Abstract
Rab4 regulates recycling from early endosomes. We investigated the role of the rab4 effector rabaptin-5α and its putative partner γ1-adaptin in membrane recycling. We found that rabaptin-5α forms a ternary complex with the γ1–σ1 subcomplex of AP-1, via a direct interaction with the γ1-subunit. The binding site for γ1-adaptin is in the hinge region of rabaptin-5α, which is distinct from rab4- and rab5-binding domains. Endogenous or ectopically expressed γ1- adaptin localized to both the trans-Golgi network and endosomes. Co-expressed rabaptin-5α and γ1-adaptin, however, co-localized in a rab4-dependent manner on recycling endosomes. Transfection of rabaptin-5α caused enlarged endosomes and delayed recycling of transferrin. RNAi of rab4 had an opposing effect on transferrin recycling. Collectively, our data show that rab4-GTP acts as a scaffold for a rabaptin-5α– γ1-adaptin complex on recycling endosomes and that interactions between rab4, rabaptin-5α and γ1-adaptin regulate membrane recycling.
Keywords: γ1-adaptin/endosomes/rab4/rabaptin-5α/receptor recycling
Introduction
Many proteins are internalized from the cell surface by AP-2 and clathrin-mediated endocytosis. After delivery to early endosomes (EEs), receptor–ligand complexes dissociate, and cargo proteins are sorted away from each other and either recycled back to the plasma membrane or delivered to other endosomal compartments. Vectorial transport between organelles critically relies on hetero-oligomeric protein assemblies that regulate budding of transport vesicles and specify targeting, docking and fusion (see Guo et al., 2000). The temporal and spatial assembly of these conserved protein complexes is therefore essential to maintain compartmental integrity. Indeed, the recent past has witnessed the identification of an increasing number of proteins that are involved in regulating the orderly flow of membrane through EEs (see Zerial and McBride, 2001).
Several rab GTPases are localized to EEs (see Zerial and McBride, 2001). Rab5 controls EE fusion, while rab4 and rab11 regulate receptor recycling pathways from EEs back to the cell surface or Golgi complex. The function of rab proteins in membrane transport has been worked out best for rab5 that exerts its activity through the coordinated recruitment of effector proteins such as hVPS34, EEA-1, rabaptin-5 and rabenosyn-5 (see Zerial and McBride, 2001). These may serve to tether incoming vesicles onto endosomes and control the activity of SNARE proteins involved in membrane fusion. Rab4 is localized onto membranes of early endocytic compartments and regulates membrane recycling (van der Sluijs et al., 1992b). We previously identified rabaptin-4 (Nagelkerken et al., 2000) that, like rabaptin-5 (Vitale et al., 1998) and rabaptin-5β (de Renzis et al., 2002), interacts with rab4 and rab5. Rab4 and rabaptin-4 co-localize on enlarged endosomal structures that are enriched in cellubrevin, a marker for recycling endosomes (REs) (Nagelkerken et al., 2000). Both rabaptin-4 and rabaptin-5β contain a 33 amino acid internal deletion within the C-terminus. Rabaptin-4 and rabaptin-5 are 96% identical, are expressed in the same cells and it remains to be established whether they are functionally equivalent. Korobko et al. (2002) recently identified rabaptin-5γ and rabaptin-5δ, two novel rabaptin variants with internal deletions in the N-terminus. Since rabaptin-5γ, rabaptin-5δ and rabaptin-4 are splice variants of rabaptin-5, we changed the name rabaptin-4 to rabaptin-5α. It is thought that bivalent rab4 and rab5 effector proteins such as rabaptins and rabenosyn-5 cooperatively link rab5- and rab4-containing microdomains on EEs (de Renzis et al., 2002) to strengthen the coupling between these domains.
Rabaptin-5 and γ-synergin bind directly to GGA2 and γ1-adaptin (Hirst et al., 2000; Shiba et al., 2002). The structural basis for the interaction involves binding of γ-synergin to a shallow hydrophobic groove surrounded by basic residues in the β-sandwich fold of γ-adaptin (Kent et al., 2002; Nogi et al., 2002). The function of these interactions in membrane transport, however, has not been explored. Given the role of rab4, we investigated the function of rabaptin-5α and γ1-adaptin in membrane recycling through the endosomal system. We found that rabaptin-5α directly and functionally associates with γ1-adaptin on REs via binding to rab4.
Results
Rabaptin-5α directly binds γ1-adaptin
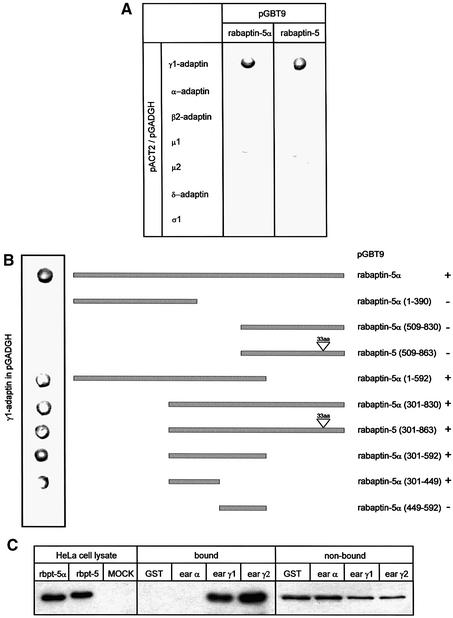
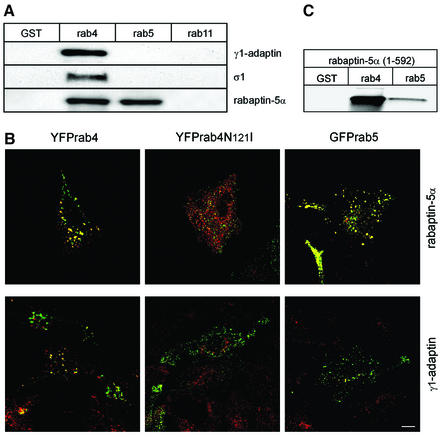
Rabaptins are bivalent cytoplasmic effectors of rab4 and rab5 that are recruited to EEs by the active form of the two GTPases (Stenmark et al., 1995; Vitale et al., 1998; Nagelkerken et al., 2000). Rabaptin-5α and rabaptin-5 differ in the most C-terminal coiled-coil region and, because the γ-adaptin-binding site on rabaptin-5 was not known, we first determined in yeast two-hybrid assays whether rabaptin-5α (like rabaptin-5) interacted with the heterotetrameric adaptor complex AP-1. As shown in Figure 1A, rabaptin-5α binds selectively to the γ-subunit of AP-1. The same results were obtained with rabaptin-5, suggesting that the C-terminus of the rabaptins is not required for γ1-adaptin binding. Mutation analysis also ruled out that the DPF motif at position 388 is involved in binding (not shown). Using rabaptin-5α truncations, we then mapped the binding domain for γ1-adaptin to amino acid residues 301–449 (Figure 1B). The ear–hinge region of α-adaptin contains an interaction platform involved in binding endocytic accessory proteins. For γ1-adaptin, only one protein, γ-synergin, is known to interact with its ear region and to be recruited to membranes (Page and Robinson, 1995). Two-hybrid assays (Supplementary figure 1 available at The EMBO Journal Online) showed that rabaptin-5α also binds to the ear–hinge of γ1-adaptin and of γ2-adaptin. γ2-adaptin interacts with the σ1 chain of AP-1 and with σ1B, implying the existence of an AP-1-related complex (see Boehm and Bonifacino, 2001) that is functionally distinct from AP-1 (Zizioli et al., 1999). We confirmed the two-hybrid results in pull-down assays of brain extract proteins where rabaptin-5α was specifically isolated by glutathione beads carrying γ1- and γ2-adaptin (Figure 1C).
Fig. 1. Rabaptin-5α(301–449) directly interacts with the ear of γ-adaptin. (A) Yeast two-hybrid assays with rabaptin-5α and rabaptin-5 showing a specific and direct interaction with γ1-adaptin. (B) Summary of two-hybrid assays with rabaptin-5α truncations showing that the domain required for γ1-adaptin binding is between amino acids 301 and 449. Binding is indicated by +. (C) Binding of rabaptin-5α to GST fusions containing the ear domain of γ1-adaptin and γ2-adaptin. GST fusion proteins were incubated with detergent-solubilized brain proteins. Bound proteins were analysed by western blot using a rabaptin-5α antibody. As size control, we used detergent lysates prepared from HeLa cells in which rabaptin-5α and rabaptin-5 were transfected with the vaccinia T7 RNA polymerase system.
Rabaptin-5α and γ1-adaptin co-localize on endosomes
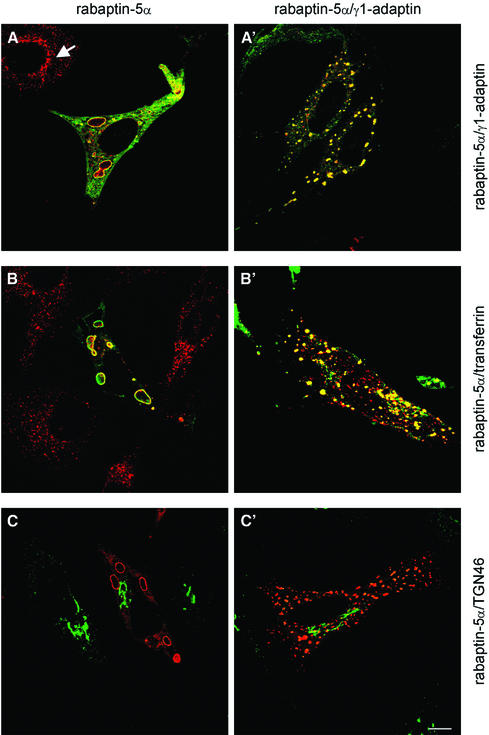
γ-adaptin is associated predominantly with the trans-Golgi network (TGN) (Robinson, 1990) and also with EEs (Stoorvogel et al., 1996). We therefore investigated whether rabaptin-5α and γ1-adaptin localized to one of these compartments. HeLa cells were transfected either with only His-rabaptin-5α or with His-rabaptin-5α and γ1-adaptin, and then processed for immunofluorescence (IF) microscopy. His-rabaptin-5α extensively co-localized with endogenous γ1-adaptin on enlarged intracellular structures of variable size, which correlated with the expression level of rabaptin-5α. Not all γ1-adaptin co-localized with rabaptin-5α. We also found γ1-adaptin labelling in the juxtanuclear region (cf. Figure 4E), as in non-transfected cells where γ1-adaptin was localized mainly in the perinuclear area (Figure 2A, arrow). In double-transfected cells, His-rabaptin-5α and γ1-adaptin were both located to cytoplasmic structures that were somewhat smaller than those in His-rabaptin-5α single transfectants (Figure 2A′). Western blots revealed that co-transfection of γ1-adaptin did not affect expression of rabaptin-5α (Supplementary figure 2). The structures on which γ1-adaptin and rabaptin-5α co-localized were endosomes, since the majority of endocytosed transferrin (Tf) was present in them (Figure 2B and B′). These structures were devoid of LAMP-1 (not shown), suggesting that they represented EEs/REs. Since γ1-adaptin is also on the TGN, we labelled the single- (Figure 2C) and double-transfected cells (Figure 2C′) with an antibody against TGN46. The majority of TGN46 is on the TGN, while a small pool was found on the endosomal structures that are positive for rabaptin-5α and γ1-adaptin. These probably represented TGN46 molecules that are cycling between the plasma membrane and the TGN, a pathway that intersects the EE (Ghosh et al., 1998).
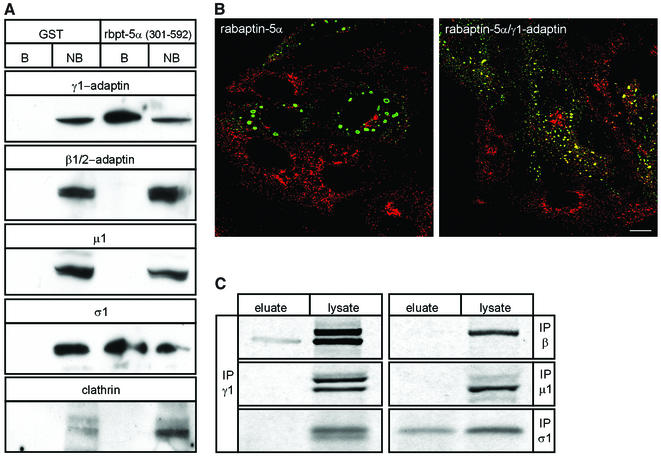
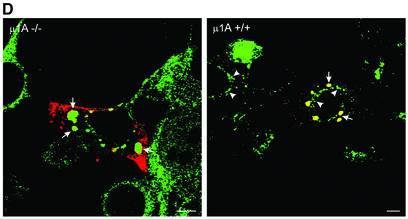
Fig. 4. γ1–σ1 AP-1 subcomplex is retained on rabaptin-5α beads. (A) GST or GST–rabaptin-5α(301–592) were incubated with glutathione–Sepharose and a HeLa cell extract. The western blot of bound (B) and non-bound (NB) fractions was probed with antibodies against γ1, β1/2, σ1, µ1 and clathrin. (B) IF of clathrin and rabaptin-5α in cells transfected with rabaptin-5α or co-transfected with rabaptin-5α and γ1-adaptin. Cells were permeabilized, fixed and labelled with a rabbit antibody against rabaptin-5α (green) and X-22 against clathrin (red). (C) HeLa cells were labelled with [35S]methionine and the lysate was incubated with GST–rabaptin-5α(301–592). Bound material was eluted with glutathione. Eluate and cell lysate were incubated with antibodies against γ1 (100/3), β1/2 (100/1), µ1 and σ1, and protein A beads. Immunoprecipitates were resolved by SDS–PAGE and analysed by phosphorimaging. To detect σ1 in the GSH eluate, gels were exposed 10 times longer than for γ1-adaptin (σ1 contains four times less methionine residues than γ1-adaptin). (D) Rabaptin-5α and γ1-adaptin co-localize on endosomes in the absence of intact AP-1. µ1A–/– and µ1A+/+ fibroblasts were transfected with rabaptin-5α-pcDNA3.1His. For γ1-adaptin labelling in µ1A+/+ fibroblasts, cells were extracted with saponin before fixation. Note that γ1-adaptin in these cells is not only present on the structures containing rabaptin-5α (arrows), but also in the TGN area (arrow heads). Saponin treatment of µ1A–/– cells removed all γ1-adaptin labelling (not shown). Cells were fixed, and labelled with a rabbit antibody against rabaptin-5α (red) and mouse γ1-adaptin antibody (Transduction labs) (green). Bar, 10 µm.
Fig. 2. Rabaptin-5α–γ-adaptin complex is localized on endosomes. HeLa cells were transfected with rabaptin-5α-pcDNA3.1His (left panels) or with rabaptin-5α and γ1-adaptin-pcDNA3 (right panels). Transfected cells were labelled for rabaptin-5α (green) and γ1-adaptin (red). Note the co-localization of rabaptin-5α and γ1-adaptin in transfected cells, and the distinct localization of γ1-adaptin in non-transfected cells (arrow) (A and A′). Cells were incubated with Alexa594-Tf for 60 min at 37°C and subsequently labelled with anti-Xpress antibody and Alexa488-conjugated IgG (B and B′). The TGN marker TGN46 does not relocate to enlarged endosomes. Transfected cells were labelled for rabaptin-5α (red) and TGN46 (green) (C and C′). Bar, 10 µm.
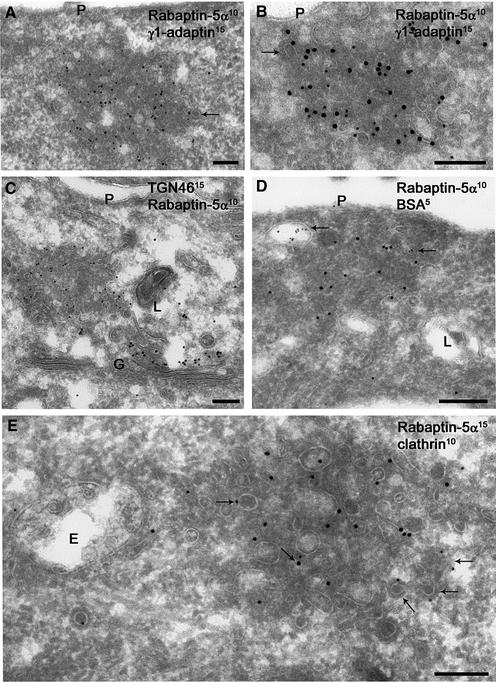
We next extended the IF results with immunogold electron microscopy (IEM). Although IF microscopy gave the impression that the rabaptin-5α-positive endocytic structures were single organelles which occasionally contained γ1-adaptin in their lumen (Figure 2), it is clear from IEM that the rabaptin-5α- and γ1-adaptin-positive membranes actually represent clusters of small tubules and vesicles (Figure 3). These structures are reminiscent of REs by their diameter and electron density. They differ from REs in control cells in that the membranes are clustered and less regular in size. Clearly, the rabaptin-5α- and γ1-adaptin-containing membranes have a very different morphology from that of the electron-luscent EEs and multivesicular bodies (de Wit et al., 1999) (Figure 3E). In the single (Figure 3A) and double (Figure 3B) transfectants, rabaptin-5α and γ1-adaptin co-localized on these tubulovesicular endosomes, which did not label for TGN46 (Figure 3C). Labelling for TGN46 also showed that the morphology of the TGN was normal in transfected cells, and that a pool of γ1-adaptin but no rabaptin-5α localized to the TGN (not shown). To rule out that rabaptin-5α in addition to recycling tubules also associated with primary endocytic vesicles, cells were allowed to internalize bovine serum albumin (BSA) conjugated to 5 nm gold for 10 min. BSA–gold primarily follows the pathway to the lysomes and rarely enters recycling tubules (de Wit et al., 1999). As shown in Figure 3D, BSA–gold-containing vesicles were not labelled for rabaptin5α. Some of the membranes of rabaptin-5α-positive clusters displayed a typical clathrin coat (arrows in Figure 3A and B). Indeed, double labelling revealed that some of the rabaptin-5α-positive membranes also labelled for clathrin (Figure 3E).
Fig. 3. Rabaptin-5α and γ-adaptin co-localize to tubulo-vesicular membrane clusters. Ultrathin cryosections of HeLa cells transfected with rabaptin- 5α-pcDNA3.1His (A,C and E) or with rabaptin-5α and γ1-adaptin-pcDNA3 (B and D). Double labelling of rabaptin-5α (10 nm gold) and γ1-adaptin (15 nm gold). Rabaptin-5α is associated with clusters of vesicular tubular membranes, where it co-localized with endogenous (A) and overexpressed γ1-adaptin (B). The overall density and diameter of rabaptin-5α-positive membranes is reminiscent of recycling tubules. The dense cytosol between the membranes indicates the presence of high concentrations of cytosolic protein. Note that some of the membranes display a coating typical of the presence of clathrin (arrows in A and B). Double labelling of TGN46 (15 nm gold) and rabaptin-5α (10 nm gold) revealed that rabaptin-5α-positive membranes do not overlap with TGN membranes (C). Rabaptin-5α- (15 nm gold) positive membranes do not contain internalized BSA–5 nm gold. Arrows point to compartments that contain the endocytic marker 10 min after internalization (D). Double labelling of rabaptin-5α (15 nm gold) and clathrin (10 nm gold). Some of the membranes within or associated with rabaptin-5α-positive membranes also stained for clathrin (arrows). Note that the nearby endosomal vacuole (E) has a normal morphology (E). G = Golgi complex, L = lysosome. Bar, 200 nm.
Localization of rabaptin-5α–γ1-adaptin is not dependent on heterotetrameric AP-1
To investigate whether rabaptin-5α-associated γ1-adaptin is part of heterotetrameric AP-1, we tested for the presence of AP-1 subunits in pull-down experiments with GST–rabaptin-5α(301–592) and HeLa extract proteins. As shown in Figure 4A, γ1-adaptin and the σ1 subunit were detected on GST–rabaptin-5α(301–592) beads. Associa tion of σ1 with the beads was mediated by γ1-adaptin, because the σ1-subunit did not bind directly to rabaptin-5α (Figure 1A). To our surprise, β1-adaptin and µ1-adaptin were not retrieved on the beads. Since β1-adaptin contains a clathrin-binding motif (Shih et al., 1995), we also assayed for clathrin. Consistent with the absence of β1-adaptin, we did not detect clathrin (Figure 4A). γ1-adaptin has been reported to interact directly with clathrin in vitro (Doray and Kornfeld, 2001). Possibly the two clathrin boxes in the hinge of γ1-adaptin are hindered sterically by GST–rabaptin-5α(301–592) under the conditions of the pull-down assay. Alternatively, the efficiency of binding of rabaptin-5α to γ1-adaptin and of γ1-adaptin to clathrin might be insufficient to detect γ1-adaptin-bound clathrin on rabaptin-5α beads. We next sought a morphological correlate for the result of the pull-down experiment using cells transfected with rabaptin-5α (Figure 4B). IF showed abundant clathrin labelling, including in the TGN area. Very little clathrin, however, was found on the rabaptin-5α-labelled endosomes. Co-transfection with γ1-adaptin increased clathrin localization on rabaptin-5α-containing endosomes.
To confirm the presence of a γ1–σ1 subcomplex, we incubated GST–rabaptin-5α(301–592) with metabolically labelled HeLa cell lysates. Bound proteins were co-eluted with GST–rabaptin-5α(301–592) using glutathione and immunoprecipitated with antibodies against AP-1 subunits. All subunits of newly synthesized AP-1 were co-immunoprecipitated with γ1-adaptin from the cell lysate, but not from the eluate of the rabaptin beads (Figure 4C). The subunits that were immunoprecipitated from the eluate were γ1-adaptin and σ1-adaptin. In agreement with the existence of a γ1-adaptin pool that is not included in intact AP-1, we found a small (<0.05% of total γ1-adaptin) but reproducible peak of γ1-adaptin at ∼120 kDa by size exclusion chromatography of mouse fibroblast cytosol (Supplementary figure 3). The majority of γ1-adaptin and α-adaptin (not shown) eluted at the position of intact AP-1 and AP-2 (Meyer et al., 2000), respectively.
We then examined if intact AP-1 is required for co-localization of rabaptin-5α and γ1-adaptin on endosomes, using fibroblasts derived from µ1A -adaptin ‘knockout’ mice. These cells contain heterotrimeric AP-1 complexes as well as dimeric γ1–β1-adaptin complexes that are not recruited onto membranes (Meyer et al., 2000). µ1A-deficient-cells and µ1A-deficient cells retransfected with µ1A cDNA were transfected with rabaptin-5α and double-labelled with antibodies against rabaptin-5α and γ1-adaptin. IF documented (Figure 4D) that overexpression of rabaptin-5α caused enlarged endosomes that were decorated with rabaptin-5α and γ1-adaptin in the µ1A-deficient as well as in the rescued cells. This showed that intact heterotetrameric AP-1 is not essential for the localization of rabaptin-5α and γ1-adaptin on endosomes, and that the endosomal localization of rabaptin-5α and γ1-adaptin is not a unique feature of HeLa cells. Pre-permeabilization of µ1A-deficient cells retransfected with µ1A cDNA revealed that γ1-adaptin also localized to the TGN area (Figure 4D, right panel, arrowheads). In µ1A-deficient cells, pre-permeabilization removes all γ1-adaptin (not shown), confirming that γ1-adaptin fails to bind tightly to membranes in these cells (Meyer et al., 2000).
Rab4 regulates localization of rabaptin-5α–γ1-adaptin on endosomes
Because rab4 and rab5 recruit rabaptin-5α to EEs, it was interesting to investigate whether they might serve as a spatial cue to localize the complex to endosomal membranes. To test biochemically whether rab4 and rab5 bound the rabaptin-5α–γ1-adaptin complex, we performed pull-down assays with GTPγS-loaded rab proteins and pig brain cytosol (Figure 5A). Rabaptin-5α, γ1-adaptin and σ1-chain were only retrieved on GST–rab4 beads, while rabaptin-5α, expectedly, was also captured on rab5 beads. Importantly, this experiment corroborated the presence of an endogenous rabaptin-5α–γ1-adaptin–σ1-chain complex in brain cytosol. We next checked with IF whether rabaptin-5α–γ1-adaptin complex localized in a rab4-dependent manner to endosomes. Upon co-transfection of yellow fluorescent protein (YFP)–rab4 or green fluorescent protein (GFP)–rab5 with rabaptin-5α, we found a significant fraction of γ1-adaptin co-localizing with rab4, but not with rab5 (Figure 5B). For technical reasons, we could not detect rabaptin-5α–γ1-adaptin simultaneously in this experiment. In Figure 2A, however, we already showed that most of the transfected rabaptin-5α localized with γ1-adaptin on endosomes. In agreement with the pull-down assays, transfected rabaptin-5α also co-localized with rab5 (Figure 5B). Control transfections with YFP–rab4-N121I, a dominant-negative rab4 mutant, and rabaptin-5α showed mutually exclusive distributions of YFP–rab4-N121I and γ1-adaptin.
Fig. 5. Rab4 recruits rabaptin-5α–γ1-adaptin. Rab4, but not rab5 and rab11 interacts with the endogenous rabaptin-5α–γ1–σ1 complex. (A) GST, GST–rab4, GST–rab5 and GST–rab11 were isolated on glutathione–Sepharose, loaded with GTPγS and incubated with pig brain cytosol. Bound fractions were immunoblotted with antibodies against γ1-adaptin, σ1 and rabaptin-5α. (B) Localization of γ1-adaptin to endosomes depends on rab4. HeLa cells were transfected with His-rabaptin-5α in combination with YFP–rab4, dominant-negative YFP–rab4-N121I or GFP–rab5. Cells were labelled with a monoclonal antibody against γ1-adaptin (100/3) (red) and a rabbit antibody against rabaptin-5α (green). Bar, 10 µm. (C) Rab4 is the major binding partner of the N-terminal binding domain on rabaptin-5α. GST fusion proteins were isolated on glutathione–Sepharose, loaded with GTPγS and incubated with 35S-labelled rabaptin-5α(1–592). Bound material was eluted with glutathione, resolved by SDS–PAGE and quantitated by phosphorimaging.
The principal binding domains for rab4 and rab5 on rabaptins are in the N- and C-termini, respectively (Vitale et al., 1998; Nagelkerken et al., 2000). We and others previously reported an additional putatitive binding site for rab5 in the N-terminus of rabaptins (Vitale et al., 1998; Nagelkerken et al., 2000). Although the morphological and biochemical experiments suggested that rab5 binding to rabaptin-5α was not important for recruitment of rabaptin-5α–γ1-adaptin complex, we nevertheless re-examined the issue of rab5 binding to the N-terminus of rabaptin-5α. Quantitation of pull-down assays (Figure 5C) with GTPγS-loaded rab4 or rab5 and 35S-labelled rabaptin-5α(1–592) revealed 10-fold more binding to rab4 than to rab5. Collectively, the IF and binding data show that rab4 is important for the localization of the rabaptin-5α–γ1-adaptin complex to endosomes.
Rabaptin-5α and γ1-adaptin regulate Tf recycling
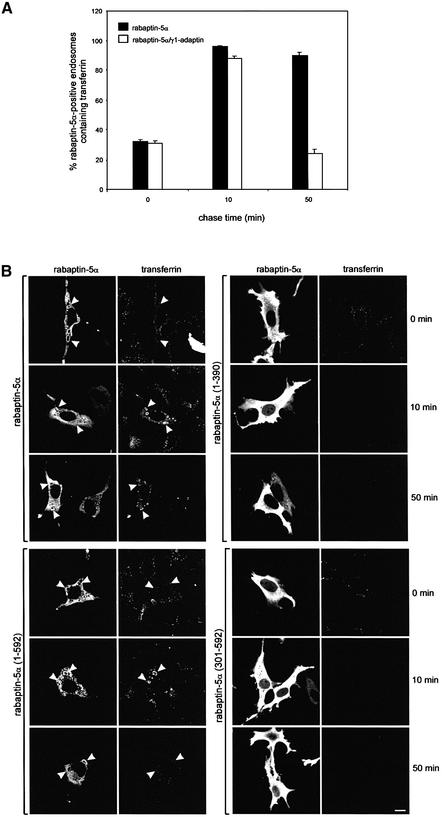
To address the function of the rabaptin-5α and γ1-adaptin complex, we investigated Tf kinetics in cells expressing His-rabaptin-5α or His-rabaptin-5α and γ1-adaptin. Cells grown were incubated for 30 min with 15 µg/ml Alexa-Tf at 16°C to accumulate the ligand in EEs. The cells were then washed and subsequently chased at 37°C in the presence of 50 µM desferroxamine. Fixed cells were labelled with an antibody against rabaptin-5α and we quantitated the number of rabaptin-5α-positive endosomes containing Tf at 0, 10 and 50 min chase. Within 10 min of chase, Tf reached the REs that were decorated with the XPRESS antibody (confocal images of the pulse–chase experiment are shown in Supplementary figure 4). As shown in Figure 6A, at the beginning of the chase, the number of rabaptin-5α-positive endosomes containing Tf is the same for the single and double transfectants. At 50 min chase, however, the single rabaptin-5α transfectants had four times more rabaptin-5α-labelled endosomes containing Tf than the double transfectants. The kinetics of Tf recycling in cells transfected with both rabaptin-5α and γ1-adaptin were comparable with those of non-transfected cells and cells transfected with only γ1-adaptin (not shown). To extend these results, we performed recycling experiments in cells expressing rabaptin-5α truncations (Figure 6B). This figure shows that the delayed exit of Tf from EEs is only seen in cells expressing rabaptin-5α(1–592) containing both the γ1-adaptin- and rab4-binding sites (Supplementary figure 5). Expression of either rab4- or γ1-adaptin binding domains did not affect Tf recycling. In fact, it appeared that Tf recycling was even slightly faster than in non-transfected cells. Collectively, the overexpression experiments reveal a functional link between rabaptin-5α and γ1-adaptin in the exit of Tf from endosomal structures.
Fig. 6. Transfected rabaptin-5α delays Tf recycling. (A) HeLa cells were transfected with rabaptin-5α-pcDNA3.1His or with rabaptin-5α-pcDNA3.1His and γ1-adaptin-pcDNA3. The cells were incubated with 15 µg/ml Alexa-Tf at 16°C for 30 min, and subsequently chased at 37°C. Cells were fixed after different periods of time and stained for rabaptin-5α. Quantitation of the fraction of rabaptin-5α-positive endosomes containing Alexa488-Tf at 0, 10 and 50 min of chase. Error bars denote the standard deviation (n = 10). (B) Rab4 and γ1-adaptin interaction domains in rabaptin-5α are required to retard Tf recycling. HeLa cells were transfected with rabaptin-5α(1–390), rabaptin-5α(301–592), rabaptin-5α(1–592) and rabaptin-5α. The cells were subjected to the pulse–chase protocol as above, fixed and labelled with anti-Xpress followed by Alexa488-labelled anti-mouse IgG. Note that only the rabaptin-5α truncation containing both rab4- and γ1-adaptin-binding sites retarded Tf recycling. Bar, 10 µm.
Knockdown of rabaptin-5α and rab4 by RNAi
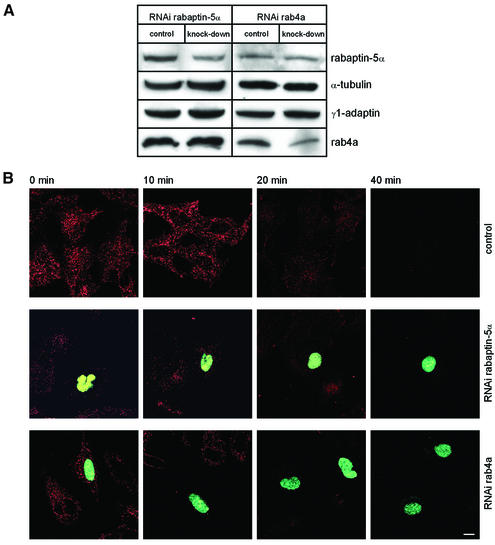
To analyse the function of the rab4–rabaptin-5α– γ1-adaptin interaction in a more subtle way, we used RNA interference (RNAi) to lower expression of two components of the complex. Since γ1-adaptin is essential for cell growth and development (Zizioli et al., 1999), we chose to target rab4a and rabaptin-5α for knockdown experiments. RNAi was carried out using the novel pKoen vector that directs synthesis of small interfering RNAs. pKoen contains a nuclear localization signal at the C-terminus of GFP which provides a convenient means to identify transfectants. HeLa cells were transfected with either pKoen (mock), pKoen-rab4a or pKoen-rabaptin-5α. Quantitation of western blots (Figure 7A) showed that expression of rab4a and rabaptin-5α was reduced by 30 and 28%, respectively. After correction for transfection efficiency (see below), we calculate that expression of rab4a and rabaptin-5α per cell was reduced >90%. Parallel dishes with cells on coverslips were subjected to Alexa-Tf pulse–chase experiments as in Figure 6. Quantitation of cells expressing GFP revealed that in 35% of the cells, rab4a was affected, while this was 25% for rabaptin-5α. RNAi of rab4a and rabaptin-5α produced distinct phenotypes as compared with non-transfected cells on the same coverslips (Figure 7B). Knocking down rab4a did not appear to affect internalization of Alexa-Tf at 16°C; however, during the chase at 37°C, the tracer was cleared faster from cells with knocked-down rab4a than from control cells. A different phenotype was observed in cells with knocked-down rabaptin-5α. Internalization of Alexa-Tf was severely reduced in those cells. Since rabaptins are required for rab5 function (Gournier et al., 1998), the drastic phenotype most probably reflects perturbation of a transport step proximal to rab4-dependent recycling. Although the low intracellular level of Tf at the end of the pulse complicated analysis of recycling, we noticed that the tracer could be chased from the cells at 37°C.
Fig. 7. RNAi of rab4a and rabaptin-5α. (A) HeLa cells were transfected with pKoen (mock), pKoen-rab4a or pKoen-rabaptin-5α for 3 days. Western blots were probed with antibodies against rab4a, rabaptin-5α and tubulin (loading control). In neither case was expression of the other two components of the complex affected. (B) Cells were incubated with 15 µg/ml Alexa-Tf at 16°C for 30 min, chased for different periods of time at 37°C, and processed for fluorescence microscopy. Cells with knocked-down rab4a or rabaptin-5α contain GFP in the nucleus. RNAi of rab4a accelerates exit of Tf from the cells, while reduced rabaptin-5α expression caused a strong inhibition of Tf internalization.
Discussion
Little is known about the proteins that regulate recycling from endosomes. We previously showed that rab4 is an important factor in this process (van der Sluijs et al., 1992b). We now report that the rab4 effector rabaptin-5α interacts with γ1-adaptin and that a complex of rab4– rabaptin-5α–γ1-adaptin is involved in exocytosis of Tf. Others recently reported binding of γ-adaptin to rabaptin-5 and γ-synergin (Hirst et al., 2000; Shiba et al., 2002), the function of which, however, had not been explored.
The association between rabaptin-5α and γ1-adaptin was direct and involved amino acids 301–449 in the hinge region of rabaptin-5α, and the ear domain of γ1-adaptin. Shiba et al. (2002) recently reported that the major binding domain for γ1-adaptin on rabaptin-5 was between amino acids 546 and 728, while Mattera et al. (2003) showed that the interaction with GGA2 required the 439FGPLV443 sequence in rabaptin-5. Our results are in agreement with the latter study because this recognition determinant is in the hinge of rabaptin-5α. The rabaptin-5α–γ1-adaptin complex was localized on membranes with the typical morphology of REs. These also contained the majority of internalized Tf, and labelled for rab11 (not shown), both of which are hallmarks of REs. The endosomal localization of the complex depended on the rab4-binding domain in rabaptin-5α, and expression of a dominant-negative rab4 mutant that does not bind rabaptin-5α inhibits localization of the complex to endosomes. Since rab4 is localized in the endosomal system, but not the TGN, this also explains why the complex was absent from the TGN. Thus rab4-GTP might serve as a docking site for the rabaptin-5α– γ1-adaptin complex and provides a spatial cue for the association of a pool of γ1-adaptin on EEs/REs. Although we confirmed that rabaptin-5α is recruited to endosomes by rab5, we found no evidence for a role for rab5 in localizing γ1-adaptin to endosomal membranes.
Several lines of evidence suggest that the interaction is functionally relevant. First, overexpression of rabaptin-5α or a rabaptin-5α truncation mutant containing both rab4- and γ1-adaptin-binding domains caused enlarged endosomes and a delay in Tf recycling. Interestingly, epsinR, another ligand of the γ1-adaptin ear domain, recently was shown to co-localize with internalized Tf, and also to generate enlarged perinuclear structures upon transfection (Mills et al., 2003). Because rabaptin-5α stabilizes rab4-GTP (Nagelkerken et al., 2000), its overexpression is predicted to recapitulate some of the effects seen after overexpression of rab4. Indeed, we found in earlier studies that overexpression of rab4a delayed clearance of internalized Tf from cells (van der Sluijs et al., 1992b). An additional small effect of overexpressed rabaptin-5α commenced, since there was a short delay in Tf transfer to the REs (Figure 6B). Secondly, RNAi of rab4 and rabaptin-5α affected the Tf receptor (TfR) cycle. As suggested by rab4a overexpression studies, knockdown of rab4a enhanced recycling. The phenotype of rabaptin-5α knockdown experiments was more complex, because rabaptin-5α, rabaptin-5 and rabaptin-5δ have identical sequences in regions that can be used for silencing. This strongly suggested that the latter two can also be targeted by our constructs. Importantly, since rabaptin-5 is necessary for rab5 function, its knockdown may reduce transfer of endocytosed Tf to EEs during the pulse (Figure 7B). This provides very low loading of the endosomal system with Tf, which technically makes it difficult to measure downstream (rab4-dependent) recycling as there is already very little Tf at the beginning of the chase. Although subtle differences in recycling might go unnoticed due to the low signals, we observed that Tf was chased-out from these cells.
AP-1 mediates transport between endosomes and the TGN (Meyer et al., 2000), and γ1-adaptin has been suggested to be important for the formation of clathrin-coated vesicles, returning TfR to the cell surface via REs (Stoorvogel et al., 1996). Because rab4 and rabaptin-5α distributions partially overlap with rab5 on EEs and with rab11 and cellubrevin on REs (Nagelkerken et al., 2000; Sönnichsen et al., 2000), our findings agree with earlier observations (van der Sluijs et al., 1992b) and support a negative regulatory role for rab4 in transport between EEs and REs. Rabaptin-5α might stabilize a transient γ1–σ1 intermediate during AP-1 assembly on rab4-containing endosomes. This model has similarities to the formation of COP-I and COP-II vesicles, where coats are assembled by stepwise targeting of subcomplexes (Lowe and Kreis, 1995; Matsuoka et al., 1998). Heterotetrameric AP complexes were imagined as rock-solid building blocks of the vesicle coat due to their stability in vitro. This picture changed after the crystal structure of the AP-2 complex was compared with functional analyses. These data indicate that the C-terminal two-thirds of the µ-adaptin flip out of the complex exposing the sorting signal-binding domain to the membrane and the cytoplasmic tails of cargo proteins. The same mechanism may occur in AP-1, if the µ-adaptin cargo-binding domain flips out, then the complex will be held together just by the γ1–β1 interaction, weakening complex stability (Ghosh and Kornfeld, 2003).
The inhibition of Tf exit from REs and their enlarged appearance by IEM and IF microscopy suggest that overexpressed rabaptin-5α disrupts AP-1-dependent budding of recycling vesicles from endosomes. In agreement with this, overexpression of epsinR perturbs cathepsin-D trafficking from the TGN and generates enlarged perinuclear compartments that are thought to arise due to inhibition of coated vesicle formation (Mills et al., 2003). Since both epsinR and rabaptin-5α bind to the γ1-adaptin ear, we conclude that normal AP-1 function requires carefully controlled interactions of AP-1 with accessory proteins. Possibly, overexpression of proteins binding to the γ1-adaptin ear causes dissociation of AP-1. Formation of a dimeric γ1–σ1 AP-1 complex is consistent with the crystal structure of the highly homologous AP-2 complex. This tetrameric adaptor conceptually is a heterodimer of tightly bound α–σ2 and β2–µ2 subcomplexes (Collins et al., 2002). Phylogenetic analysis of coat protein sequences also indicates that multisubunit adaptor complexes evolved from a dimeric complex consisting of a large and small subunit, which would thus form the smallest functional unit (see Boehm and Bonifacino, 2001). Because β-adaptins contain the essential clathrin-binding site, dissociation of the AP-1 complex explains the reduced amount of clathrin on the endosomal membranes, and is in agreement with the absence of clathrin, µ1 and β1 in rabaptin-5α pull-down assays. µ1 and β1 are the AP-1 subunits known to interact with sorting motifs of cargo proteins (see Boehm and Bonifacino, 2001). Dissociation of AP-1 into dimers would therefore prevent binding of this AP-1 subcomplex with its endosomal cargo proteins. Clathrin localization to endosomes could be restored by transfection of γ1-adaptin, which also reduced the size of this compartment. Since the γ1-adaptin hinge contains two clathrin-binding motifs, these probably served to recruit cytoplasmic clathrin, bypassing the need for β1-adaptin in budding. This minimal coat, however, does not have the ability to sort cargo proteins since it is missing the µ1-subunit. The present study reveals for the first time the existence and a biological activity of an AP-1-subcomplex. Although we found a minute pool of cytosolic γ1-adaptin that is not present in AP-1, it is not known how much of the γ1–σ1 dimer is present on endosomal membranes. Importantly, the transfection experiments in µ1A–/– fibroblasts showed that these cells retained the ability to localize rabaptin-5α and γ1-adaptin on endosomes even when there is no intact AP-1.
Rabaptin-5 was shown previously to bind to GGA2 and γ1-adaptin (Hirst et al., 2000; Shiba et al., 2002). GGAs localize to the TGN and possibly endosomes, and play a role in cargo sorting and transport from the TGN to endosomes as they recognize acidic dileucine sorting signals, and facilitate transport of the mannose-6-phosphate receptors from the TGN to endosomes (see Dell’Angelica and Payne, 2001). The significance of the rabaptin-5–γ-adaptin and rabaptin-5–GGA interactions, however, requires further investigations, particularly with respect to rab5 function at EEs and GGA2 function in transport from the TGN to endosomes. Interestingly, rabenosyn-5 regulates transport of lysosomal enzymes between the Golgi complex and endosomes (Nielsen et al., 2000) as does its binding partner Vps45p in yeast (Peterson et al., 1999). Conceivably, rab5 and some of its effectors may have additional roles beyond EE fusion. Since rab5 can functionally replace Ypt5 in fission yeast (Armstrong et al., 1993) and because Vps21p is required for sorting of vacuolar proteins (Horazdovski et al., 1994), this would be consistent with a pathway between the Golgi and endosomes regulated by rab5 and a subset of its effectors. Indeed, while this manuscript was under review, Mattera et al. (2003) identified a divalent interaction between GGA–rabaptin-5 and the rab5 guanine nucleotide exchange protein rabex5. Rabaptin-5 was shown to reduce clathrin binding to GGAs, while the novel complex is thought to link formation of transport vesicles at the TGN with endosome tethering events.
Several rabaptin mRNAs have been found that, with the exception of rabaptin-5β, are generated by alternative splicing (Korobko et al., 2002). The available antibodies, however do not allow a distinction to be made between the endogenous proteins, and functional differences between them have not been reported. It is therefore tempting to speculate that a redundancy of bivalent effector proteins might strengthen the coupling between rab5 effector domains and rab4 effector domains that control entry to and exit from endocytic organelles, respectively. In support of this, preliminary results (P.van der Sluijs, unpublished) show that rabaptin-5α and rabaptin-5 heterodimerize in vitro. In summary, our findings show a functional link between rab4 and a γ1–σ1 subcomplex of AP-1 and may help to obtain a better understanding of endosomal membrane dynamics.
Materials and methods
Cell culture and transfection
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS), and µ1A+/+ and µ1A–/– mouse fibroblasts were maintained as described (Meyer et al., 2000). Cells were grown to 20% confluency and transfected using FuGENE 6 (Boehringer Mannheim, Almere, The Netherlands) or calcium phosphate. Cells received 5 mM sodium butyrate 18 h before experiments to induce expression of cytomegalovirus (CMV)-driven constructs. For some experiments, HeLa cells were transfected with the recombinant T7 RNA polymerase vaccinia system. For RNAi knockdown experiments of rab4a and rabaptin-5α, transfected cells were harvested and lysed after 24, 48, 72, 96 and 120 h. Western blots of detergent lysates were analysed for expression and showed that silencing was achieved after 3 days.
Antibodies
A rabbit antibody against rabaptin-5α was generated by injecting animals with purified His-rabaptin-5α(11–388). Antibodies against rab4a/b, rab4a, tubulin and the His6 Xpress epitope have been described before (van der Sluijs et al., 1992a; Mohrmann et al., 2002). Rabbit antibodies against µ1 and σ1 were generously provided by Linton Traub; the mouse monoclonal antibody X-22 was given by Frances Brodsky. Polyclonal antibodies against γ1-adaptin and clathrin were generous gifts of Scottie Robinson and Silvia Corvera, respectively. The following antibodies were from the indicated sources, sheep anti-TGN46 (Serotec, Oxford, UK), mouse monoclonals 100/3 against human γ1 and 100/2 against β1/2 (Sigma, Dordrecht, The Netherlands), goat anti-clathrin heavy chain (Santa Cruz, Heerhugowaard, The Netherlands) and mouse monoclonal anti-γ1-adaptin (Transduction Laboratories, Breda, The Netherlands). Horseradish peroxidase (HRP)- and fluorescently labelled reagents were from sources previously described (Mohrmann et al., 2002).
Plasmids
Recombinant fusion proteins were expressed from pGEX plasmids (Pharmacia, Roosendaal, The Netherlands). Rabaptin constructs were cloned in pcDNA3 and pcDNA3.1His (Invitrogen, Leek, The Netherlands). Adaptin two-hybrid plasmids were generously provided by Juan Bonifacino and Kazuhisa Nakayama. Truncation constructs were generated by ligating PCR fragments in the appropriate plasmids. Rab4, rab4-N121I and rab5 were cloned in pEYFP-C3 and pEGFP-C2 (Westburg, Leusden, The Netherlands), respectively. The RNAi plasmid pKoen was constructed as follows: a BglII–BamHI fragment from pEGFP was excised to remove the BglII and HindIII sites. A nuclear localization signal was introduced at the C-terminus of EGFP by ligating the oligonucleotides 5′-ACGATCCAAAAAAGAAGAGAAAGGTAGATC CAAAAAAGAAGAGAAAGGTAT-3′ (forward) and 5′-ATACCTTT CTCTTCTTTTTTGGATCTACCTTTCTCTTCTTTTTTGGATCGT-3′ (reverse) in the ScaI site. Finally, the H1 promotor from pSuper (Brummelkamp et al., 2002) was excised with PvuII and inserted in the blunted AseI site of the plasmid. Oligos used for RNAi of rabaptin-5α and rab4a (available on request) were inserted between the BglII and HindIII sites of pKoen. All synthetic cDNAs were verified by restriction analysis and dye termination sequencing.
GST pull-down assays
Detergent extracts were pre-cleared for 2 h at 4°C with 20 µg of GST and 60 µl of 50% (v/v) glutathione beads (Pharmacia). Cleared extract was incubated at 4°C with 20 µg of C-terminal GST–adaptin or GST– rabaptin-5α(301–592) and 60 µl of 50% (v/v) glutathione beads. After 2–4 h, beads were pelleted, washed five times with phosphate-buffered saline (PBS), 0.1% NP-40, and boiled in reducing Laemmli sample buffer. Samples were resolved on 6 or 12.5% SDS–polyacrylamide minigels, and analysed by western blot. Detection was with enhanced chemiluminescence reagent (Boehringer Mannheim). Pull-down assays with GST–rab fusions were carried out as described (Nielsen et al., 2000). For some binding experiments, GST–rabaptin-5α(301–592) was incubated with a detergent lysate of [35S]methionine-labelled HeLa cells as described below. Bound proteins were eluted with 50 mM glutathione and subjected to immunoprecipitation. Bound 35S-labelled proteins were detected by phosphorimaging on a STORM 860.
Metabolic labelling and immunoprecipitation
HeLa cells were labelled with 1 mCi/ml of 35S Promix and lysed in 1 ml of buffer A (1% Triton X-100, 50 mM Tris–HCl pH 7.4 or 8, 0.3 M NaCl, 0.1% BSA). Detergent lysates were cleared in a microfuge (10 min at 15 000 r.p.m.) and the lysates were used for immunoprecipitation. A 15 µl aliquot of protein A–Sepharose was washed with PBS, 1% BSA and subsequently incubated with either 10 µl of anti-γ1 100/3 or anti-β1/2 100/1, or 5 µl of anti-µ1 or anti-σ1 antibody at 4°C for 2 h. Beads with bound antibodies were incubated with 500 µl of cell lysate at 4°C for 2 h. The beads were then washed three times with buffer A followed by two washes with buffer A without Triton X-100, and bound material was boiled for 5 min in 20 µl of reducing Laemmli sample buffer. Samples were resolved by SDS–PAGE, and detection of bound 35S-labelled proteins was by phosphorimaging.
Fluorescence microscopy
Cells were grown for 24–48 h on coverslips and incubated for 30 min in serum-free α- minimal essential medium (MEM), washed once with uptake medium (α–MEM, 20 mM HEPES–KOH, 0.5% BSA) and incubated with 25 µg/ml human Tf labelled with Alexa488 or Alexa 594 at 37°C. Cells were first fixed with 2.5% paraformaldehyde (PFA) for 15 min at 37°C, followed by 30 min of 2.5% PFA at room temperature. For pulse–chase experiments, cells were incubated with 15 µg/ml Alexa-labelled Tf for 30 min at 16°C. After one wash with α-MEM, 20 mM HEPES–KOH 0.5% BSA, the cells were re-incubated with chase medium (α-MEM, 20 mM HEPES–KOH, 0.5% BSA, 50 µM desferroxamine) for different periods of time, fixed and subjected to confocal IF microscopy (Mohrmann et al., 2002). For γ1-adaptin labelling in µ1A-retransfected µ1A–/– mouse fibroblasts, and clathrin labelling in HeLa cells, cells were extracted with saponin before fixation (Bucci et al., 1992). A semi-quantitative estimate of the effect of rabaptin-5α on Tf kinetics was made by selecting five rabaptin-5α-positive endosomes (per cell and per chase time) and counting how many of those contained Tf. This analysis was performed in 10 cells per time point and in duplicate transfections.
Immunoelectron microscopy
Cells were fixed and processed for IEM as described (de Wit et al., 1999). For labelling of TGN46, glutaraldehyde was omitted from the fixative. BSA conjugated to 5 nm colloidal gold (final OD520 = 5) was used to mark primary endocytic vesicles. Cells were incubated with serum-free MEM containing 1% FCS and BSA–5 nm gold in a CO2 incubator at 37°C. After 10 min, cells were quickly rinsed with ice-cold serum-free MEM, and fixed.
Miscellaneous methods
Yeast two-hybrid assays, purification of GST fusion proteins (Nagelkerken et al., 2000) and preparation of detergent extracts (Page et al., 1999) were performed as described. Gel filtration of mouse fibroblast cytosol on a Sepharose 200 column was carried out as before (Meyer et al., 2000). In vitro transcription–translation reactions were performed as described by the vendor (Promega, Haarlem, The Netherlands) and immediately used in GST pull-down experiments.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Linton Traub, Scottie Robinson, Silvia Corvera, Juan Bonifacino, Kazuhisa Nakayama and Frances Brodsky for generously providing antibodies and cDNAs, and Reuven Agami for pSuper. We are indebted to Marc van Peski and Rene Scriwanek for their help in preparing the figures. This research was supported by NWO-ALW and NWO-MW (P.v.d.S.) and by the DFG (P.S.).
References
- Armstrong J.A., Craighead,M.W., Watson,R., Ponnambalam,S. and Bowden,S. (1993) S.pombe ypt5: a homologue of the rab5 endosome fusion regulator. Mol. Biol. Cell, 4, 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M. and Bonifacino,J.S. (2001) Adaptins: the final recount. Mol. Biol. Cell, 12, 2907–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- Bucci C., Parton,R., Mather,I., Stunnenberg,H., Simons,K. and Zerial,M. (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell, 70, 715–728. [DOI] [PubMed] [Google Scholar]
- Collins B.M., McCoy,A.J., Kent,H.M., Evans,P.R. and Owen,D.J. (2002) Molecular architecture and functional model of the endocytic AP2 complex. Cell, 109, 523–535. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E.C. and Payne,G.S. (2001) Intracellular cycling of lysosomal enzyme receptors: cytoplasmic tails tales. Cell, 106, 395–398. [DOI] [PubMed] [Google Scholar]
- de Renzis S., Sönnichsen,B. and Zerial,M. (2002) Divalent rab effectors regulate the sub-compartmental organization and sorting function of early endosomes. Nat. Cell Biol., 4, 124–133. [DOI] [PubMed] [Google Scholar]
- de Wit H., Lichtenstein,Y., Geuze,H., Kelly,R.B., van der Sluijs,P. and Klumperman,J. (1999) Synaptic vesicles form by budding from tubular extensions of sorting endosomes in PC12 cells. Mol. Biol. Cell, 10, 4163–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B. and Kornfeld,S. (2001) γ-subunit of the AP-1 adaptor complex binds clathrin: implications for cooperative binding in coated vesicle assembly. Mol. Biol. Cell, 12, 1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P. and Kornfeld,S. (2003) AP-1 binding to sorting signals and release from clathrin-coated vesicles is regulated by phosphorylation. J. Cell Biol., 160, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R.N., Mallet,W.G., Soe,T.T., McGraw,T.E. and Maxfield,F.R. (1998) An endocytosed TGN38 chimeric protein is delivered to the TGN after trafficking through the endocytic recycling compartment in CHO cells. J. Cell Biol., 142, 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier H., Stenmark,H., Rybin,V., Lippe,R. and Zerial,M. (1998) Two distinct effectors of the small GTPase rab5 cooperate in endocytic membrane fusion. EMBO J., 17, 1930–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Sacher,M., Barrowman,J., Ferro-Novick,S. and Novick,P. (2000) Protein complexes in transport vesicle targeting. Trends Cell Biol., 10, 251–255. [DOI] [PubMed] [Google Scholar]
- Hirst J., Lui,W.W.Y., Bright,N.A., Totty,N., Seaman,M.N.J. and Robinson,M.S. (2000) A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans Golgi network and the vacuole/lysosome. J. Cell Biol., 149, 67–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovski B.F., Busch,G.R. and Emr,S. (1994) VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J., 13, 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent H.M., McMahon,H.T., Evans,P.R., Benmarah,A. and Owen,D.J. (2002) γ-adaptin appendage domain: structure and binding site for eps15 and γ-synergin. Structure, 10, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Korobko E.V., Kiselev,S.L. and Korobko,I.V. (2002) Multiple rabaptin-5-like transcripts. Gene, 292, 191–197. [DOI] [PubMed] [Google Scholar]
- Lowe M. and Kreis,T.E. (1995) In vitro assembly and disassembly of coatomer. J. Biol. Chem., 270, 31364–31371. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Orci,L., Amherdt,M., Bednarek,S., Hamamoto,S., Schekman,R. and Yeung,T. (1998) COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell, 93, 263–275. [DOI] [PubMed] [Google Scholar]
- Mattera R., Arighi,C.N., Lodge,R., Zerial,M. and Bonifacino,J.S. (2003) Divalent interaction of the GGAs with the rabaptin-5–rabex-5 complex. EMBO J., 22, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Zizioli,D., Lausmann,S., Eskelinen,E.,L., Hamann,J., Saftig,P., von Figura,K. and Schu,P. (2000) µ1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J., 19, 2193–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills I.G., Praefcke,G.J.K., Vallis,Y., Peter,B.J., Olesen,L.E., Gallop,J.L., Butler,P.J.G., Evans,P.R. and McMahon,H.T. (2003) Epsin R: an AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol., 160, 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann K., Leijendekker,R., Gerez,L. and van der Sluijs,P. (2002) Rab4 regulates transport to the apical plasma membrane in Madin–Darby canine kidney cells. J. Biol. Chem., 277, 10474–10481. [DOI] [PubMed] [Google Scholar]
- Nagelkerken B., van Anken,E., van Raak,M., Gerez,L., Mohrmann,K., van Uden,N., Holthuizen,J., Pelkmans,L. and van der Sluijs,P. (2000) Rabaptin4, a novel effector of rab4a, is recruited to perinuclear recycling vesicles. Biochem. J., 346, 593–601. [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis,S., Utenwiler-Joseph,S., Miaczynska,M., Dewitte,F., Wilm,M., Hoflack,B. and Zerial,M. (2000) Rabenosyn-5, a novel rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol., 151, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi T. et al. (2002) Structural basis for the accessory protein recruitment by the γ-adaptin ear domain. Nat. Struct. Biol., 9, 527–531. [DOI] [PubMed] [Google Scholar]
- Page L.J. and Robinson,M.S. (1995) Targeting signals and subunit interactions in coated vesicle adaptor complexes. J. Cell Biol., 131, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L., Sowerby,P.J., Lui,W.W.Y. and Robinson,M.S. (1999) γ-synergin: an EH domain-containing protein that interacts with γ-adaptin. J. Cell Biol., 146, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.R., Burd,C.G. and Emr,S.D. (1999) Vac1p coordinates rab and phosphoinositol 3-kinase signaling in a vps45-dependent vesicle docking/fusion at the endosome. Curr. Biol., 9, 159–162. [DOI] [PubMed] [Google Scholar]
- Robinson M.S. (1990) Cloning and expression of γ-adaptin, a component of clathrin coated vesicles associated with the Golgi apparatus. J. Cell Biol., 111, 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Takatsu,H., Shin,H.W. and Nakayama,K. (2002) γ-adaptin interacts directly with rabaptin-5 through its ear domain. J. Biochem., 131, 327–336. [DOI] [PubMed] [Google Scholar]
- Shih W., Galusser,A. and Kirchhausen,T. (1995) A clathrin-binding site in the hinge of the β2 chain of mammalian AP-2 complexes. J. Biol. Chem., 270, 31083–31090. [DOI] [PubMed] [Google Scholar]
- Sönnichsen B., De Renzis,S., Nielsen,E., Rietdorf,J. and Zerial,M. (2000) Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of rab4, rab5 and rab11. J. Cell Biol., 149, 901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Vitale,G., Ullrich,O. and Zerial,M. (1995) Rabaptin-5 is a direct effector of the small GTPase rab5 in endocytic membrane fusion. Cell, 83, 423–432. [DOI] [PubMed] [Google Scholar]
- Stoorvogel W., Oorschot,V. and Geuze,H.J. (1996) A novel class of clathrin coated vesicles budding from endosomes. J. Cell Biol., 132, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P., Hull,M., Huber,L.A., Male,P., Goud,B. and Mellman,I. (1992a) Reversible phosphorylation–dephosphorylation determines the localization of rab4 during the cell cycle. EMBO J., 11, 4379–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P., Hull,M., Webster,P., Goud,B. and Mellman,I. (1992b) The small GTP binding protein rab4 controls an early sorting event on the endocytic pathway. Cell, 70, 729–740. [DOI] [PubMed] [Google Scholar]
- Vitale G., Rybin,V., Christoforidis,S., Thornqvist,P.O., McCaffrey,M., Stenmark,H. and Zerial,M. (1998) Distinct rab-binding domains mediate the interaction of rabaptin-5 with GTP-bound rab4 and rab5. EMBO J., 17, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M. and McBride,H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol., 2, 107–117. [DOI] [PubMed] [Google Scholar]
- Zizioli D., Meyer,C., Guhde,G., Saftig,P., von Figura,K. and Schu,P. (1999) Early embryonic death of mice deficient in γ-adaptin. J. Biol. Chem., 274, 5385–5390. [DOI] [PubMed] [Google Scholar]