA) | Yu et al. (2000) |
| SYPT1 | MS1343, ΔrsxP::tet | This study |
| SYAK1 | MS1343, ΔrsxA::kan | This study |
| SYBK1 | MS1343, ΔrsxB::kan | This study |
| SYCK1 | MS1343, ΔrsxC::kan | This study |
| SYDK1 | MS1343, ΔrsxD::kan | This study |
| SYGK1 | MS1343, ΔrsxG::kan | This study |
| SYET1 | MS1343, ΔrsxE::tet | This study |
| SYNT1 | MS1343, Δnth::tet | This study |
| JHRA1 | MS1343, ΔrseA nadB3140::Tn10, Tetr | This study |
| JHRB1 | MS1343, ΔrseB nadB3140::Tn10, Kanr | This study |
| JHRC1 | MS1343, rseC::Ω nadB3140::Tn10, Kanr, Chlr | This study |
| JHRC2 | GC4468, rseC::Ω nadB3140::Tn10, Kanr, Chlr | This study |
| JHRBC1 | MS1343, ΔrseBC::Ω nadB3140::Tn10, Kanr, Chlr | This study |
| JHRMC1 | MC1392, rseC::Ω nadB3140::Tn10, Kanr, Chlr, Tetr | This study |

Fig. 1. Constitutive mutants with elevated soxS expression. Deep red colonies that express the soxS gene on McConkey plates in the absence of oxidants were selected from the Tn10-insertional mutant pool in the MS1343 (soxSp::lacZ) background. The β-galactosidase activity was determined at early exponential phase and is presented in Miller units, as described in the text.
To identify the mutated genes, the regions of Tn10 insertion were cloned and sequenced. Three mutations (MC1323, MC1353 and MC1392) were all mapped at the same region, called the ydg locus, showing high homology in sequence and gene (operon) structure to the rnf (Rhodobacter nitrogen fixation) genes of Rhodobacter capsulatus (Figure 2A). We named this locus rsx (reducer of SoxR) and used the same alphabetical numbering in accordance with the rnf genes in R.capsulatus. In all three mutations, Tn10 was found inserted at the C-terminal part of the rsxB gene. One remaining mutation (MC1306) was mapped in the rseB gene of the rpoE-rseABC operon encoding a sigma factor (σE) and its regulators (Figure 2A).
Fig. 2. The gene structure of the rsx and rpoE-rse operons. (A) The map of rsxPABCDGE, nth and rpoE-rseABC genes in comparison with the rnf genes of R.capsulatus. Homologous gene pairs are indicated with the same shadings and bi-directional arrows. The sites of Tn10 insertions are indicated. (B) The [4Fe–4S] binding motifs in RsxB and RsxC. Two consecutive CX2CX2CX3CP motifs are highlighted. (C) Conserved regions among gene products of E.coli rseC (Eco RseC), R.capsulatus rnfF (Rca RnfF) and H.influenzae (Hin) P44060. The conserved cysteine residues are presented in bold.
Both RsxB and RsxC contain two ferredoxin-like motifs of C-X2-C-X2-C-X3-CP, a putative binding site of the [4Fe–4S] cluster (Figure 2B). The homologous genes in Hemophilus influenzae and R.capsulatus also contain these ferredoxin-like motifs (Kumagai et al., 1997). RsxC, which is longer by 220 amino acids than R.capsulatus RnfC, contains an NADH-binding motif that is also conserved in other RnfC homologs. The RnfABCDGE proteins are known to be required for nitrogen fixation in R.capsulatus, most probably in transferring electrons to nitrogenase as a membrane-bound complex (Schmehl et al., 1993; Jouanneau et al., 1998). The gene arrangement in the E.coli rsx locus matches well with that in R.capsulatus rnf except that E.coli lacks an rnfH homolog and instead contains a non-homologous gene rsxP preceding rsxA and nth downstream of rsxE. It has been reported previously that the nth gene encoding endonuclease III is co-transcribed with the upstream rnf homolog genes (Gifford and Wallace, 2000).
The hydrophobicity prediction suggests that RsxA, RsxD and RsxE contain >6 transmembrane domains, whereas RsxB and RsxG contain one hydrophobic domain at each N-terminus. Only RsxC lacks any hydrophobic patch. The topology of RsxA (YdgL) and RsxE (YdgQ) has been examined previously to reveal that they contain six transmembrane domains with both ends of RsxA protruding into the periplasm and both ends of RsxE protruding into the cytoplasm (Sääf et al., 1999). In R.capsulatus, RnfA has also been demonstrated to be a membrane protein with six transmembrane domains. RnfB and RnfC have been shown to locate at the periphery of the membrane, stabilizing each other (Kumagai et al., 1997). Therefore, it is most likely that Rsx proteins in E.coli form a multisubunit complex in the membrane.
The rseABC genes constitutes an operon with rpoE encoding σE, which transcribes its target genes in response to cell envelope stress. RseA, an anti-σE factor located in the inner membrane, binds and negatively regulates σE. RseB is an accessory factor that binds to RseA in the periplasmic face and negatively regulates σE. No particular role for RseC has been assigned (Missiakas and Raina, 1997; De Las Peñas et al., 1997; Ades et al., 1999). RseC is predicted to be a membrane protein with two transmembrane segments at the C-terminus. We found that rseC is homologous to the N-terminal half of the rnfF gene in the fdxC–fdxN gene cluster located upstream of the rnfA gene in divergent orientation in R.capsulatus (Figure 2A and C). They share many residues in common, including the cysteines that also appear in two hypothetical proteins (P44020 and P44060) from H.influenzae.
Effect of rsx and rse mutations is mediated via SoxR
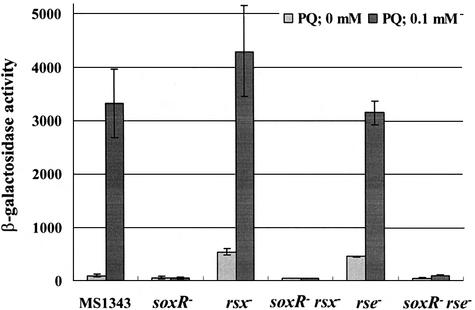
To test whether the elevated soxS expression in rsx::Tn10 and rse::Tn10 mutants in the absence of oxidative stress is mediated via SoxR, a soxR deletion allele (soxR9::cat from BM900, Table I) was introduced into MC1392 (rsx::Tn10) and MC1306 (rse::Tn10) mutants by P1 transduction. In double mutants, the level of soxS expression decreased to the low level observed in the soxR mutant (BM900) in the absence or presence of paraquat (Figure 3). This epigenetic effect of soxR mutation indicates that the rsx and rse mutations caused a constitutive phenotype through the action of SoxR. To investigate whether the effect of rsx and rse mutations is specific for the soxRS system and not for other oxidant-responsive system such as oxyR, we introduced rsx::Tn10 and rse::Tn10 alleles from MC1392 and MC1306 into the oxyS::lacZ fusion strain. The expression level of oxyS did not change by introducing these alleles, confirming that the effect of these mutations is specific for the soxRS system (data not shown).
Fig. 3. SoxR-mediated soxS activation in rsx and rse mutants. Expression of soxSp-driven β-galactosidase was determined for wild-type (MS1343) and various mutants [BM900(soxR–), MC1392(rsx–), BMC9-23(soxR– rsx–), MC1306(rse–) and BMC9-06(soxR– rse–)]. Cells were grown in LB to early exponential phase (A600 = 0.2), and then either left untreated or treated with paraquat (PQ, 0.1 mM) for 60 min, followed by β-galactosidase activity assay. The mean value from four independent experiments is presented.
Effect of each gene in rsx and rse operons on soxS expression
The rsxPABCDGE (ydgKLMNOPQ) and nth genes have been suggested to constitute an operon transcribed from two promoters immediately upstream of rsxP and rsxA (Gifford and Wallace, 2000). We confirmed the location of a promoter in front of the rsxA gene by S1 mapping. This promoter is stronger than the upstream one, whose start site we did not determine, by >5-fold as judged by S1 analysis (data not shown).
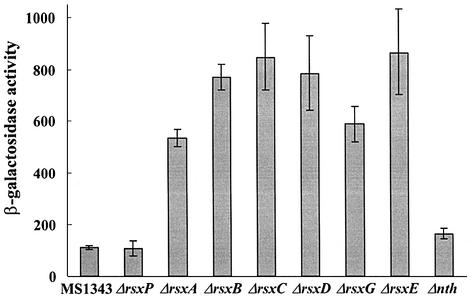
The original Tn10 insertions at rsx (MC1323, 1353 and 1392) and at rse (MC1306) loci most probably have caused polar effects on the genes located downstream of the insertion site. Therefore, we constructed in-frame deletion mutants of each gene by swapping the entire coding region from the start to stop codons with an antibiotic resistance gene in-frame, avoiding the polar effect on downstream genes. When each in-frame deletion mutant of rsxA, B, C, D, G and E was constructed in the reporter strain MS1343, we found that the mutations all enhanced soxS expression to a level comparable with that observed in the original rsx::Tn10 mutants (Figure 4). On the other hand, deletion of rsxP or nth did not cause any significant change in soxS expression. Introduction of each of the rsxA, B, C, D, G and E genes on a multicopy vector to each corresponding deletion mutant decreased soxS expression, confirming the effect of each gene (data not shown). Since six proteins are all required to maintain the low level of soxS expression, it is most likely that RsxA, B, C, D, G and E proteins might all act together in reducing SoxR, possibly as a complex, as their homologs may do in R.capsulatus in reducing nitrogenase.
Fig. 4. Effect of each ORF in the rsx operon on soxS expression. An in-frame deletion of each gene in the rsx operon was constructed in the MS1343 background to create SYPT1 (ΔrsxP::tet), SYAK1 (ΔrsxA:: kan), SYBK1 (ΔrsxB::kan), SYCK1 (ΔrsxC::kan), SYDK1 (ΔrsxD:: kan), SYGK1 (ΔrsxG::kan), SYEK1 (ΔrsxE::kan) and SYNT1 (Δnth::tet). All strains were grown to an OD600 of 0.5 and their basal β-galactosidase activities were measured. The mean value from three independent experiments is presented.
We then examined the role of each gene in the rseABC locus. When we introduced individual mutant alleles of rseA, rseB, rseC and rseBC into MS1343, only the rseC mutation (JHRC1, Table I) enhanced soxS expression (Figure 5A, lanes 1–4). Deletion of both rseB and rseC gave results similar to rseC mutation (lane 5). Since Tn10 was inserted at the rseB gene in the original mutant, the effect of mutation must be due to its polar effect downstream of the rseC gene. To confirm this, we performed a complementation experiment, introducing the wild-type rseB or rseC gene on a multicopy vector to the ΔrseBC mutant. As expected, only the wild-type rseC gene reduced soxS expression in the ΔrseBC mutant to the wild-type level. These results clearly demonstrated that RseC functions in keeping the level of soxS expression low, most probably via keeping SoxR in a reduced (inactive) state.
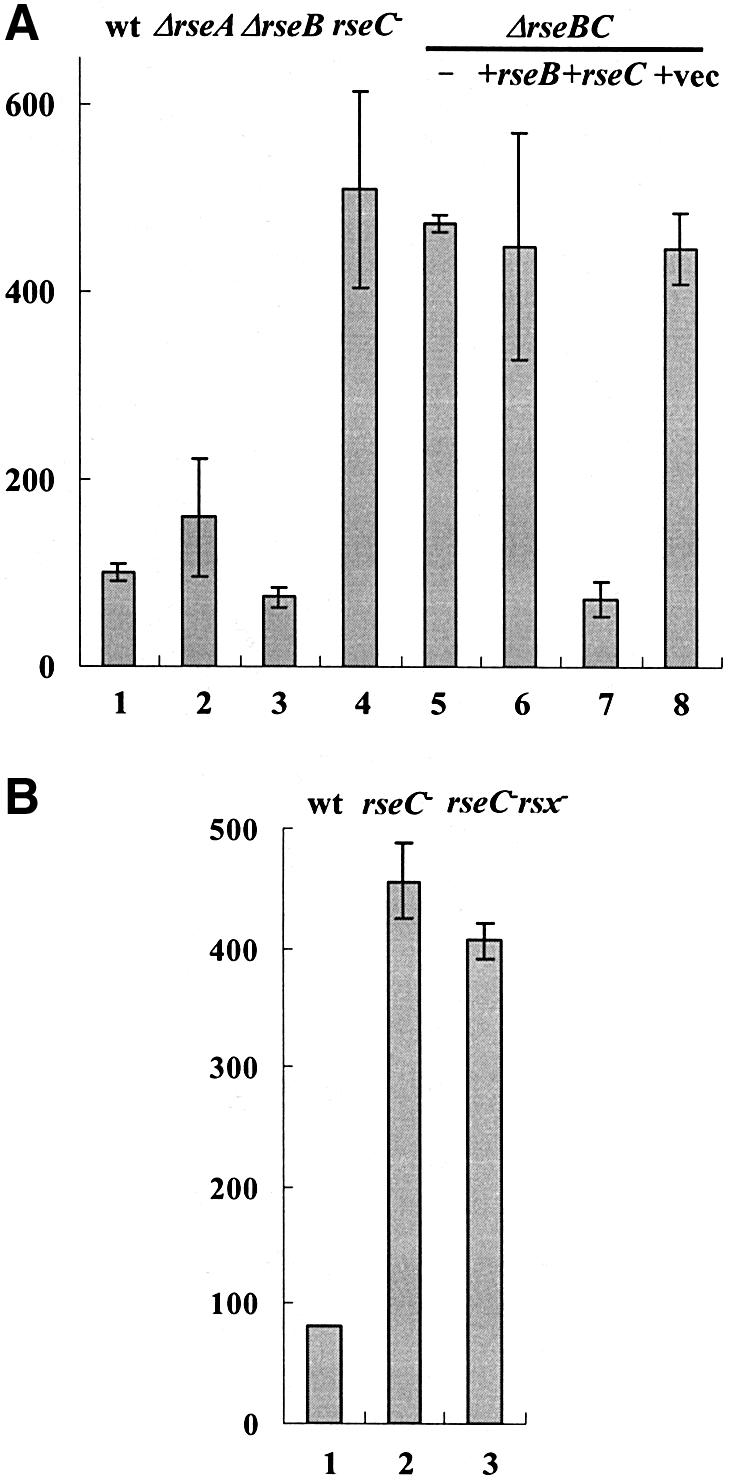
Fig. 5. Effect of each ORF in the rse operon and double mutations of rsx and rseC on soxS expression. (A) An in-frame deletion of each gene in the rse operon was constructed in the MS1343 (wt) background to create JHRA1 (ΔrseA, lane 2), JHRB1 (ΔrseB, lane 3), JHRC1 (ΔrseC–, lane 4) and JHRBC1 (ΔrseBC, lane 5) mutants. For complementation experiments, rseB and rseC genes were expressed in ΔrseBC cells on multicopy vector pTrc99A (Pharmacia) controlled by the fused trc promoter (lanes 6 and 7, respectively). Complementation with the parental pTrc99A vector was performed in parallel (lane 8). (B) The double mutant JHRMC1 (rsx– rseC–) created by transducing the original rsxB::Tn10 mutant allele of MC1392 to JHRC1 (rseC–) was measured for soxS expression along with MS1343 (wt) and JHRC1 (rseC–). The basal β-galactosidase activity was measured at OD600 = 0.5. The mean values from three independent experiments are presented.
To test whether RseC protein works together with or independently of Rsx proteins in modulating SoxR activity, we constructed a double mutant (JHRMC1, Table I) containing rsxB::Tn10 and rseC– alleles in the MS1343 background. The double mutation did not cause any additive increase in soxS expression (Figure 5B), suggesting that rseC works together with rsx gene products in the same pathway in modulating SoxR activity.
Redox state of SoxR in rsx and rseC mutants
To assess the redox state of SoxR in rsx and rseC mutants, we performed EPR spectroscopic analysis using whole cells. EPR spectroscopy has been used to analyze the redox state of the overproduced proteins containing an iron–sulfur cluster in intact cells, since it can determine the unpaired electron of the Fe–S cluster (Johnson et al., 1985), and has been applied successfully to monitor the redox state of SoxR in vivo (Ding and Demple, 1997; Gaudu et al., 1997). The [2Fe–2S] cluster of SoxR produces a characteristic EPR spectrum in its reduced form ([2Fe–2S]+), which disappears on oxidation to [2Fe–2S]2+ (Hidalgo et al., 1995). Since only a small amount of SoxR exists in wild-type cells (<100 molecules per cell), spectroscopic observation in vivo requires the overproduction of SoxR. We overproduced SoxR protein in wild-type and rsxC or rseC mutants, and confirmed that similar amounts of SoxR were present in the soluble fraction, as judged by SDS–PAGE. The X-band EPR spectra from these cells were recorded at 96 K as described in Materials and methods.
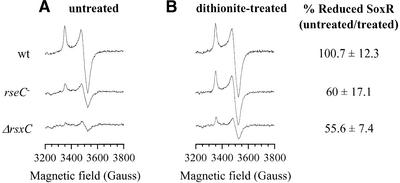
SoxR in wild-type cells demonstrated its characteristic spectrum as a reduced form (Figure 6A). The intensity of the EPR signal was significantly decreased in rseC and more in rsxC mutants. Since only the reduced form of the [2Fe–2S] cluster produces the EPR signal, the data demonstrated clearly that the amount of reduced SoxR decreased significantly in rsxC and rseC mutants compared with the wild-type. When we used sodium dithionite, a strong reductant, to convert all the [2Fe–2S] clusters in SoxR to the reduced form, the EPR signal from the wild-type sample did not change significantly, whereas those from mutant samples increased significantly (Figure 6B). Assuming that the integrated value of the EPR signal from the dithionite-treated sample represents total [2Fe–2S] clusters of SoxR in each strain, we calculated the percentage of reduced SoxR in each cell type by taking the ratio of signal values from untreated versus dithionite-treated samples. The data indicate that only ∼60 and 56% of the [2Fe–2S] clusters in overproduced SoxR exist as reduced forms in rseC and rsxC mutants, respectively, whereas nearly all SoxR are reduced in the wild-type. The signal intensity of the fully reduced (dithionite-treated) SoxR in mutant cells was lower than that of the wild type. Normalizing the integrated values to the same amount of SoxR polypeptides overproduced, the mutant signals corresponded to ∼50 and 30% of the wild-type level in rseC and rsxC mutants, respectively. We interpret these data to suggest that the [2Fe–2S] cluster in SoxR might be unstable in the mutants, with a more drastic effect in the rsxC mutant. This is in accordance with a previous observation that some SoxRc mutant proteins that exist in more oxidized forms tend to lose iron more easily (Gaudu et al., 1997). Overall, the EPR analysis demonstrated clearly that in rsx and rseC mutants, SoxR exists more in the oxidized form, agreeing with the prediction from the genetic data that the elevated soxS expression in the mutants is due to the activated (oxidized) SoxR. Thus we propose that the rsxABCDGE and rseC gene products are required in maintaining the reduced state of SoxR.
Fig. 6. EPR analysis of SoxR overproduced in rsx and rseC mutant cells. Wild-type (GC4468), MS11(ΔrsxC::kan) or JHRC2 (rseC–) cells harboring pTac1-SoxR were grown to OD600 = 0.5 and induced with 1 mM IPTG for 2 h at 25°C. Cells were harvested and resuspended to an equal cell density in 50 mM MOPS pH 7.6, 0.2 M KCl and 0.2 M LiCl2. The concentrated cell suspension was then transferred directly to an EPR tube pre-cooled in liquid nitrogen. Sodium dithionite was mixed immediately before EPR measurement, as described in the text. The EPR signals were obtained at 96 K. The signal from cells containing control plasmid (pTac1) was subtracted from each measurement. The percentage of reduced SoxR in each cell type was determined by double integration of the signals, and taking the ratio of values from untreated versus dithionite-treated samples. Average values from three independent experiments are presented with standard deviations.
Effect of rsx and rse mutations on re-reduction of SoxR after oxidative stress
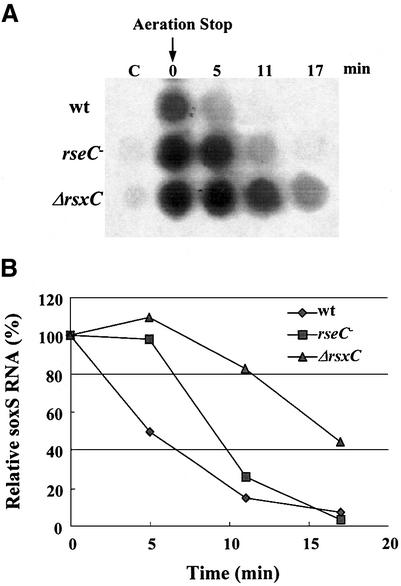
The [2Fe–2S] cluster of SoxR is oxidized rapidly on exposure to redox-cycling agents when the O2 supply is sufficient. Limiting the O2 supply by stopping aeration leads to rapid re-reduction of SoxR, allowing it to return to its inactive (reduced) state within 10 min as monitored by EPR spectroscopy (Ding and Demple, 1997). The rapid change in SoxR activity can be monitored by measuring the level of soxS mRNA due to its very short half-life in vivo. If RsxABCDGE and RseC function in reducing SoxR, mutations in rsx or rseC will retard the rate of re-reduction of SoxR, causing the elevated soxS mRNA level to persist for longer. We monitored the decay rate of soxS mRNA after stopping aeration in the presence of paraquat in wild-type, rsxC and rseC mutants. The soxS mRNA decreased to ∼15% of the level within 10 min in the wild-type cell. In rsxC or rseC mutants, the soxS mRNA decreased more slowly than the wild-type (Figure 7), consistent with the proposal that Rsx and RseC proteins function in reducing SoxR.
Fig. 7. Re-reduction kinetics of SoxR monitored by the change in soxS mRNA level. (A) Wild-type, JHRC1 (rseC–) and MC1393 (ΔrsxC::kan) cells were treated with 0.1 mM paraquat at OD600 = 0.2 for 30 min with vigorous shaking to activate SoxR to its full level. Activation was interrupted by limiting the O2 supply by stopping shaking. Cells were removed at 0, 5, 11 and 17 min after the shaker stopped. Total RNA was prepared and analyzed by dot-blot hybridization using a specific probe for soxS mRNA. (B) The intensities of the hybridized dots were quantified by autoradiographic image analyzer (Fuji, FLA-2000) and plotted relative to the value (100%) at the 0 time point.
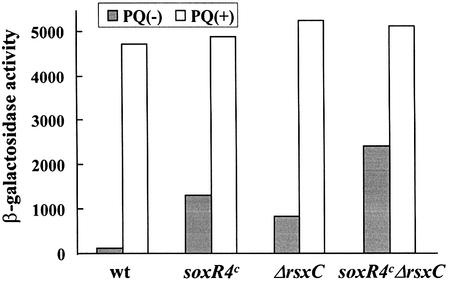
Additive effect of rsx and soxR4c constitutive mutations
Several constitutive mutants of SoxR, whose C-termini just downstream of the [2Fe–2S] site are truncated, have been reported (Nunoshiba and Demple, 1994; Gaudu and Weiss, 1996). One of those mutants, soxR4c (soxR4::cat), whose C-terminal 19 amino acids were replaced with 49 residues from the cat cassette, was examined in conjunction with the rsx mutant. The level of soxS expression in the soxR4c mutant was slightly higher than but comparable with the level observed in rsxC mutants (Figure 8). In the soxR4cΔrsxC double mutant, the basal level of soxS expression was elevated further to about half the level of full induction, exhibiting an additive effect of rsx and soxR4c mutation. These results suggest that the mechanism for the increased oxidation of SoxR in the Δrsx mutant is different from that in the soxR4c mutant.
Fig. 8. Additive effect of soxRc and rsx constitutive mutations. The soxR constitutive mutant allele from BW847 was transduced into MS1343 (wt), resulting in BM847 (soxR4c). BM847 was transduced further with the ΔrsxC allele from MC1393, resulting in a soxR4cΔrsxC double mutant (BMC847-93). MS1343 (wt), BM847, MC1393 and BMC847-93 cells were grown to an OD600 = 0.2, treated or not with paraquat (0.1 mM) for 1 h and assayed for β-galactosidase activity.
Discussion
The redox potential of SoxR at pH 7.6 has been estimated to be about –285 mV (Ding et al., 1996; Gaudu and Weiss, 1996). More than 40% and up to 95% of SoxR exists as a form containing reduced [2Fe–2S] during aerobic growth (Hidalgo et al., 1997). Since the cytoplasmic redox potential of E.coli has been estimated to be in the range of –260 to –280 mV (Gilbert, 1990; Hwang et al., 1995), it is very likely that SoxR in vivo is actively maintained in the reduced state, implying the presence of a specific reducing system for [2Fe–2S] of SoxR. In this work, we have presented evidence that rsx and rseC gene products constitute the components of a reducing system for SoxR.
The estimated redox potential of –340 mV for NAD(P)H/ NAD(P)+ suggests that electrons may be transferred from NAD(P)H to SoxR in the reducing system. The structural similarity between the E.coli rsxABCDGE and R.capsulatus rnfABCDGE gene products supports the prediction that the Rsx proteins are likely to convey electron transfer from NAD(P)H to SoxR as Rnf proteins might do in mediating electrons from NADH to nitrogenase in R.capsulatus (Schmehl et al., 1993; Kumagai et al., 1997). Unlike E.coli, R.capsulatus is able to fix dinitrogen, which is a process requiring high energy and high potential electrons. RnfB of R.capsulatus contains potential binding sites for a [2Fe–2S] and two [4Fe–4S] clusters, whereas RnfC has potential binding sites for two [4Fe–4S] clusters, NAD(H) and FMN (Kumagai et al., 1997). Considering the similarity of RsxB and RsxC to RnfB and C, it is very likely that RsxB and C may function as the core of the reductase system, taking part in transferring electrons to SoxR, instead of nitrogenase which is absent in E.coli. The purified RsxC exhibits NADPH-dependent cytochrome c reducing activity in vitro even in the absence of SoxR, implying that it can transfer electrons from NADPH (M.-S.Koo and J.-H.Lee, unpublished results). Whether RsxB and RsxC function as the core components of the reductase for SoxR, forming a complex with other Rsx proteins, and whether it consumes NADPH or NADH need to be elucidated in the future.
Recently, Kobayashi and Tagawa (1999) reported that E.coli cells overproducing SoxR have SoxR-enhanced cytochrome c reductase activity in the presence of NADPH, and they isolated the responsible protein from the cytoplasm. Since the rate of cytochrome c reduction by the protein was enhanced ∼7- to 10-fold when SoxR was added, the protein was inferred to be SoxR reductase. Interestingly, the molecular weight of the protein is similar to that of RsxC. However, the identity of the protein is uncertain since its coding gene has not been characterized. Our result does not exclude the possibility that there still remain reducing systems other than the Rsx–RseC system. In the absence of Rsx and RseC proteins, a back-up system may operate to reduce SoxR, as hinted at by the slow return of the elevated soxS mRNA to the uninduced level in the rsx and rseC mutants.
The level of constitutive soxS expression in rsx and rseC mutants was lower than the fully induced level, being ∼15–20% of the full induction. The redox potential of SoxR is comparable with or slightly more negative than that of the cytoplasm. Thus in the absence of the reduction system, SoxR will exist as a partially but not fully oxidized form. In light of this, it is understandable that the basal constitutive level of SoxS expression in the mutants reflects this partially oxidized (activated) state of SoxR. Under condition of excess superoxide generation, all SoxR will be converted into the completely oxidized state, and thus it makes sense that paraquat treatment further increases the soxS expression in the mutant to the maximum level comparable with the full induction level in the wild-type (Figures 3 and 8). The observation that the double mutation of rsx and soxR4c increased soxS expression additively (to ∼50% of full induction) leads us to exclude the possibility that the C-terminally modified soxR4c mutation caused hindrance of reduction, by limiting access to the reducing system composed of Rsx proteins (Nunoshiba and Demple, 1994; Gaudu and Weiss, 1996). The mechanism for SoxR activation by the soxR4c mutation could include a shift in redox equilibrium to a more oxidized form or a conformational change to an active form. If there exist other reducing path(s) for SoxR separate from the Rsx system, it is also possible that the soxR4c mutation could hinder the reduction mediated not by Rsx but by other system(s). Whether other soxRc mutants would behave similarly to soxR4c in terms of interaction with the Rsx system remains an intriguing question to be addressed. The soxRc mutations, whose effect is not additive with that of rsx, might reveal a possible mechanism for the Rsx system to convey electrons to the oxidized SoxR.
Although RsxB and RsxC may play a central role as the core electron mediators in reducing SoxR, we propose that they are part of the reductase complex in the cytoplasmic membrane composed of several Rsx proteins plus RseC. The membrane localization of RsxA, RsxE and RseC has been demonstrated previously (Missiakas et al., 1997; Sääf et al., 1999). We observed the distrubution of RsxB, C and G in the membrane fraction, as predicted (J.-H.Lee and S.-Y.Rah, in preparation). As an analogy to the observation that the membrane-bound RnfB and C proteins interact with each other (Kumagai et al., 1997; Jeong and Jouaneau, 2000), we hypothesize that Rsx proteins might also form a membrane-bound complex. The finding that rseC and rsx mutations are not additive, and that rseC is homologous to rnfF in R.capsulatus that is required for nitrogen fixation (Schmehl et al., 1993), led us to propose that they may act in the same electron transfer pathway and probably function within the same membrane-bound complex. Whether any of the components in the reducing complex interacts with SoxR and whether RseC interacts with any of the Rsx proteins need to be elucidated in the future.
Materials and methods
Bacterial strains and culture conditions
Bacterial strains used in this study are listed in Table I. Cells were grown at 37°C in Luria–Bertani (LB) broth with vigorous aeration. Appropriate antibiotics were added at 100 µg/ml (ampicillin), 34 µg/ml (chloramphenicol), 50 µg/ml (kanamycin) and 20 µg/ml (tetracycline). Isopropyl-β-d-thiogalactopyranoside (IPTG) and paraquat were added at concentrations described in the text.
Construction of soxSp::lacZ reporter strain
A DNA fragment containing the soxS promoter (from nucleotide –135 to +65 relative to the transcription start site) was amplified by PCR using the primer pair SoxSF (TACTTTCATAGAATTCCAGCGCCGAT, EcoRI site underlined) and SoxSR (TATTCTAGGAGGATCCAAAAGACTAC, BamHI site underlined). The fragment was cloned into the EcoRI–BamHI site of pRS415 (Simons et al., 1987) to direct the expression of lacZ under the control of the soxS promoter (soxSp). The recombinant pRS415 containing the soxSp::lacZ fusion gene was transformed into the wild-type E.coli GC4468. It was then infected with phage λRZ5, allowing homologous recombination with the plasmid to occur in vivo. The recombinant phages were then allowed to lysogenize GC4468 cells via insertion through the att site in the chromosome. Single copy lysogens were screened by pale blue color of the colonies on X-Gal plates containing ampicillin, and selected on the basis of the lowest basal level of β-galactosidase activity. The resulting strain (MS1343) containing the soxSp::lacZ fusion gene on the chromosome was isolated and used as the wild-type reporter strain for the rest of this study.
β-galactosidase assay
Cells were grown in LB medium to an optical density of 0.2 at 600 nm and then either left untreated or treated with various concentrations of paraquat for 0.5–1 h at 37°C. β-galactosidase activity was assayed by adding o-nitrophenyl-β-d-galactopyranoside (ONPG) after permeabilization of the cell with SDS–chloroform (Miller, 1972).
Random insertional mutagenesis by mini-Tn10
A random mutation library was constructed with mini-Tn10 as described by Kleckner et al. (1991). To avoid the mutation in the soxRS locus itself, we constructed the mutant library in the ΔsoxRS strain (BW829, Δsox-8::cat). BW829 cells were infected with λNK1323 carrying the mini-Tn10. Mutant colonies with Tn10 insertion were selected on plates containing tetracycline and chloramphenicol. They were pooled and their genes were transduced into MS1343 using the P1vir transduction system (Sternberg and Mauer, 1991). Transductants that exhibit constitutive phenotype for soxS expression, forming blue or red colonies on X-Gal or MacConkey plates in the absence of any oxidants, were isolated. The mutated locus was cloned and identified by Southern hybridization using a Tn10-specific probe. The precise location of the Tn10 insertion was determined by nucleotide sequencing.
Sequence analysis of the rsx locus
Nucleotide sequences encompassing the rsxPABCDGE and nth genes (DDBJ/EMBL/GenBank accession No. AE000258) were compared with the public database using BLAST and CLUSTAL W programs. The localization and topology of RsxABCDGE and RseC proteins were predicted using the SOSUI (http://sosui.proteome.bio.tuat.ac.jp/sosuimenu0.html) and TMHMM programs (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Construction of deletion mutants
Two methods were used to construct deletion mutants: plasmid integration/resolution (Koh and Roe, 1995) and open reading frame (ORF) replacement with linear DNA (Yu et al., 2000). MC1393 (ΔrsxC::kan), MC1394 (ΔrsxBC::kan) and SYBK1 (ΔrsxB) strains were constructed by plasmid integration and resolution. The NruI–NheI internal fragment of rsxC and the NarI–NheI internal fragment of rsxBC were replaced with a Kanr cassette from pUC4-KIXX (Pharmacia) to generate MC1393 and MC1394, respectively. To generate SYBK1 (ÄrsxB), an internal 411 bp between two NruI sites in rsxB was deleted in-frame, with a Kanr block inserted at the SspI site upstream of the rsxP promoter to provide the selection marker (Koo, 2001; Rah, 2002). Co-integrates were generated in the CP367 (polAts) strain, and the desired mutants were selected as described in Koh and Roe (1995). The mutated gene was then transferred into the wild-type strain GC4468 or MS1343 by P1 transduction. The constructs were confirmed by Southern hybridization and PCR.
In-frame ORF replacement was performed as described by Yu et al. (2000) to create SYPT1 (ΔrsxP::tet), SYAK1 (ΔrsxA::kan), SYCK1 (ΔrsxC::kan), SYDK1 (ΔrsxD::kan), SYGK1 (ΔrsxG::kan), SYET1 (ΔrsxE::tet) and SYNT1 (Δnth::tet) mutants (Rah, 2002). Either the Kanr or the Tetr cassette was amplified by PCR with a 50 nucleotide flanking sequence of the target gene. The start and stop codons of the target gene were replaced precisely with the start and stop codons of the Kanr or Tetr genes. Amplified linear DNA was introduced by electroporation into DY330, which contains the exo, bet and gam genes of λ phage regulated by the λcI repressor to enhance recombination frequency of the linear DNA. The gene replacement by homologous recombination was selected on antibiotic plates and confirmed by PCR and Southern hybridization. The mutated alleles were transferred to GC4468 or MS1343 strains by P1 transduction.
The rse mutants (JHRA1, JHRB1, JHRC1 and JHRBC1) were created by P1 transduction of mutant alleles from CAG22977(ΔrseA), CAG22974(ΔrseB), CAG22682 (rseC–) and CAG22681 (ΔrseBC) into MS1343 (De Las Peñas et al., 1997).
Electron paramagnetic resonance (EPR) spectroscopy of SoxR
Wild-type (GC4468), MS11(ΔrsxC::kan) and JHRC2 (rseC–) cells containing pTac1 vector or pTac1-SoxR, which overproduces SoxR under the control of the tac promoter (Koo, 2001), were grown to OD600 of 0.5 in LB containing ampicillin (100 µg/ml). SoxR expression was induced with 1.0 mM IPTG for 2 h at 25°C. Cells from 200 ml of culture were harvested at 4°C and resuspended in 7 ml of ice-cooled EPR buffer (50 mM MOPS pH 7.6, 0.2 M LiCl2 and 0.2 M KCl). Following measurement of OD600 of the whole cell resuspension, cells were re-harvested at 4°C and suspended in 0.7–1 ml of EPR buffer, aiming to produce equal cell density among samples. Aliquots of 0.3 ml of cell suspension were then transferred to a pre-cooled EPR tube, and X-band EPR spectra were recorded at 96 K on a Bruker model EMX spectrometer (Bruker, Germany), equipped with a continuous flow N2 temperature controller (Bruker, model BVT3000). To obtain EPR spectra of fully reduced SoxR in cells, the concentrated whole-cell suspensions were treated with 210 mM (final concentration) sodium dithionite. A 1 M solution of sodium dithionite was prepared anaerobically in 50 mM MOPS buffer pH 7.6, and aliquots of 80 µl were transferred to EPR tubes and mixed with cell suspension immediately before spectral measurements. The expression level of SoxR in the soluble fraction of cells subjected to EPR analysis was confirmed on SDS–PAGE in a parallel experiment. A nearly equal amount of SoxR polypeptide was overproduced reproducibly in the soluble fraction to ∼5% of the total protein at maximum in wild-type and the mutants. The spectrum from cells containing control vector was subtracted from the data to obtain SoxR-specific signals. Typical EPR parameters used were as follows: temperature of 96 K, microwave frequency of 9.449 GHz, microwave power of 10 mW, modulation frequency of 100 kHz and modulation amplitude of 1 mT.
Isolation and dot-blot analysis of soxS mRNA
To induce soxS mRNA, GC4468 cells were treated with 0.1 mM paraquat for 30 min at an OD600 of 0.2, under vigorous shaking. Shaking was then stopped to limit aeration, and cells were taken at various time points after the shaker stopped. Total RNAs were prepared using the Ultraspec™-II total RNA isolation kit (Biotecx Laboratories, Inc.) as recommended by the manufacturer, except that cells were pre-treated with lysozyme (4 mg/ml) in 50 mM glucose, 25 mM Tris–HCl pH 8.0 and 10 mM EDTA for 2–5 min on ice. RNA (30 µg) was loaded on a Hybond-N+ membrane (Amersham Pharmacia Biotech) equilibrated with 10× SSC by applying a vacuum in a dot-blotting apparatus (Hoefer) and immobilized by UV cross-linking. Hybridization was performed with a soxS-specific probe labeled by random priming with [α-32P]dATP (Sambrook et al., 1989).
Acknowledgments
Acknowledgements
We are grateful to Drs B.Weiss, B.Demple and C.A.Gross for their kind donation of strains. This work was supported by a research grant (R03-2001-00047) from the Korea Science and Engineering Foundation to J.-H.R. J.-H.L. was the recipient of a BK21 fellowship for post-doctoral fellows from the Ministry of Education and Human Resources.
References
- Ades S.E., Connolly,L.E., Alba,B.M. and Gross,C.A. (1999) The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-α factor. Genes Dev., 13, 2449–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile-Cuevas C.F. and Demple,B. (1991) Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res., 19, 4479–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A.Z., Bradner,J.E. and O’Halloran,T.V. (1995) DNA-bend modulation in a repressor-to-activator switching mechanism. Nature, 374, 371–375. [DOI] [PubMed] [Google Scholar]
- Bauer C.E., Elsen,S. and Bird,T.H. (1999) Mechanisms for redox control of gene expression. Annu. Rev. Microbiol., 53, 495–523. [DOI] [PubMed] [Google Scholar]
- De Las Peñas A., Connolly,L. and Gross,C.A. (1997) The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol. Microbiol., 24, 373–385. [DOI] [PubMed] [Google Scholar]
- Ding H. and Demple,B. (1997) In vivo kinetics of a redox-regulated transcriptional switch. Proc. Natl Acad. Sci. USA, 94, 8445–8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., Hidalgo,E. and Demple,B. (1996) The redox state of the [2Fe–2S] clusters in SoxR protein regulates its activity as a transcription factor. J. Biol. Chem., 271, 33173–33175. [DOI] [PubMed] [Google Scholar]
- Gaudu P. and Weiss,B. (1996) SoxR, a [2Fe–2S] transcription factor, is active only in its oxidized form. Proc. Natl Acad. Sci. USA, 93, 10094–10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudu P. and Weiss,B. (2000) Flavodoxin mutants of Escherichia coli K-12. J. Bacteriol.. 182, 1788–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudu P., Moon,N. and Weiss,B. (1997) Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe–S centers of SoxR in vivo. J. Biol. Chem., 272, 5082–5086. [DOI] [PubMed] [Google Scholar]
- Gifford C.M. and Wallace,S.S. (2000) The genes encoding endonuclease VIII and endonuclease III in Escherichia coli are transcribed as the terminal genes in operons. Nucleic Acids Res., 28, 762–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert H.F. (1990) Molecular and cellular aspects of thiol–disulfide exchange. Adv. Enzymol. Relat. Areas Mol. Biol., 63, 69–172. [DOI] [PubMed] [Google Scholar]
- Hausladen A., Privalle,C.T., Keng,T., DeAngelo,J. and Stamler,J.S. (1996) Nitrosative stress: activation of the transcription factor OxyR. Cell, 86, 719–729. [DOI] [PubMed] [Google Scholar]
- Hidalgo E. and Demple,B. (1994) An iron–sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J., 13, 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo E. and Demple,B. (1997) Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J., 16, 1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo E., Bollinger,J.M.,Jr, Bradley,T.M., Walsh,C.T. and Demple,B. (1995) Binuclear [2Fe–2S] clusters in the Escherichia coli SoxR protein and role of the metals in transcription. J. Biol. Chem. 270, 20908–20914. [DOI] [PubMed] [Google Scholar]
- Hidalgo E., Ding,H. and Demple,B. (1997) Redox signal transduction: mutations shifting [2Fe–2S] centers of the SoxR sensor-regulator to the oxidized form. Cell, 88, 121–129. [DOI] [PubMed] [Google Scholar]
- Hwang C., Lodish,H.F. and Sinskey,A.J. (1995) Measurement of glutathione redox state in cytosol and secretory pathway of cultured cells. Methods Enzymol., 251, 212–221. [DOI] [PubMed] [Google Scholar]
- Johnson M.K., Morningstar,J.E., Cecchini,G. and Ackrell,B.A. (1985) In vivo detection of a three iron cluster in fumarate reductase from Escherichia coli. Biochem. Biophys. Res. Commun., 16, 131–136. [DOI] [PubMed] [Google Scholar]
- Jeong H.S. and Jouanneau,Y. (2000) Enhanced nitrogenase activity in strains of Rhodobacter capsulatus that overexpress the rnf genes. J. Bacteriol., 182, 1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau Y., Jeong,H.-S., Hugo,N., Meyer,C. and Willison,J.C. (1998) Overexpression in Escherichia coli of the genes from Rhodobacter capsulatus. Characterization of two-membrane-bound iron–sulfur protein. Eur. J. Biochem., 251, 54–64. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Benger,J. and Gottesman,S. (1991) Uses of transposons with emphasis on Tn10. Methods Enzymol., 204, 139–181. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. and Tagawa,S. (1999) Isolation of reductase for SoxR that governs an oxidative response regulon from Escherichia coli. FEBS Lett., 451, 227–230. [DOI] [PubMed] [Google Scholar]
- Koh Y.S. and Roe,J.H. (1995) Isolation of a novel paraquat-inducible (pqi) gene regulated by the soxRS locus in Escherichia coli. J. Bacteriol., 177, 2673–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo M.S. (2001) Characterization of protein factor regulating superoxide sensor SoxR in Escherichia coli. PhD Thesis, Seoul National University, Seoul, Korea.
- Kumagai H., Fujiwara,T., Matsubara,H. and Saeki,K. (1997) Membrane localization, topology and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry, 36, 5509–5521. [DOI] [PubMed] [Google Scholar]
- Lazazzara B.A., Beinert,H., Khoroshilova,N., Kennedy,M.C. and Kiley,P.J. (1996) DNA binding and dimerization of the Fe–S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem., 271, 2762–2768. [DOI] [PubMed] [Google Scholar]
- Liochev S.I. and Fridovich,I. (1992) Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc. Natl Acad. Sci. USA, 89, 5892–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Missiakas D. and Raina,S. (1997) Protein folding in the bacterial periplasm. J. Bacteriol., 179, 2465–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiakas D., Mayer,M.P., Lemaire,M., Georgopoulos,C. and Raina,S. (1997) Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol., 24, 55–71. [DOI] [PubMed] [Google Scholar]
- Nunoshiba T. and Demple,B. (1994) A cluster of constitutive mutations affecting the C-terminus of the redox-sensitive SoxR transcriptional activator. Nucleic Acids Res., 22, 2958–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunoshiba T., de Rojas-Walker,T., Wishnok,J.S., Tannenbaum,S.R. and Demple,B. (1993) Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc. Natl Acad. Sci. USA, 90, 9993–9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten C.E., Outten,F.W. and O’Halloran,T.V. (1999) DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli.J. Biol. Chem., 274, 37517–37524. [DOI] [PubMed] [Google Scholar]
- Pomposiello P.J. and Demple,B. (2001) Redox-operated genetic switches; the SoxR and OxyR transcription factors. Trends Biotechnol., 19, 109–114. [DOI] [PubMed] [Google Scholar]
- Popescu C.V., Bates,D.M., Beinert,H., Munck,E. and Kiley,P.J. (1998) Mossbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 13431–13435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rah S.Y. (2002) Characterization of resPABCDGE operon regulating SoxR in Escherichia coli. MSc. Thesis, Seoul National University, Seoul, Korea.
- Sääf A., Johansson,M., Wallin,E. and von Heijne,G. (1999) Divergent evolution of membrane protein topology: the Escherichia coli RnfA and RnfE homologues. Proc. Natl Acad. Sci. USA, 96, 8540–8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schmehl M., Jahn,A., Meyer zu Vilsendorf,A., Hennecke,S., Masepohl,B., Schuppler,M., Marxer,M., Oelze,J. and Klipp,W. (1993) Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet., 241, 602–615. [DOI] [PubMed] [Google Scholar]
- Simons R.W., Houman,F. and Kleckner,N. (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene, 53, 85–96. [DOI] [PubMed] [Google Scholar]
- Sternberg N.L. and Mauer,R. (1991) Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol., 204, 18–43. [DOI] [PubMed] [Google Scholar]
- Storz G. and Imlay,J.A. (1999) Oxidative stress. Curr. Opin. Microbiol., 2, 188–194. [DOI] [PubMed] [Google Scholar]
- Tsaneva I.R. and Weiss,B. (1990) soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol., 172, 4197–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. and Weiss,B. (1991) Two divergently transcribed genes, soxR and soxS control a superoxide response regulon of Escherichia coli. J. Bacteriol., 173, 2864–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Dunham,W.R. and Weiss,B. (1995) Overproduction and physical characterization of SoxR, a [2Fe–2S] protein that governs an oxidative response regulon in Escherichia coli.J. Biol. Chem., 270, 10323–10327. [DOI] [PubMed] [Google Scholar]
- Yu D., Ellis,M.H., Lee,E.-C., Jenkins,N.A., Copeland,N.G. and Court,D.L. (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA, 97, 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]