Abstract
Eukaryotic RNA polymerase II transcribes precursors of mRNAs and of non-protein-coding RNAs such as snRNAs and snoRNAs. These RNAs have to be processed at their 3′ ends to be functional. mRNAs are matured by cleavage and polyadenylation that require a well-characterized protein complex. Small RNAs are also subject to 3′ end cleavage but are not polyadenylated. Here we show that two newly identified proteins, Pti1p and Ref2p, although they were found associated with the pre-mRNA 3′ end processing complex, are essential for yeast snoRNA 3′ end maturation. We also provide evidence that Pti1p probably acts by uncoupling cleavage and polyadenylation, and functions in coordination with the Nrd1p-dependent pathway for 3′ end formation of non-polyadenylated transcripts.
Keywords: cleavage and polyadenylation uncoupling/Pti1p/Ref2p/snoRNA 3′ end formation/yeast
Introduction
mRNA 3′ end formation is achieved in two coupled reactions; cleavage followed by polyadenylation of the precursor (for a recent review see Edmonds, 2002). A large protein complex comprising the cleavage and polyadenylation factors CF IA (cleavage factor IA), CPF (cleavage and polyadenylation factor), Nab4p and Nab2p has been identified which can recapitulate the processing reaction in vitro on synthetic pre-mRNA substrates (Minvielle-Sebastia et al., 1994, 1998; Kessler et al., 1996, 1997; Ohnacker et al., 2000; Hector et al., 2002). CF IA is a tetrameric factor that consists of Rna14p, Rna15p, Pcf11p and Clp1p (Minvielle-Sebastia et al., 1997; and references therein). CPF contains multiple subunits ranging in size from 150 to 20 kDa. The first definition of this factor (Ohnacker et al., 2000) assigned the nine following polypeptides to it: Yhh1p/Cft1p (150 kDa; Stumpf and Domdey, 1996; Preker et al., 1997), Ydh1p/Cft2p (105 kDa; Preker et al., 1997; Zhao et al., 1997), Ysh1p/Brr5p (100 kDa; Chanfreau et al., 1996; Jenny et al., 1996; Preker et al., 1997), Pta1p (90 kDa; Preker et al., 1997), Pap1p (64 kDa; Lingner et al., 1991; Preker et al., 1997), Mpe1p [58 kDa; Vo et al., 2001; note that this polypeptide was referred to originally as Pfs1p in Preker et al. (1997) and Ohnacker et al. (2000)], Pfs2p (53 kDa; Preker et al., 1997; Ohnacker et al., 2000), Fip1p (50 kDa; Preker et al., 1995) and Yth1p (26 kDa; Barabino et al., 1997). Recently, pre-mRNA 3′ end processing could be reconstituted in vitro with CF IA and CPF purified from protein A-tagged subunits, the latter containing six additional polypeptides compared with previous descriptions of the factor. They were identified as Swd2p, Glc7p, Ssu72p, Pti1p, Ref2p and a 20 kDa protein encoded by the YDL094c open reading frame (ORF) (Dichtl et al., 2002). Among them, only Ref2p has been suggested to be directly involved in pre-mRNA 3′ end processing (hence its name, RNA end formation; Russnak et al., 1995).
Yeast small nucleolar RNAs (snoRNAs) are also subject to 3′ end cleavage to provide an entry site for exonucleases that produce mature 3′ ends by exonucleolytic trimming (Chanfreau et al., 1998a,b; Allmang et al., 1999; van Hoof et al., 2000; Perumal and Reddy, 2002). In some instances, 3′ entry sites are generated by recognition of a specific stem structure by the RNA endonuclease Rnt1p (Chanfreau et al., 1998a,b). However, many mono- and polycistronic snoRNAs do not bear a Rnt1p-specific signal in their 3′ region. Recently, some cis-acting elements and trans-acting factors have been identified in yeast that participate in 3′ end formation of at least some snRNAs and snoRNAs. The Nrd1 protein, which interacts with the C-terminal domain (CTD) of RNA polymerase II (pol II), the RNA-binding protein Nab3p, the CTD kinase Ctk1p, and Sen1p have all been shown to function in snRNA and snoRNA 3′ end maturation (Conrad et al., 2000; Steinmetz et al., 2001). More surprisingly, mutations in RNA14 and RNA15 genes that code for CF IA subunits (Minvielle-Sebastia et al., 1994) have been described to inhibit poly(A)-independent 3′ end processing of U2 and U5 pre-snRNAs and also of different box C/D and box H/ACA pre-snoRNAs (Fatica et al., 2000; Morlando et al., 2002). However, the CF IA-complementary factor CPF (Barabino et al., 2000; Ohnacker et al., 2000; Dichtl and Keller, 2001) has been proposed not to participate in this processing since none of the CPF mutants tested so far were affected in small RNA 3′ end processing (Dichtl et al., 2002; Morlando et al., 2002). Together, these results suggested that CF IA might work in combination with a set of proteins distinct from CPF, for instance the Nrd1p-associated polypeptides, to achieve poly(A)-independent 3′ end formation of sn(o)RNAs (Steinmetz et al., 2001; Morlando et al., 2002). However, we show here that two newly discovered CPF-associated proteins, Pti1p and Ref2p, are involved in cleavage-dependent/polyadenylation-independent 3′ end formation of snoRNAs, therefore demonstrating that most of the pre-mRNA 3′ end processing machinery participates in independently transcribed snoRNA 3′ end formation.
Results
PTI1 is a multicopy suppressor of a pcf11 temperature-sensitive mutant
The four components of CF IA are encoded by the RNA14, RNA15, PCF11 and CLP1 genes. Mutant alleles of each of these genes are deficient for cleavage and polyadenylation in vitro (Minvielle-Sebastia et al., 1994, 1997; Amrani et al., 1997; our unpublished data). To extend our knowledge of the reaction, we sought multicopy suppressors of the pcf11-2 temperature-sensitive mutant. We transformed pcf11-2 cells with a multicopy plasmid-borne genomic library and looked for temperature-resistant clones at 37°C. About 30% of the thermoresistant clones tested carried inserts sharing the putative ORF YGR156w called PTI1 in the Saccharomyces cerevisiae genome database (SGD; http://genome-www.stanford.edu/Saccharomyces/). After subcloning PTI1 individually into a new multicopy vector (pFL44; Bonneaud et al., 1991), we could still observe suppression of the pcf11-2 temperature-sensitive phenotype (Figure 1A). Therefore, overexpression of PTI1 was sufficient to allow growth at 37°C of the otherwise temperature-sensitive pcf11-2 allele. PTI1 is an essential gene that codes for a predicted protein of 425 amino acids, well conserved in eukaryotes. Peculiar features were identified in Pti1p such as an N-terminal RRM-type RNA-binding domain and several potential phosphorylation sites. Remarkably, database searches assigned the 64 kDa subunit of cleavage stimulation factor (CstF) as a putative mammalian homologue of Pti1p (see SGD). Also, Pti1p showed significant similarities with the CF IA subunit Rna15p, which has been found itself to be very similar to CstF-64K (Takagaki and Manley, 1994).
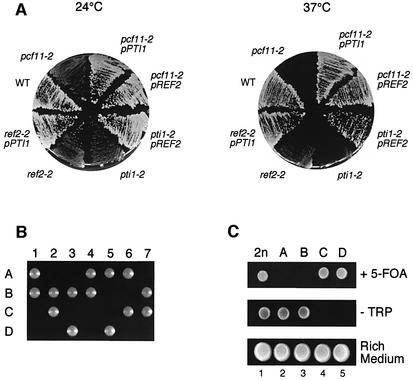
Fig. 1. Phenotypic analysis of pcf11-2, pti1-2, ref2-2 and ref2Δ mutants. (A) Temperature sensitivity of the pcf11-2 mutant can be suppressed by overexpression of PTI1 or REF2 cloned on a multicopy plasmid (pPTI1 and pREF2, respectively). In addition, PTI1 and REF2 are multicopy suppressors of ref2-2 and pti1-2, respectively. (B) REF2 is essential for viability in the W303 yeast strain background. Tetrads from a REF2/ref2Δ::TRP1 heterozygote were dissected on rich medium and incubated at 22°C for at least a week. Two spores only were viable under these conditions, even after a prolonged incubation time (3 weeks). (C) Sporulation of a diploid strain heterozygous for the REF2::TRP1 disruption (lane 1, 2n) gave two spores bearing the deletion which were viable on medium lacking tryptophan provided they were transformed with a plasmid-borne wild-type REF2 gene (lanes 2 and 3, spores A and B, middle row). Those spores were unable to grow on a 5-fluoro-orotic acid (5-FOA)-containing medium since the URA3-marked complementing plasmid was lost when cells were cultivated in the presence of this drug (lanes 2 and 3, upper row). In contrast, the two non-disrupted spores were inviable on medium lacking tryptophan but were not affected by 5-FOA (lanes 4 and 5, spores C and D, middle and upper rows, respectively).
We next screened two-hybrid libraries with PTI1 as bait. This technique allows for the detection of protein–protein interactions in vivo (Fields and Song, 1989). Among the preys obtained, the most significant clones corresponded to PTA1 and REF2 genes (10 independent clones for both genes with seven different inserts each time). Pta1p has already been identified as an intrinsic subunit of CPF (Preker et al., 1997), whereas Ref2p has been described originally as an enhancer of usage of weak poly(A) sites (Russnak et al., 1995). Consistently, multicopy suppression of pcf11-2 temperature sensitivity was also obtained with REF2 at 37°C (Figure 1A). These genetic data suggested that PTI1, like REF2 and PTA1, could code for proteins involved in mRNA 3′ end formation.
We subsequently investigated whether PTI1 and REF2 were essential for cell viability in the wild-type strain W303. We confirmed that disruption of the PTI1 ORF led to lethality (data not shown; see also SGD). Unexpectedly, we also found REF2 to be essential for vegetative growth at all temperatures on rich medium, which is the opposite finding to previous analyses of the gene (Russnak et al., 1995; Winzeler et al., 1999). Indeed, diploid heterozygotes REF2/ref2Δ::TRP1 were sporulated, and tetrad analysis gave rise to two viable, tryptophan-auxotroph spores (therefore carrying the non-disrupted wild-type REF2 gene) and two lethal spores (Figure 1B). The non-viable spores corresponded to the ref2::TRP1 disruptants since they can survive on medium lacking trytophan provided they are rescued by the plasmid-borne REF2 gene (Figure 1C, spores A and B; see also Materials and methods). This discrepancy could be explained by the different genetic background used in our study. In addition, it should be recalled here that only one-third of the REF2 gene was replaced by the LEU2 marker in Russnak et al. (1995). This deletion resulted in a slow growth phenotype that could therefore be attributed to the production of a truncated, albeit still partially functional protein. These results demonstrated that REF2 as well as PTI1 were essential for viability of W303.
Pti1p and Ref2p are not essential for pre-mRNA 3′ end processing in vitro
It has been shown that extracts prepared from pre-mRNA 3′ end processing mutants generally exhibit cleavage and/or polyadenylation defects as tested in vitro (e.g. Minvielle-Sebastia et al., 1994; Preker et al., 1997). We thus generated conditional lethal mutants by subjecting PTI1 and REF2 to mutagenic PCR to gain more insight into their potential role in mRNA 3′ end formation. The temperature-sensitive alleles pti1-2 and ref2-2 obtained were unable to grow at 37°C but survived at 24°C, although exhibiting a slow growth phenotype (Figure 1A). Interestingly, overexpression of PTI1 and REF2 suppressed cell lethality of ref2-2 and pti1-2 at 37°C, respectively, suggesting again that they have related functions (Figure 1A). We then prepared extracts competent for pre-mRNA 3′ end processing in vitro from wild-type and mutant pti1-2 and ref2-2 strains as described (Minvielle-Sebastia et al., 1994). Unexpectedly, we found pti1-2 and ref2-2 mutant extracts to be as active in cleavage and polyadenylation as the wild-type extract (data not shown). The putative role of Pti1p and Ref2p in mRNA maturation was investigated further by seeking their association with the 3′ end processing complex. We performed tandem affinity purification (TAP method; Rigaut et al., 1999) of CPF from PFS2-TAP- and MPE1-TAP-expressing strains as both Pfs2p and Mpe1p have been identified as CPF subunits (Ohnacker et al., 2000; Vo et al., 2001). Mass spectrometry analysis of the factors revealed that both Pti1p and Ref2p were associated with CPF (our data, not shown), as has been suggested recently in a large-scale analysis of yeast protein complexes (Gavin et al., 2002).
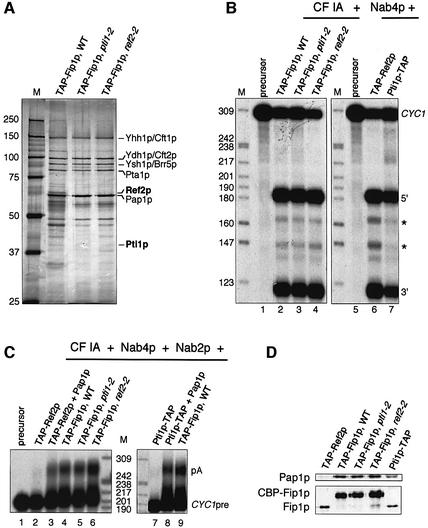
We subsequently purified CPF from the wild-type and pti1-2 and ref2-2 mutant strains in which the Fip1p subunit was N-terminally TAP tagged (see Materials and methods). The factors obtained showed band patterns on protein gels similar to each other and to those previously described following purification by alternative methods (Figure 2A; Preker et al., 1997; Ohnacker et al., 2000; Vo et al., 2001; Dichtl et al., 2002). However, Pti1-2p and Ref2-2p were not detectable by western blotting in the mutant factors whereas they were present in the processing extracts (see Supplementary data available at The EMBO Journal Online). They nevertheless were assayed in vitro for pre-mRNA 3′ end cleavage in combination with wild-type CF IA and recombinant Nab4p (Figure 2B) and for polyadenylation in association with CF IA, Nab4p and Nab2p (Figure 2C; Minvielle-Sebastia et al., 1998; Hector et al., 2002). As seen with extracts, efficient cleavage of the CYC1 pre-mRNA (Figure 2B, lanes 2–4) and polyadenylation of the pre-cleaved substrate CYC1pre (Figure 2C, lanes 4–6) could be observed with either the wild-type or pti1-2 and ref2-2 purified factors. These activities were retained even at higher temperature (34°C; data not shown), strongly supporting that Pti1p and Ref2p were not essential for pre-mRNA 3′ end processing in vitro.
Fig. 2. Reconstitution of pre-mRNA 3′ end processing activity with purified factors. (A) Silver-stained gel of purified CPF obtained from TAP-tagged Fip1p in the wild-type (WT) or pti1-2 and ref2-2 mutant background. M, molecular weight marker, in kDa. (B) Cleavage assays with purified CF IA, recombinant Nab4p and the different TAP factors purified as described in Materials and methods. Lanes 1 and 5, unreacted iso-1-cytochrome c precursor (CYC1); 5′, 3′, upstream and downstream cleavage products, respectively. Cryptic cleavage products are indicated by asterisks (Minvielle-Sebastia et al., 1998). (C) Polyadenylation of the CYC1 pre-cleaved RNA (CYC1pre) which ends at the natural polyadenylation site. Assays were performed with purified CF IA, recombinant Nab2p, Nab4p and Pap1p (where indicated), and the different TAP factors purified as described in Materials and methods. Lane 1, unreacted CYC1pre precursor; lane 3, 10 ng of Pap1p were added; lane 8, 20 ng of Pap1p were added. pA, polyadenylated products. M, labelled MspI-digested pBR322 serves as molecular weight markers, in number of nucleotides. (D) Western blot analysis of the different TAP factors. The membranes were probed with polyclonal antibodies to either Pap1p (at a 1:2000 dilution) or Fip1p (diluted at 1:10 000). CBP-Fip1p refers to as the calmodulin-binding peptide-tagged Fip1 protein obtained after TEV protease cleavage during TAP purification (Rigaut et al., 1999).
To test more directly whether Pti1p- and Ref2p-associated components are competent for mRNA 3′ end formation, we purified the Pti1p-TAP and TAP-Ref2p factors (see Materials and methods). The isolated complexes were assayed for cleavage and polyadenylation in vitro. Although they appeared similarly active in cleavage as a TAP-Fip1p wild-type CPF (Figure 2B, compare lane 6 and 7 with lane 2), they failed to polyadenylate the CYC1pre substrate unless recombinant poly(A) polymerase (Pap1p) complemented them in the assays (Figure 2C, compare lanes 2 and 3, and 7 and 8). In comparison, TAP-Fip1p complexes purified from the wild-type or mutant pti1-2 or ref2-2 strains were proficient on their own (Figure 2C, lanes 4–6 and lane 9). The inefficient polyadenylation activity could be explained at least in part by the very low levels of Pap1p detected by western blotting of TAP-Ref2p and Pti1p-TAP complexes compared with all three TAP-Fip1p complexes (Figure 2D). These results showed that TAP-Ref2p and Pti1p-TAP factors could associate with CF I in a complex that could at least specifically cleave an mRNA precursor. However, Pti1p and Ref2p essential function did not seem to be related to 3′ end formation of this class of RNA precursors. As pol II also transcribes precursors of small RNAs, we investigated whether mutations in PTI1 and REF2 would affect snoRNA 3′ end maturation.
snoRNA 3′ end formation is impaired in pti1 and ref2 mutants
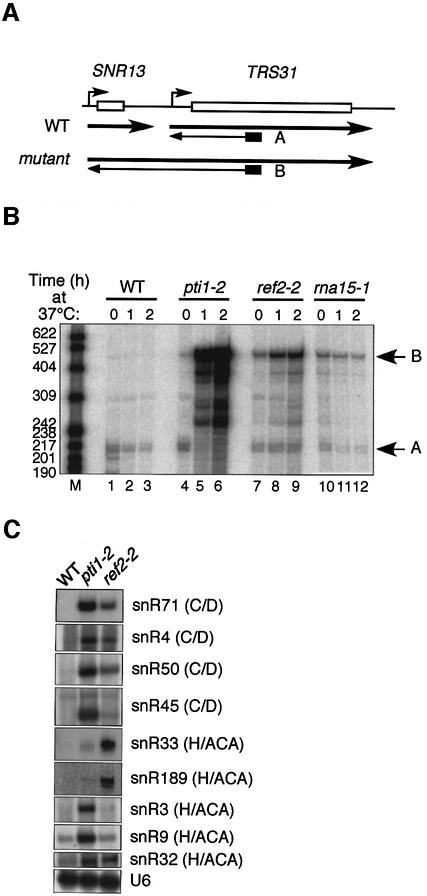
In yeast, snoRNAs are transcribed by pol II, in most cases as independent, either mono- or polycistronic units. In some instances, the 5′ end of the snoRNA corresponds to the 5′ end of the primary transcript and carries a trimethylguanosine cap (Chanfreau et al., 1998a; Fatica et al., 2000). However, it also happens that the 5′ end is matured by endonucleolytic cleavage followed by exonucleolytic trimming. Likewise, the 3′ end is formed by exonucleolytic degradation of the precursor after cleavage. The entry sites for exonucleases are generated by the RNA endonuclease Rnt1p in the 5′ portion of the precursor, but this is very often not the case for the 3′ end (Chanfreau et al., 1998a; Allmang et al., 1999; van Hoof et al., 2000). Recent studies implicated CF IA but not CPF as essential for 3′ end cleavage of small stable RNAs based on the analysis of several mutants in both factors (Fatica et al., 2000; Morlando et al., 2002). To test whether pti1-2 and ref2-2 mutations may affect 3′ end formation of snoRNAs, we performed primer extension analysis of transcripts from the SNR13-TRS31 region of the genome as described previously (Figure 3A; Steinmetz et al., 2001; Morlando et al., 2002). An oligonucleotide complementary to the TRS31 ORF revealed extended transcripts by reverse transcription of total RNA extracted from an rna15 mutant strain after shift to 37°C (Figure 3B, lanes 10–12, product B) but not from the wild-type strain (Figure 3B, lanes 1–3, product A). This indicated that Rna15p was involved in vivo in box C/D snR13 3′ end formation as described (Morlando et al., 2002). Interestingly, the strong rna15-1 mutant allele tested here was impaired already at permissive temperature (24°C; Figure 3B, lane 10). More strikingly, the pti1-2 strain showed a rapid and dramatic accumulation of extended product B after shift to the restrictive temperature (Figure 3B, lanes 4–6). Likewise, the ref2-2 strain exhibited product B accumulation although to a lesser extent (Figure 3B, lanes 7–9). In the pti1-2 mutant only, product A was undetectable after shift to 37°C, suggesting that initiation of transcription at the TRS31 promoter was almost entirely abolished by promoter occlusion (Greger et al., 2000). Unlike with pti1-2 and ref2-2, extended product B did not accumulate in the rna15-1 mutant, which is also impaired in mRNA 3′ end formation, probably because the TRS31 mRNA was not polyadenylated properly and was thus degraded (Figure 3B).
Fig. 3. Reverse transcription analysis of extended transcripts of some snoRNAs in wild-type and mutant strains. (A) Schematic representation of the SNR13-TRS31 region of the chromosome. ‘A’ refers to reverse transcription products in a wild-type strain, and ‘B’ to snR13 3′-extended transcripts. (B) Polyacrylamide gel electrophoresis of reverse transcripts from wild-type (WT), pti1-2, ref2-2 and rna15-1 strains 0, 1 and 2 h after shift to 37°C. ‘A’ and ‘B’ are as depicted in (A); M, molecular weight markers in number of nucleotides. (C) Reverse transcription analysis of several box C/D and H/ACA snoRNAs in wild-type and pti1-2 and ref2-2 mutant strains. The pol III-transcribed U6 RNA is given as a normalization control for the reverse transcription.
Several other snoRNAs, including box C/D snR71, snR4, snR50 and snR45, and box H/ACA snR33, snR189, snR3, snR9 and snR32, also showed readthrough transcripts in pti1-2 and ref2-2 mutants (Figure 3C). Surprisingly, ref2-2 mutant reproducibly displayed a more pronounced effect on the accumulation of the 3′-extended forms of box H/ACA snR33 and snR189 than that observed with the pti1-2 mutant, whereas snR32 3′ end formation looked equally affected by both mutations. The reason for these differences currently is unclear, but this suggests that Pti1p and Ref2p have no redundant function in snoRNA 3′ end formation. Therefore, CPF-associated Pti1p and Ref2p are probably generally required for 3′ end formation of independently transcribed snoRNAs.
Overexpression of Pti1p uncouples cleavage and polyadenylation
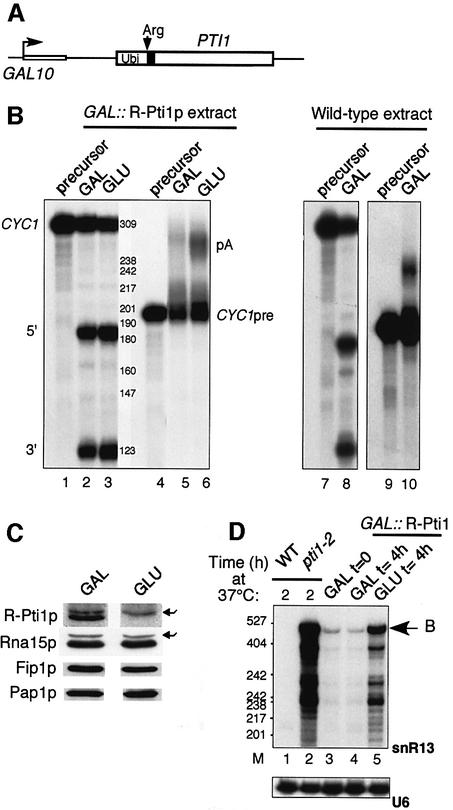
Our results suggested that the same global protein machinery, i.e. CF I + CPF, is required for pol II-transcribed polyadenylated and non-polyadenylated RNA 3′ end maturation. Since polyadenylation of snoRNAs is detrimental to their function (Fatica et al., 2000), this apparatus must thus discriminate between mRNA and snoRNA precursors. This may be achieved in part through recognition of cis-acting elements specific for both precursors (Fatica et al., 2000; Edmonds, 2002). However, trans-acting factors might be essential to mediate uncoupling of cleavage and polyadenylation. To address whether Pti1p, which generally exhibited the strongest snoRNA 3′ end formation defect in its mutant form, might play such a role, we constructed a strain where expression of Pti1p is driven by the strong inducible/repressible GAL10 promoter in order to intensify its putative function in uncoupling the two reactions. Our construct also introduced a ubiquitin moiety at the N-terminus such that the protein started with the destabilizing amino acid arginine after in vivo de-ubiquitylation and therefore was rapidly degraded (Figure 4A; Billy et al., 2000). Extracts prepared from cells grown in galactose (GAL) or after a shift to a glucose-containing medium (GLU) for 4 h were assayed in vitro for pre-mRNA 3′ end processing. Cleavage of the CYC1 precursor was normal with both the GAL and GLU extracts (Figure 4B, lanes 2 and 3). However, polyadenylation of the CYC1pre transcript was significantly impaired when Pti1p was overexpressed in galactose, whereas it became normal as Pti1p expression was repressed in glucose (Figure 4B, compare lanes 5 and 6). As a control, an extract prepared from a wild-type, unmodified strain grown in galactose could process the CYC1 precursors efficiently (Figure 4B, lanes 7–10). A western blot analysis of the GAL and GLU extracts showed that Pti1p was undetectable in glucose-grown cells, whereas levels of the CF IA component Rna15p or CPF subunits such as Fip1p and even the poly(A) polymerase Pap1p were not affected by the ubiquitin system in either growth condition (Figure 4C). Depletion of Pti1p in vivo correlated with the same 3′ end formation defect as observed with the temperature-sensitive allele pti1-2 (Figure 4D, lanes 1–5). The inhibitory effect of Pti1p overexpression on polyadenylation was also examined with an extract prepared from a strain where the destabilizing ubiquitin-arginine conditional system was omitted in order to avoid possible side effects of the method (Figure 5A; Lafontaine and Tollervey, 1996). In this situation, Pti1p overexpression affected polyadenylation even more dramatically (Figure 5B, lanes 5 and 6) whereas cleavage remained unaffected (Figure 5B, lanes 2 and 3). Low levels of Pti1p were still visible on western blots in the GLU extracts but nevertheless allowed polyadenylation to proceed (Figure 5C). Here again, neither CF IA subunit Rna15p nor CPF-associated Pap1p levels declined after the glucose shift (Figure 5C). It is worth recalling here that other CPF subunits subjected to identical conditional expression have been reported to show opposite effects, i.e. inhibition of polyadenylation following repression of protein expression (e.g. Ysh1p and Yth1p; Jenny et al., 1996; Barabino et al., 2000). Together with the data obtained with TAP-Ref2p and Pti1p-TAP complexes on polyadenylation (Figure 2C), these results suggested that Pti1p, and probably Ref2p, may function in snoRNA 3′ end formation by uncoupling cleavage and polyadenylation.
Fig. 4. Conditional expression of PTI1 affects 3′ end formation of pol II-transcribed RNAs. (A) A destabilizing ubiquitin-arginine moiety was fused to Pti1p and the hybrid construct was expressed under the control of the inducible GAL10 promoter. The fusion was reintroduced at the PTI1 chromosomal locus (Billy et al., 2000). (B) Extracts prepared from GAL::R-Pti1p-expressing cells grown in galactose (GAL) or after a 4 h shift to glucose (GLU) were tested for cleavage of the CYC1 precursor (lanes 1–3) and polyadenylation of the CYC1pre RNA (lanes 4–6). A wild-type extract made from an unmodified BMA64 wild-type strain grown in galactose was tested for cleavage (lanes 7 and 8) and polyadenylation (lanes 9 and 10) as above. (C) Immunoblot analysis of the GAL and GLU extracts (20 µg) with polyclonal antibodies to Pti1p (1:500), Rna15p (1:10 000), Fip1p (1:10 000) and Pap1p (1:2000). The arrowheads point to irrelevant polypeptides that cross-react with antibodies to Pti1p and Rna15p. (D) Analysis of snR13 extended transcripts in the wild-type, pti1-2 and GAL::R-Pti1p conditional mutant strains. The reverse transcription was performed on RNAs extracted from wild-type and pti1-2 cells after a shift to 37°C for 2 h (lanes 1 and 2, respectively), or from GAL::R-Pti1p-expressing cells grown in galactose (lane 3, GAL t = 0) then re-inoculated in either galactose or glucose medium for 4 h (lanes 4 and 5, respectively). ‘B’ refers to snR13 extended transcripts as diagrammed in Figure 3A. U6 serves as a normalization control for the reverse transcription. M, molecular weight markers in number of nucleotides.
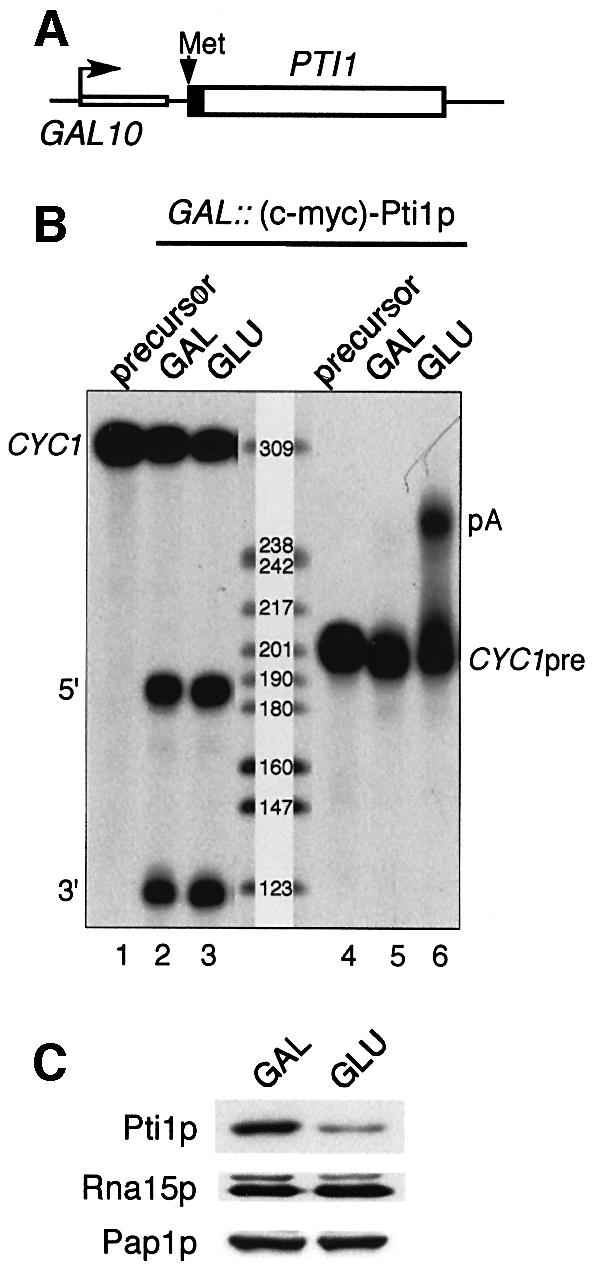
Fig. 5. Conditional expression of Ptip1 under the GAL promoter. (A) Schematic representation of the hybrid construct introduced at the PTI1 chromosomal locus (Lafontaine and Tollervey, 1996). The Pti1 protein is tagged at the N-terminus with the c-myc epitope. Its expression is induced in galactose-containing medium and repressed in glucose. The black box represents the c-myc epitope tag. (B) Cleavage and polyadenylation assays were performed with extracts prepared with the GAL::(c-myc)-Pti1p-expressing strain as in Figure 4B. (C) Immunoblot analysis of the GAL and GLU extracts with monoclonal antibodies to the c-myc epitope tag (1:2000), and with polyclonal antibodies to Rna15p and Pap1p as in Figure 4C.
The Nrd1p- and Pti1p-dependent snoRNA 3′ end formation pathways are connected
A recent study highlighted the role of the RNA-binding protein Nrd1p and its associated partners Nab3p, Sen1p and Ctk1p in snRNA and snoRNA 3′ end formation. Nrd1p interacts with the pol II CTD whose integrity is also essential for Nrd1-dependent 3′ end maturation (Steinmetz et al., 2001). The nrd1 and sen1 mutant alleles exhibited termination defects in snoRNA transcription that resulted in readthrough snoRNA transcripts similar to those found with pti1-2 and ref2-2. Moreover, nab3, ctk1 and CTD truncation mutants also accumulated extended transcripts, supporting their involvement as a complex in an Nrd1p-dependent sn- and snoRNA 3′ end formation pathway (Steinmetz et al., 2001). The amounts of NRD1 mRNA and protein are inversely proportional as a consequence of a regulatory mechanism involving Nrd1p binding to a responsive element located near the 5′ end of its message. In addition, a northern blot analysis revealed that the NRD1 mRNA is more abundant in nrd1, sen1 and nab3 mutant strains after temperature shift, showing that factors involved in the Nrd1-dependent pathway of sn(o)RNA formation coordinate Nrd1p expression (Steinmetz et al., 2001). To extend the overlap between the Nrd1p-dependent and Ptip1/Ref2p-dependent pathways for 3′ end formation of non-polyadenylated pol II transcripts, we analysed the effects of pti1-2 and ref2-2 mutations on NRD1 mRNA levels at restrictive temperature. Remarkably, as previously observed with the nrd1 mutant strain, NRD1 mRNA was more abundant at 37°C in pti1-2 than in the wild-type strain (Figure 6A). Surprisingly, the ref2-2 mutation showed no significant effect. As controls, the nab3-11 temperature-sensitive mutant exhibited similar NRD1 message accumulation (Figure 6A), and extended SNR13 transcripts could be observed at a restrictive temperature for nab3-10 and nab3-11 conditional alleles (Figure 6B, lanes 1–4). Therefore, the NRD1 gene regulation involves components of both the Nrd1p-dependent and Pti1p-dependent 3′ end formation of snoRNA, very probably through a coordinated mechanism.

Fig. 6. NRD1 mRNA autoregulation is affected by mutations in the PTI1 gene. (A) Northern blot of NRD1 mRNA in the pti1-2, ref2-2, nab3-11 and wild-type strains with RNA extracted after a 2 h shift to 37°C. The blot with the pol III-transcribed SCR1 was used as a loading control. (B) Analysis of snR13 extended transcripts in the nab3-10 and nab3-11 conditional mutants. The reverse transcription was performed on RNAs extracted from nab3-10 and nab3-11 cells cultivated at permissive temperature (24°C; lanes 1 and 3) or after a shift to 37°C for 2 h (lanes 2 and 4). ‘A’ and ‘B’ are as diagrammed in Figure 3A. U6 serves as a normalization control for the reverse transcription. M, molecular weight markers in number of nucleotides.
Discussion
We identified Pti1p and Ref2p as new components associated with the pre-mRNA 3′ end processing complex. However, they did not appear to be essential for mRNA 3′ end maturation. Instead, they were found to be required primarily for the 3′ end formation of snoRNAs. We do not know at the moment whether Pti1p and Ref2p also direct 3′ end maturation of the other class of pol II-transcribed small RNAs, the snRNAs. PTI1 and REF2 were found as multicopy suppressors of a mutant in the CF IA subunit pcf11-2. This is consistent with CF IA’s role in small RNA 3′ end formation since it has been reported previously that this factor also contributes to snoRNA and snRNA 3′ end maturation (Fatica et al., 2000; Morlando et al., 2002). Therefore, we can anticipate that pti1 and ref2 will also be impaired in snRNA 3′ end formation. We currently are testing this possibility.
REF2 was discovered originally in a screen to identify mutants defective in pre-mRNA 3′ end processing (Russnak et al., 1995). A ref2 mutant strain showed a decrease in the use of a CYC1 poly(A) site located within an artificial intron that had been positioned inside the HIS4 reporter gene. This intron was spliced out efficiently in the ref2Δ mutant background, not in the wild-type, therefore allowing HIS4 to be functional. However, in vitro pre-mRNA 3′ end processing was not dramatically affected in ref2Δ extracts. One can reconcile our results with those published by Platt and co-workers by postulating that some readthrough products resulting from the ref2 deletion (that did not rise to a lethal phenotype in this study) could lead to the production of the HIS4 gene product and therefore allow growth on medium containing the histidine precursor.
Pti1p and Ref2p have been found as integral constituents of CPF by mass spectrometry analysis of factors purified from cells where different CPF subunits have been tagged (Dichtl et al., 2002; Gavin et al., 2002; our results). Until now, all of the already characterized CPF subunits have shown altered pre-mRNA 3′ end processing activity when mutant alleles of their genes were analysed (Edmonds, 2002; and references therein). Only recently, a new polypeptide stably associated with CPF, Ssu72p, revealed its role in balancing elongation and termination of transcription by pol II of pre-mRNAs. The authors reported that in vitro pre-mRNA 3′ end processing and transcription termination of a few snoRNAs were not affected in the ssu72-2 mutant allele (Dichtl et al., 2002). However, one cannot exclude that an extension of this analysis to more snoRNA transcription units would reveal a more general defect in transcription termination.
Our results showed that neither pti1-2 nor ref2-2 mutants were deficient in pre-mRNA 3′ end processing. Instead, pti1-2 and ref2-2 showed a major 3′ end form ation defect of independently transcribed snoRNAs. Surprisingly, we found that the levels of readthrough products accumulating after shift to the non-permissive temperature varied significantly between pti1-2 and ref2-2 depending upon the snoRNA tested (see Figure 3C). Interestingly, it seems to be independent of the class of small RNA to which the snoRNAs tested belong (box C/D or H/ACA). More frequently, larger amounts of extended transcripts accumulated in the pti1-2 mutant compared with ref2-2, and in one case only, both mutants exhibited equivalent amount of transcripts (snR32). This phenomenon cannot be attributed to the strength of the alleles tested since the same mutants were employed throughout this study. We would rather favour the model where Pti1p and Ref2p have non-overlapping functions in the cell and thus affect snoRNA 3′ end formation differently when mutated. They could perhaps define alternative classes of snoRNAs that would not be related to their belonging to box C/D or H/ACA classes of snoRNAs but rather to the 3′ end processing elements located in the downstream regions of their transcription units.
Our results showed that Pti1p and Ref2p represent the first CPF subunits whose primary function is related to small RNA 3′ end processing. These results also suggested that Pti1p, and very probably Ref2p, function in the uncoupling of cleavage and polyadenylation during small RNA 3′ end formation. We propose that a factor containing already characterized CPF subunits and at least Pti1p and Ref2p could specifically recognize the 3′ end formation signals for non-polyadenylated pol II transcripts in combination with cleavage/polyadenylation factor CF IA. We could tentatively refer to this factor as snCF for small nuclear RNA cleavage factor. snCF might co-exist in yeast cells with CPF and CF IA, and constitute, when associated with CF IA, the cleavage-dependent/polyadenylation-independent 3′ end processing apparatus. This complex could perform the primary cleavage event of snoRNA precursors which would occur without further polyadenylation due to the presence of Pti1p and possibly additional factors. Since 3′ end formation and export are coupled events, this uncoupling could prevent snoRNAs from being exported. How polyadenylation is inhibited is not yet understood. However, the factors purified by reverse TAP tagging of either Pti1p or Ref2p can cleave when combined with CF IA but do not polyadenylate a pre-mRNA substrate, very probably because poly(A) polymerase barely associates with these complexes. Thus, our data would suggest that inhibition of polyadenylation might arise from partial to complete exclusion of Pap1p from CPF and gives rise to the snCF complex.
Recently, PTI1 was found as a partial suppressor of a ctk1Δ cold-sensitive mutant (Ctk1p is the catalytic subunit of the CTDK-I kinase that phosphorylates the pol II CTD) (Skaar and Greenleaf, 2002). It has been proposed in this study that Pti1p could be complexed in an alternative form of CF IA where Pti1p would substitute for Rna15p. This assumption was based on the sequence similarity between the two proteins. However, we never found Pti1p associated with CF IA but with the snCF factor. Moreover, our two-hybrid data showed interactions of Pti1p with Pta1p and Ref2p, both components of snCF/CPF. Skaar and collaborators have also shown that mutations in PTI1 affected poly(A) site choice of some transcripts in vivo, and draw the conclusion that the pol II CTD is phosphorylated by CTDK-I to couple transcription to pre-mRNA 3′ end processing by a specific Pti1p-associated complex. This is consistent in part with our results demonstrating that pti1-2 (and ref2-2) inhibited transcription termination by pol II. However, we favour a primary role for Pti1p in snoRNA 3′ end formation mainly for the following reasons. First, our data demonstrated that Pti1p and Ref2p are not essential for pre-mRNA 3′ end processing in vitro (Figure 2) but mutations in their genes affect snoRNA 3′ end formation in vivo (Figure 3). Secondly, the presence of Pti1p and Ref2p in snCF correlated with the exclusion of Pap1p from the complex (Figure 2), which is consistent with the fact that polyadenylation of snoRNAs is detrimental to their accumulation (Fatica et al., 2000). This observation was strengthened by the observation that only the polyadenylation step was inhibited upon Pti1p overexpression (Figures 4 and 5). This has never been demonstrated so far for other 3′ end processing components. Finally, we have observed an overlap between the Pti1p-dependent and the Nrd1-dependent 3′ end processing pathways for non-polyadenylated pol II transcripts, as mutations in PTI1 deregulated NRD1 mRNA levels (Figure 6). Interestingly, ctk1Δ cells also showed increased snoRNA readthrough products (Steinmetz et al., 2001). Therefore, PTI1 overexpression very probably compensated for the termination defect observed in ctk1Δ cells by Skaar and Greenleaf (2002).
In coordination with CF IA/snCF, the Nrd1p-dependent pathway for small RNA maturation could link termination of transcription to snoRNA 3′ end processing. More work will be necessary before we can recapitulate Rnt1-independent snoRNA 3′ end cleavage in vitro. We assume this will be possible starting from the association of CF IA and snCF. However, other as yet uncharacterized factors perhaps will be required.
Materials and methods
Yeast strains
The pcf1-2 (Amrani et al., 1997), pti1-2 and ref2-2 mutants were derived from W303 (ade2-1 leu2-3,112 ura3-1 trp1-1 his3-11,15 can1-100) (Thomas and Rothstein, 1989). REF2 disruption was carried out by replacing the fragment ranging from –2 to +1582 (according to the start codon) in BMA64–2n diploid strain with the TRP1 marker according to Baudin et al. (1993). Independent diploids were dissected on rich medium, and 32 tetrads were allowed to grow at 22°C for at least 3 weeks. Only two viable, tryptophan-auxotroph spores were recovered. After inspection of the two non-viable spores under the microscope, we observed that they started to germinate but stopped growing after about five divisions. Mutant alleles of the PTI1 and REF2 genes were generated by mutagenic PCR and re-integrated at their chromosomal locus as described (Vo et al., 2001). The C-terminal PTI-TAP-expressing strain was constructed in BMA64 (ade2-1 leu2-3,112 ura3-1 trp1Δ his3-11,15 can1-100; Baudin-Baillieu et al., 1997) according to Rigaut et al. (1999). The N-terminal TAP-Fip1p-expressing strains were constructed either in the wild-type BMA64 strain (YSD10) or in the mutant pti1-2 (LM135) or ref2-2 (LM137) isogenic strains according to S.Dheur, F.Voisinet-Hakil and L.Minvielle-Sebastia (in preparation). The TAP-Ref2p strain (YSD26) was constructed in BMA64. These N-terminally modified genes were constructed by integration at the chromosomal locus of a cassette adding in-frame a TAP tag identical to that described by Séraphin and coworkers and according to the same experimental procedure (Rigaut et al., 1999). All the modified strains showed growth characteristics similar to an untagged wild-type strain. To generate the GAL::R-Pti1p-expressing strain (LM141), the PTI1 ORF was fused to the ubiquitin-arginine coding sequence and expressed under the control of the GAL10 promoter according to Billy et al. (2000). The GAL::(c-myc)-PTI1 strain (LM145) was constructed following a method previously described (Lafontaine and Tollervey, 1996). Strains YPN100 (nab3-10) and YPN102 (nab3-11) carried mutations in the NAB3 gene as described (Conrad et al., 2000) and were a generous gift of Maurice S.Swanson (University of Florida).
Two-hybrid screens
For the two-hybrid assays, the PTI1 ORF was cloned into the pGBD9 vector. This construct was able to complement the pti1-2 mutant defect at non-permissive temperature, demonstrating that the fusion was functional. Strain J693 was first transformed with pGBD9-PTI1 and then with either the DHFII (F.Lacroute, unpublished material) or FRYL (Fromont-Racine et al., 1997) genomic libraries. Among 2 × 106 transformants obtained, 80 positive clones were sequenced; 60 carried inserts of type A (Fromont-Racine et al., 1997). Two genes, PTA1 and REF2, were found repeatedly, with 19 independent clones for each gene and seven different inserts each time.
Purification of TAP-tagged factors and in vitro 3′ end processing assays
TAP-tagged factors were purified from 2 l cultures according to Rigaut et al. (1999), except that extracts were prepared using a previously described spheroplast procedure (Butler and Platt, 1988). The TAP factors were dialysed and kept frozen at –80°C. Mass spectrometry analysis was carried out as published (Vo et al., 2001). Cleavage and polyadenylation assays were performed essentially as described (Minvielle-Sebastia et al., 1998). In brief, cleavage assays were performed with 0.5 µl of purified CF IA (Minvielle-Sebastia et al., 1998), 30 ng of recombinant Nab4p and either 1 µl of TAP-Fip1p (WT), 1.3 µl of TAP-Fip1p (pti1-2), 1 µl of TAP-Fip1p (ref2-2), 2 µl of TAP-Ref2p or 8 µl of Pti1p-TAP factors. Polyadenylation assays were performed as for cleavage, but 80 ng of recombinant Nab2p were added to control poly(A) tail synthesis (Hector et al., 2002).
Primer extension assays
All primer extension analyses were carried out according to George and Kadonaga (1996), with 10 µg of total RNA. Extended products were separated by gel electrophoresis on 6% polyacrylamide–8.3 M urea gels and analysed by autoradiography.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are indebted to Maurice S.Swanson for the generous gift of strains and reagents, and helpful comments on the manuscript. We thank Pierre Legrain and Micheline Fromont-Racine for the kind gift of the FRYL two-hybrid library, all the members of our laboratories for critical reading of the manuscript, and Katell Bathany and Geneviève Demaison for their technical assistance. This work was supported by the CNRS (ATIPE), the Ministère de la Recherche Scientifique, La Fondation pour la Recherche Médicale/Fondation BNP-Paribas (to L.M.-S.) and La Ligue Nationale contre le Cancer (to L.M.-S. and F.W.). S.D. was supported by a post-doctoral fellowship from La Ligue Nationale contre le Cancer and by the CNRS (Chercheur associé). L.T.A.V. was the recipient of a fellowship from the Ministère de la Recherche Scientifique. We wish to dedicate this work to the memory of Professor Michel Blot.
References
- Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Minet,M., Wyers,F., Dufour,M.-E., Aggerbeck,L.P. and Lacroute,F. (1997) PCF11 encodes a third protein component of the yeast cleavage and polyadenylation factor I. Mol. Cell. Biol., 17, 1102–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S.M., Hübner,W., Jenny,A., Minvielle-Sebastia,L. and Keller,W. (1997) The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev., 11, 1703–1716. [DOI] [PubMed] [Google Scholar]
- Barabino S.M., Ohnacker,M. and Keller,W. (2000) Distinct roles of two yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J., 19, 3778–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A., Guillemet,E., Cullin,C. and Lacroute,F. (1997) Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast, 13, 353–356. [DOI] [PubMed] [Google Scholar]
- Billy E., Wegierski,T., Nasr,F. and Filipowicz,W. (2000) Rcl1p, the yeast protein similar to the RNA 3′-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J., 19, 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos,O., Li,G., Labouesse,M., Minvielle-Sebastia,L. and Lacroute,F. (1991) A family of low and high copy replicative, integrative and single-stranded S.cerevisiae/E.coli shuttle vectors. Yeast, 7, 609–615. [DOI] [PubMed] [Google Scholar]
- Butler J.S. and Platt,T. (1988) RNA processing generates the mature 3′ end of yeast CYC1 messenger RNA in vitro. Science, 242, 1270–1274. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Noble,S.M. and Guthrie,C. (1996) Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF). Science, 274, 1511–1514. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Legrain,P. and Jacquier,A. (1998a) Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Rotondo,G., Legrain,P. and Jacquier,A. (1998b) Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J., 17, 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad N.K., Wilson,S.M., Steinmetz,E.J., Patturajan,M., Brow,D.A., Swanson,M.S. and Corden,J.L. (2000) A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics, 154, 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B. and Keller,W. (2001) Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J., 20, 3197–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B., Blank,D., Ohnacker,M., Friedlein,A., Roeder,D., Langen,H. and Keller,W. (2002) A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell, 10, 1139–1150. [DOI] [PubMed] [Google Scholar]
- Edmonds M. (2002) A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol., 71, 285–389. [DOI] [PubMed] [Google Scholar]
- Fatica A., Morlando,M. and Bozzoni,I. (2000) Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J., 19, 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S. and Song,O.K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain,J.C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- George C.P. and Kadonaga,J.T. (1996) Primer-extension analysis of RNA. In Krieg,P.A. (ed.), A Laboratory Guide to RNA: Isolation, Analysis and Synthesis. Wiley-Liss, New York, NY, pp. 133–136.
- Greger I.H., Aranda,A. and Proudfoot,N. (2000) Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 97, 8415–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R.E., Nykamp,K.R., Dheur,S., Anderson,J.T., Non,P.J., Urbinati,C.R., Wilson,S.M., Minvielle-Sebastia,L. and Swanson,M.S. (2002) Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J., 21, 1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- Kessler M.M., Zhao,J. and Moore,C.L. (1996) Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J. Biol. Chem., 271, 27167–27175. [DOI] [PubMed] [Google Scholar]
- Kessler M.M., Henry,M.F., Shen,E., Zhao,J., Gross,S., Silver,P.A. and Moore,C.L. (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev., 11, 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D. and Tollervey,D. (1996) One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res., 24, 3469–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Radkte,I., Wahle,E. and Keller,W. (1991) Purification and characterisation of poly(A) polymerase from Saccharomyces cerevisiae. J. Biol. Chem., 266, 8741–8746. [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J. and Keller,W. (1994) RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J., Wiederkehr,T., Strahm,Y. and Keller,W. (1997) The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in pre-messenger RNA 3′-end formation. Proc. Natl Acad. Sci. USA, 94, 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Beyer,K., Krecic,A.M., Hector,R.E., Swanson,M.S. and Keller,W. (1998) Control of cleavage site selection during mRNA 3′-end formation by a yeast hnRNP. EMBO J., 17, 7454–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M., Greco,P., Dichtl,B., Fatica,A., Keller,W. and Bozzoni,I. (2002) Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol., 22, 1379–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnacker M., Barabino,S.M., Preker,P.J. and Keller,W. (2000) The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J., 19, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal K. and Reddy,R. (2002) The 3′ end formation in small RNAs. Gene Expr., 10, 59–78. [PMC free article] [PubMed] [Google Scholar]
- Preker P.J., Lingner,J., Minvielle-Sebastia,L. and Keller,W. (1995) The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell, 81, 379–389. [DOI] [PubMed] [Google Scholar]
- Preker P.J., Ohnacker,M., Minvielle-Sebastia,L. and Keller,W. (1997) A multisubunit 3′-end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J., 16, 4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko,A., Rutz,B., Wilm,M., Mann,M. and Séraphin,B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol., 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Russnak R., Nehrke,K.W. and Platt,T. (1995) REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3′-end formation. Mol. Cell. Biol., 15, 1689–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar D.A. and Greenleaf,A.L. (2002) The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell, 10, 1429–1439. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J., Conrad,N.K., Brow,D.A. and Corden,J.L. (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature, 413, 327–331. [DOI] [PubMed] [Google Scholar]
- Stumpf G. and Domdey,H. (1996) Dependence of yeast pre-mRNA 3′-end processing on CFT1: a sequence homolog of the mammalian AAUAAA binding factor. Science, 274, 1517–1519. [DOI] [PubMed] [Google Scholar]
- Takagaki Y. and Manley,J.L. (1994) A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature, 372, 471–474. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo L.T.A., Minet,M., Schmitter,J.M., Lacroute,F. and Wyers,F. (2001) Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol. Cell. Biol., 21, 8346–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E.A. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Zhao J., Kessler,M.M. and Moore,C.L. (1997) Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem., 272, 10831–10838. [DOI] [PubMed] [Google Scholar]