Abstract
SH3 domains are protein recognition modules within many adaptors and enzymes. With more than 500 SH3 domains in the human genome, binding selectivity is a key issue in understanding the molecular basis of SH3 domain interactions. The Grb2-like adaptor protein Mona/Gads associates stably with the T-cell receptor signal transducer SLP-76. The crystal structure of a complex between the C-terminal SH3 domain (SH3C) of Mona/Gads and a SLP-76 peptide has now been solved to 1.7 Å. The peptide lacks the canonical SH3 domain binding motif P–x–x–P and does not form a frequently observed poly-proline type II helix. Instead, it adopts a clamp-like shape around the circumfence of the SH3C β-barrel. The central R–x–x–K motif of the peptide forms a 310 helix and inserts into a negatively charged double pocket on the SH3C while several other residues complement binding through hydrophobic interactions, creating a short linear SH3C binding epitope of uniquely high affinity. Interestingly, the SH3C displays ion-dependent dimerization in the crystal and in solution, suggesting a novel mechanism for the regulation of SH3 domain functions.
Keywords: crystal structure/Gads/Mona/SH3 domain dimer/SLP-76
Introduction
Discovery and characterization of Src homology 2 and 3 (SH2 and SH3) domains have been fundamental in understanding many aspects of intracellular signalling processes. These small, globular and highly versatile protein interaction modules contribute to key signal transduction events, such as regulation of cell growth and activation of immune defence cells, but also appear as structural components in a variety of systems. Mammalian genomes contain hundreds of SH3 domains [for a comprehensive overview see http://smart.embl-heidelberg.de/ (Schultz et al., 1998; Letunic et al., 2002)]. A large number of SH3 domains recognize specific proline-rich sequences which adopt the conformation of polyproline type II helices (PPII) and contain the core consensus motif P–x–x–P (Bork et al., 1997; Kay et al., 2000; Mayer, 2001; Musacchio, 2002). The SH3 domain interactions with the core motifs of their ligands are mediated through stacking of aromatic rings from residues like Trp, Tyr and Phe with the pyrrolidine ring of the prolines in the core consensus motif. Binding specificity and affinity beyond the core motif are generated through additional hydrophobic or charge interactions. Recent studies show that atypical SH3 domain binding motifs which lack the P–x–x–P core consensus exist (Manser et al., 1998; Cestra et al., 1999; Mongiovi et al., 1999; Barnett et al., 2000; Kang et al., 2000; Kato et al., 2000; Ghose et al., 2001; Lewitzky et al., 2001; Nishida et al., 2001; Kami et al., 2002).
Adaptor proteins are implicated in the transmission of many intracellular signals, in particular near the plasma membrane. They usually consist of multiple protein interaction domains (like SH2 and SH3) and/or docking sites for these domains. The hematopoietic adaptor Mona (monocytic adaptor), a member of the Grb2 family, was cloned as a yeast-two hybrid (Y2H) binding partner of the receptor kinase c-Fms/CSF1-R (Bourette et al., 1998). Simultaneously and shortly thereafter, several other groups obtained similar clones which were named Gads (Grb2-related adaptor downstream of Shc), Grap2, GrpL, Grf40, GRID and GRBLG (Liu and McGlade, 1998; Qiu et al., 1998; Asada et al., 1999; Law et al., 1999; Ellis et al., 2000). The name Mona/Gads used throughout this article takes into account the first two publications (Bourette et al., 1998; Liu and McGlade, 1998).
Mona/Gads binds to several intracellular signalling proteins, including SLP-76, LAT, Shc, GCIP, c-Cbl, HPK1 and Gab3 (Liu et al., 2000, 2001; Tomlinson et al., 2000; Xia et al., 2000; Bourgin et al., 2002; Burack et al., 2002; Yankee et al., 2003). Targeted disruption of the Mona/Gads gene locus leads to a proliferation block in CD4–CD8– thymocytes and an impaired response of thymocytes to cross-linking of their CD3 surface antigen (Yoder et al., 2001). Co-immunoprecipitation experiments further showed that the SLP-76–LAT complex induced by CD3 stimulation is disrupted in the absence of Mona/Gads protein. Tyrosine-phosphorylated LAT is recognized through the Mona/Gads SH2 domain. In contrast, the interaction with SLP-76 is mediated by the SH3C domain of Mona/Gads (Liu et al., 1999). The SLP-76 binding site for Mona/Gads SH3C has been mapped (Liu et al., 1999; Berry et al., 2002) and is practically identical to the SLP-76 region reported to bind the Grb2 SH3C domain (Lock et al., 2000; Lewitzky et al., 2001). In addition, Drk, the Drosophila homologue of mammalian Grb2 and Mona/Gads, binds two sequence motifs in the ‘Daughter of Sevenless’ (Dos) protein which strongly resemble the SLP-76 binding site of Grb2 and Mona/Gads (P–x3–R–x2–K–P) (Feller et al., 2002). Drk and Dos are both crucial players in the development of the R7 photoreceptor cell of the compound fly eye.
Several mammalian signalling proteins also contain sequences with the P–x3–R–x2–K–P motif (Berry et al., 2002; Feller et al., 2002). Examples include AMSH, which interacts with Mona/Gads in the Y2H screen (Asada et al., 1999), the B-cell adaptor SLP-65/BLNK, the Gab family of multisite docking proteins (Gab1, Gab2, Gab3) and the ubiquitin peptidase USP8 [formerly UBPY (Naviglio et al., 1998)]. Gab3 has recently been shown to form complexes with Mona/Gads during monocyte/macrophage differentiation (Bourgin et al., 2002). USP8 binds to Mona/Gads in vitro, but we have been unable to co-immunoprecipitate the endogenously expressed proteins from cell lysates (M.Lewitzky and S.M.Feller, unpublished data).
To understand the molecular basis of the Mona/Gads SH3C domain binding selectivity in more detail, a large panel of known and potential binding motifs was analysed using isothermal titration calorimetry (ITC). The binding motif with the highest affinity was subsequently cocrystallized with the SH3C domain. The three-dimensional structure of the complex reveals a novel type of SH3–peptide interaction in which the peptide clamps onto the binding site. Moreover, the SH3C forms an ion-mediated dimer within the crystal, which was subsequently also detected in solution.
Results and discussion
Analysis of known and potential Mona/Gads SH3C domain binding motifs
Table I shows the results of ITC measurements analysing different peptides for binding to Mona/Gads SH3C and two homologous SH3C domains from Grb2 and Drk (see Figure 1 for SH3 domain sequence alignment and homology). Initially, the region essential for efficient binding of Mona/Gads SH3C was determined. A previous study had raised the possibility that a relatively large motif, P–x3–R–x2–K–P–x7–P–L–D, might be necessary for mouse SLP-76 and mouse Gab1 binding to the homologous Grb2 SH3C domain (Lock et al., 2000). The P–L–D motif of mouse SLP-76 is replaced by P–F–D in human SLP-76 (Jackman et al., 1995) and P–L–E in human Gab1, but is retained in the human and mouse Gab2 and Gab3 proteins. For the Mona/Gads SH3C we did not find evidence for a role of residues distant to the P–x3–R–x2–K–P motif. Peptides of 23 and 21 amino acids derived from SLP-76 and Gab3, the in vivo binding partners of Mona/Gads (P1, P4, P5, P26; see Table I for sequences, relevant P–L–D or P–F–D motifs underlined), did not bind any better than a 13 amino acid SLP-76 peptide (P2). The Kd for the complex between P2 and Gads/Mona SH3C (amino acids 256–322) was 0.181 µM, a remarkably tight binding for SH3 domain interactions. In contrast, the affinity of P2 for the Grb2 SH3C domain was about 50-fold lower (Kd = 9.7 µM). Binding of an 11 amino acid SLP-76 peptide (P3) to Mona/Gads SH3C was somewhat weaker (Kd = 0.675 µM) than P2 binding.
Table I. Analysis of peptide binding affinities by isothermal titration calorimetry.
| Peptide | Kd (µM) | ||
|---|---|---|---|
| P1 | HKTAKLPAPSIDRSTKPPLDR | SLP-76(1) aa 225–245 (21aa) | 0.329 |
| P2 | PAPSIDRSTKPPL | SLP-76(1) aa 231–243 (13aa) | 0.181/0.118a/9.699(G3C) |
| P3 | APSIDRSTKPP | SLP-76(1) aa 232–242 (11aa) | 0.675 |
| P4 | PAPSIDRSTKPPLDRSLAPLDRE | SLP-76(1) aa 231–253 (23aa) m | 0.540 |
| P5 | PAPSIDRSTKPPLDRSLAPFDRE | SLP-76(1) aa 231–253 (23aa) | 0.574 |
| P6 | PPPPAGRNHSPLP | SLP-76(2) aa 198–210 (13aa) | NBD |
| P7 | TKPPLDRSLAPFD | SLP-76(3) aa 239–251 (13aa) | NBD |
| P8 | KPPFSDKPSIPAG | SLP-76(4) aa 260–272 (13aa) | NBD |
| P9 | HLPKIQKPPLPPT | SLP-76(5) aa 278–290 (13aa) | NBD |
| P10 | PLPLPNKPRPPSP | SLP-76(6) aa 399–411 (13aa) | NBD |
| P11 | PLPNKPRPPSPAE | SLP-76(7) aa 401–413 (13aa) | NBD |
| P12 | PAASIDRSTKPPL | SLP-76(1) aa 231–243 (mut P1→A) | 3.056 |
| P13 | PAPSIARSTKPPL | SLP-76(1) aa 231–243 (mut D→A) | 4.562 |
| P14 | PAPSIERSTKPPL | SLP-76(1) aa 231–243 (mut D→E) | 1.247 |
| P15 | PAPSIKRSTKPPL | SLP-76(1) aa 231–243 (mut D→K) | 2.587 |
| P16 | PAPSIDASTKPPL | SLP-76(1) aa 231–243 (mut R→A) | NBD |
| P17 | PAPSIDRSTAPPL | SLP-76(1) aa 231–243 (mut K→A) | NBD |
| P18 | PAPSIDRSTRPPL | SLP-76(1) aa 231–243 (mut K→R) | 8.368 |
| P19 | PAPSIDRSTKAPL | SLP-76(1) aa 231–243 (mut P2→A) | 4.118 |
| P20 | PAPSIDRSTKVPL | SLP-76(1) aa 231–243 (mut P2→V) | 1.292 |
| P21 | PDIPPPRPPKPHP | Gab1(1) aa 338–350 (13aa) | 146.242 |
| P22 | EPPPVDRNLKPDR | Gab1(2) aa 515–527 (13aa) | 0.603 |
| P23 | RNLKPDRKVKPAP | Gab1(3) aa 521–533 (13aa) | 55.792 |
| P24 | SNTPPPRPPKPSH | Gab3(1) aa 304–316 (13aa) | 11.179 |
| P25 | EPPPVNRDLKPQR | Gab3(2) aa 434–446 (13aa) | 0.387 |
| P26 | PPPVNRDLKPQRKSRPPPLDLR | Gab3(2) aa 434–456 (23aa) | 0.747 |
| P27 | NVPQIDRTKKPAV | USP8(1) aa 403–415 (13aa) | 0.940 |
| P28 | VTPTVNRENKPTC | USP8(2) aa 736–748 (13aa) | 5.767 |
| P29 | KPPVVDRSLKPGA | AMSH aa 229–241 (13aa) | 0.473 |
| P30 | KAPMVNRSTKPNS | SLP-65/BLNK aa 202–214 (13aa) | 0.921 |
| P31 | PPPPLPPKKRR | PxxPxK peptide (11 aa) | NBD+/25.891(G3N) |
| P32 | PPPPLPPRRRR | PxxPxR peptide (11 aa) | 20.739/5.404(G3N) |
| P33 | DCPPVNRKLKPKV | Dos(1) aa 638–650 (13aa) D.m. | 23.125(D) |
| P34 | GPPSVDRKCKPNA | Dos(2) aa 690–702 (13aa) D.m. | 92.460(D) |
The SH3 domain used is Gads/Mona SH3C (aa 256–322) expressed as a GST fusion protein unless indicated otherwise.
aGads/Mona SH3C aa 265–322 without GST tag and gel filtration purified was used.
(G3C), Grb2 SH3C domain; (G3N), Grb2 SH3N domain (both GST fusion proteins).
(D), Drk SH3C domain (GST fusion protein, SH3 sequence from Drosophila melanogaster).
Peptide sequences are derived from human proteins unless indicated otherwise: m, mouse; D.m., D.melanogaster.
aa, amino acids; NBD, no binding detectable; +, very low affinity, not quantifiable.
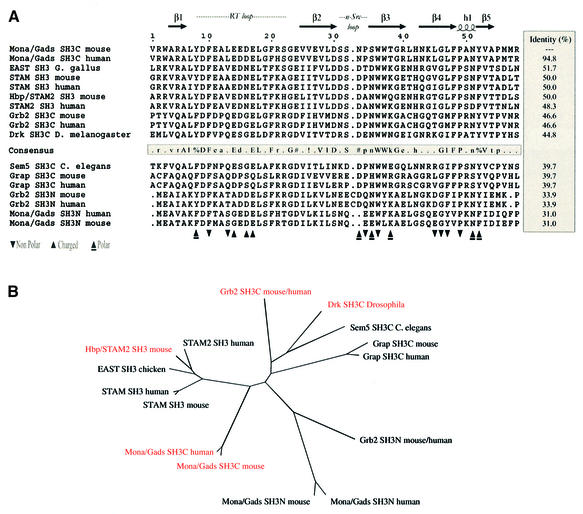
Fig. 1. (A) Sequence alignment of selected SH3 domains. The secondary structure of the Mona/Gads SH3C is indicated at the top. Triangles refer to the residues of mouse Mona/Gads SH3C interacting with the SLP-76 peptide according to the crystal structure. ‘n-Src loop’ and ‘RT loop’ highlight regions named after sequence variations of viral and cellular Src proteins. Amino acid numbering for the Mona/Gads SH3C in this figure and throughout this article follows from the Mona/Gads SH3C boundaries of the construct used for the crystal structure determination. Amino acid consensus of SH3 domains with more than 40% identity with Mona/Gads SH3C is indicated as follows: threshold for capital letters, 90% identical; threshold for lower-case letters, 50% identical; !, either Ile or Val; %, either Phe or Tyr; #, Asn, Asp, Gln or Glu. (B) The dendrogram shows how selected SH3 domains relate to each other according to amino acid sequence homology. SH3 domains reported to bind to motifs with the consensus P–x3–R–x2–K–P are coloured. Based upon the homology analysis performed with MultAlin and PHYLIP, the closest homologues of the Mona/Gads SH3C domain are the SH3 domains of HBP/STAM2, STAM and EAST. HBP/STAM2 binds to USP8 in a forced expression system (Kato et al., 2000). No structural information on this SH3 domain is available to date.
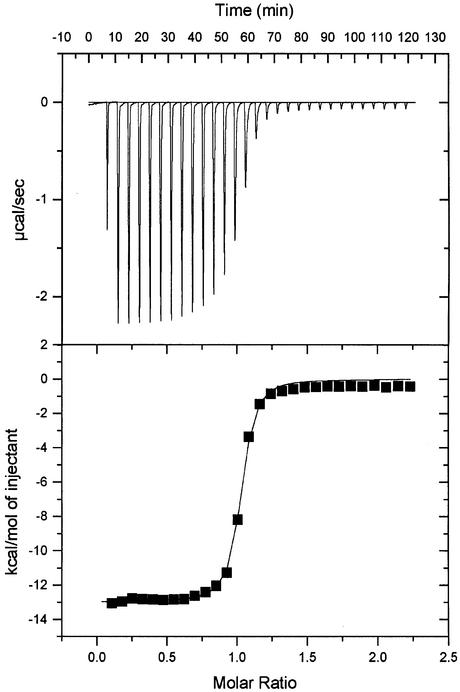
Subsequently, a Mona/Gads SH3C domain, truncated for crystallization purposes (amino acids 265–322) and purified by gel filtration chromatography after cleavage of the GST tag, was analysed by ITC. Its Kd for the SLP-76 peptide P2 was 0.118 µM (Table I; Figure 2). To our knowledge, this is the highest affinity measured in solution for an SH3 binding peptide of this length.
Fig. 2. Representative result from the ITC measurements. A SLP-76 peptide with 13 amino acids (P2) was titrated into a chamber filled with highly purified Gads/Mona SH3C (amino acids 265–322). Kd is ∼0.118 µM.
SLP-76 contains six motifs which lack the Lys residue of the P–x3–R–x2–K–P motif and instead conform to the broader consensus P–x3–R/K–x3–P. None of these bound detectably to the Mona/Gads SH3C domain (P6–P11), suggesting that a single positive amino acid flanked in the –4 and +4 positions by prolines is not sufficient for stable binding.
Replacement of single residues in the SLP-76 peptide P2 showed that mutation of Arg or Lys in the P–x3–R–x2–K–P motif to Ala (P16, P17) abolished binding to Mona/Gads SH3C. Substitution of Lys by Arg (P18) led to a 50-fold reduction in binding affinity. These findings point to a crucial role for Arg and Lys in Mona/Gads SH3C binding. A substantial loss of binding was also observed when the prolines of the P–x3–R–x2–K–P motif were changed to alanines (P12, P19). Substitution of the Pro following the Lys by Val (P20) reduced the affinity 7-fold. Replacement of the Asp preceding Arg (P–x2–D–R–x2–K–P) by Ala (P13) was similarly effective in reducing the affinity, suggesting that this position also contributes to the strong binding of the wild-type sequence P2. Changing Asp to the residue Glu (P14), which is also negatively charged, was only slightly less disruptive. Surprisingly, replacement of Asp by Lys (P15) did not disrupt binding substantially more than the replacement by Glu.
To gain further insight into the binding affinities of naturally occurring protein epitopes with a complete or partial P–x3–R–x2–K–P motif, a panel of peptides derived from SLP-65/BLNK, Gab1, Gab3, AMSH and USP8 was analysed (P21–P30).
Peptides binding with better than micromolar affinity were identified for each of these proteins (P22, P25, P27, P29, P30) and always contain the full P–x3–R–x2–K–P motif. Other binding peptides with lower affinities were also detected (P21, P23, P24, P28). With the exception of the P28 peptide derived from USP8, these share only a reduced consensus motif (R–x2–K–P). Two more typical SH3 domain binding peptides with the classical P–x–x–P–x–K and P–x–x–P–x–R motifs (P31, P32) bind at best poorly to the Mona/Gads SH3C domain.
Beyond mammalian proteins, it was recently shown that the Drosophila adaptor Drk needs two epitopes containing the motif P–x3–R–x2–K–P in the Dos protein to determine correctly the R7 cell fate in the compound eye of the fly (Feller et al., 2002). Therefore binding of Dos-derived peptides (P33, P34) to Drk SH3C was also investigated. With Kd values of 23 and 92 µM, respectively, both peptide interactions are much weaker than those observed for SLP-76 and several other mammalian sequences discussed above. Nevertheless, both sequences are clearly required for normal fly eye development. This suggests to us that an as yet unknown mechanism may generate a stable Dos–Drk complex and also emphasizes the need to investigate in vivo functions of protein epitopes with low in vitro binding affinities.
Structure of the SH3–SLP-76 peptide complex
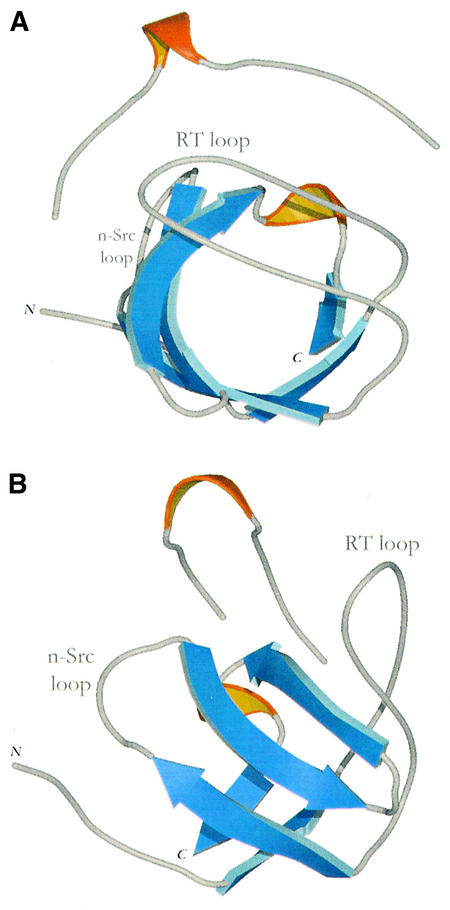
The structure of the Mona/Gads SH3C in complex with P2 was determined by multiwavelength anomalous dispersion (MAD) using a single crystal of selenomethionated Mona/Gads SH3C (Table II). The resulting density-modified map allowed unambiguous interpretation of the polypeptide chains. The SH3C domain displays the classical β-barrel-like fold typical of all known SH3s (Figure 3). It comprises five β-strands (residues 4–6, 25–30, 36–41, 44–49 and 53–55; see also Figure 2) and a 310-helical segment at residues 50–52, as defined by DSSP (Kabsch and Sander, 1983). There are two molecules in the asymmetric unit of the crystal in which residues –4 to 55 and 3 to 57, respectively, are visible (amino acids –4 to –1 result from a protease cleavage site overhang). Each domain is in a complex with a single peptide molecule docked along the circumference of the β-barrel. The bound peptides have obvious density for 12 of the 13 amino acids and an overall extended conformation with a 310 helix formed by residues 6–10.
Table II. Data reduction, phasing and refinement statistics.
| |
Peak |
Inflection |
Remote |
|---|---|---|---|
| Data collection | |||
| Resolution (Å)a | 1.69 (1.75–1.69) | ||
| Space group | P21 | ||
| Cell dimensions(Å) | a = 28.69, b = 72.07, c = 34.17 | ||
| Cell angles (°) | α = β =90, γ = 97.70 | ||
| Wavelength (Å) | 0.9786 | 0.9788 | 0.8731 |
| f′, f″ | –8.2, 7.2 | –10.1, 3.7 | –2.3, 4.0 |
| Total data | 99 470 | 104 025 | 80 439 |
| Unique data | 15 317 | 15 352 | 15 506 |
| Redundancy | 6.5 | 6.8 | 5.2 |
| Completeness (%) | 98.2 (81.4) | 98.1 (80.4) | 99.9 (99.9) |
| I/σ(I) | 28.2 (4.0) | 25.9 (3.3) | 22.2 (2.3) |
|
Rmerge (%)b |
5.5 (40.4) |
5.0 (50.6) |
4.7 (44.5) |
| Phasing |
|
|
|
| Rano (%)c | 5.7 | 4.5 | 4.9 |
| Rdisp (%)d | – | 3.4 | 4.8 |
| FOM (solve/resolve) |
0.4/0.45 |
|
|
| Refinement |
|
|
|
| Resolution range | 36.04–1.70 | ||
| No. of unique reflections | 13 807 | ||
| Rworke (Rfreef) | 17.86 (21.94) | ||
| R.m.s.d. bonds (Å) | 0.012 (0.021)g | ||
| R.m.s.d. angles (deg) | 1.375 (1.955)g | ||
| R.m.s.d. chiral (Å3) | 4.827 (5.000)g | ||
| R.m.s.d. main-chain bond B (Å2) | 0.770 (1.500)g | ||
| R.m.s.d. side-chain bond B (Å2) |
2.440 (3.000)g |
|
|
| Model quality (Ramachandran plot)h |
|
|
|
| Residues in most favoured regions (%) | 94.5 | ||
| Residues in additional allowed regions (%) |
5.5 |
|
|
| No. of atoms per asymmetric unit (average B values) (Å2) |
|
|
|
| Protein | 919 (21.3) | ||
| Peptide | 179 (23.0) | ||
| Water | 239 (37.8) | ||
| Ions (Cd) | 1 (17.8) |
FOM, figure of merit (the cosine of the mean phase error); R.m.s.d., root mean square deviation.
aValues in parentheses correspond to the highest resolution shell.
bRmerge = ΣjΣh (|Ij,h – <Ih>|)/ΣjΣh(<Ih>), where h is the unique reflection index, Ij,h is the intensity of the symmetry-related reflection and <Ih> is the mean intensity.
cRano = Σ|I(+) – I(–)|/Σ|<I>| for anomalous differences, where <I> is the average of Friedel amplitudes.
dRdisp = Σ|Iλ1 – Iλ2|/Σ|<I>| for dispersive differences, where <I> is the average amplitude at two wavelengths λ1 and λ2.
eR = Σh||Fo|h – |Fc|h|/ Σh|Fo|h, where h defines the unique reflections.
fCalculated on a random 5% of the data.
gTarget values in parentheses.
hValues from PROCHECK (Laskowski et al., 1993).
Fig. 3. Two views of the Mona/Gads SH3C structure in complex with the 13 amino acid SLP-76 peptide (P2). β-strands are coloured blue and 310 helices are shown in orange. (A) View looking down the length of the β-barrel. (B) View rotated by 90° on the vertical axis.
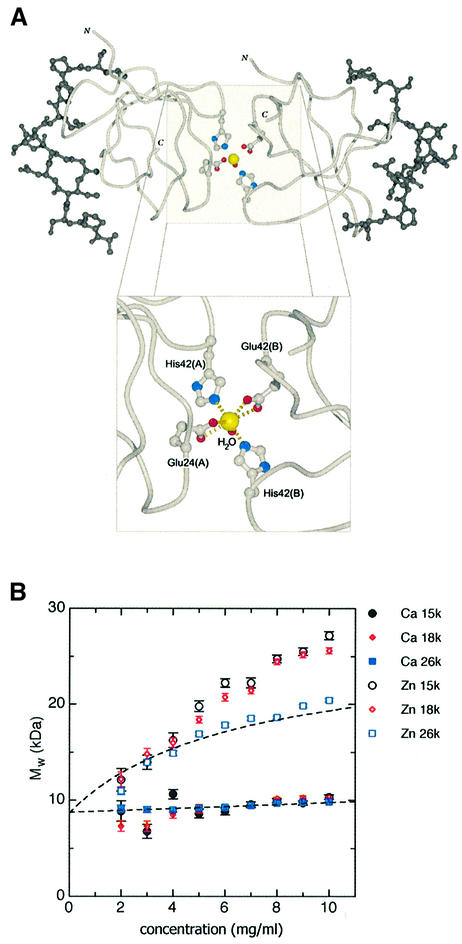
The two SH3 domains within the asymmetric unit form a dimer through the coordination of a cadmium ion present in the crystallization medium. The seven-coordinate site is arranged in a pentagonal bipyramid. Five atoms coordinate in plane with the cadmium atom (Oε1 and Oε2 of Glu24 from each SH3 domain and a water molecule) and two atoms at near-orthogonal positions to the plane on either side (Nε2 of His42 from each domain). This suggests that Mona/Gads SH3C has the potential to homodimerize in the presence of physiologically relevant ions like calcium or zinc, as discussed further later.
Model features and electrostatic potential of the SH3–peptide contact region
The buried surface area at the peptide–SH3 interface is 1179 Å2. This is nearly identical with the 1200 Å2 interaction surface described for the Hck SH3 domain in complex with the HIV Nef protein core, which binds with a Kd of 0.25 µM (Lim, 1996; Musacchio, 2002). The contact area in the Hck SH3–Nef complex not only results from the binding of a short peptide but is complemented by α-helical elements in HIV Nef interacting with the RT loop of the Hck SH3 domain. The buried surface area of the PPII-helical short linear peptide of Nef alone is only about 780 Å2 and the Kd of this peptide is 91 µM.
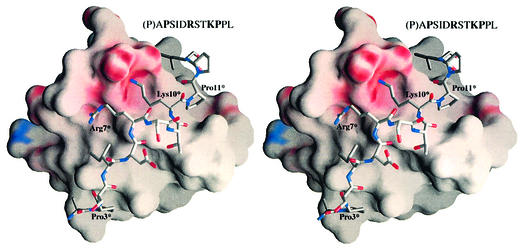
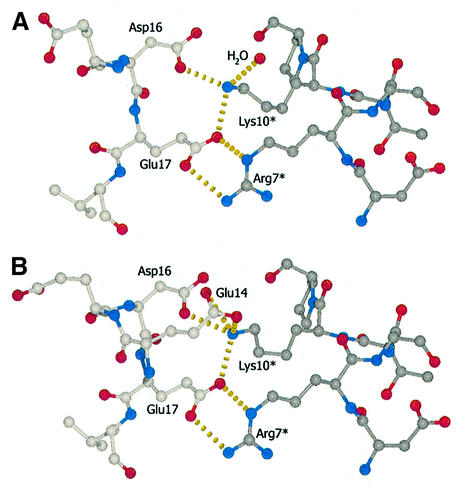
The SLP-76 peptide winds around the circumference of the barrel-like SH3C domain in a semicircular fashion. The corresponding docking area is overwhelmingly hydrophobic, except for one region of high negative electrostatic potential defining the interface of the peptide residues Arg7* and Lys10* (all SLP-76 peptide residues are denoted with an asterisk) with the SH3 domain (Figure 4). Figure 5 shows the SLP-76 peptide and interacting SH3 domain residues. Ala2* and Pro3* rest in a groove on the protein surface primarily defined by Tyr8 and Tyr52 which extend away from the body of the protein at 90° to each other. Pro3* in particular is involved in weak hydrophobic stacking with the ring of Tyr52 at an average distance of 4 Å. This could account for the loss of binding affinity upon substitution of Pro3* by Ala as described above. Ser4* is facing the solvent and is not likely to be a significant structural component during binding, while the side-chain of Ile5* is buried in the next hydrophobic groove defined by Mona/Gads residues Tyr52, Phe10 and Pro49 and complemented by the aliphatic region of Arg7* side-chain carbon atoms. The most distinctive structural feature within the otherwise extended peptide is a 310 helix formed by amino acids Arg7* to Lys10* (Figures 3 and 4). This feature depends in part upon the contribution of Asp6*, which stabilizes both Ser8* and Thr9* in the 310 arrangement through hydrogen bonds. Asp6* could conceivably be substituted by other residues that retain multiple hydrogen bonding potential without a significant effect on the structure. The latter could explain the binding characteristics of Asp6* to either Glu or Lys mutants (Table I, P14 and P15) as described above. The backbone oxygen atom of Asp6* also forms a hydrogen bond with Trp36 on the SH3C domain. Arg7* to Lys10* is a region of extensive interactions that seems to dominate the nature of the molecular interface. Arg7* hydrogen bonds with the Oε atoms of Glu17 in the SH3C. The same Glu17 is also involved in hydrogen bonding to Lys10*, along with Glu14 and Asp16. Glu14 adopts two equally stable conformations within the asymmetric unit (Figure 6). It either forms a shared hydrogen bond with both of its Oε atoms competing for the incoming hydrogen from Lys10* or faces away from it and hydrogen bonding is completed by a water molecule. Lys10* is further held in position through stacking interactions of its aliphatic side-chain atoms with the indole ring of Trp36. Pro11* is positioned facing yet another groove on the domain surface. This corroborates the ITC result obtained with peptides P19 and P20 where Pro11* is mutated to Ala and Val, respectively, resulting in reduced binding strength, presumably due to compromised hydrophobic interactions. Pro12* is exposed to the environment and does not appear to play an important role in SH3 domain docking. Binding is completed with Leu13* whose hydrophobic side-chain is positioned in a hydrophobic groove formed chiefly by Leu45 and Leu47, flanked by Asp16 and Thr38.
Fig. 4. Divergent stereo view of the electrostatic potential surface representation of the Mona/Gads SH3C in complex with the SLP-76 peptide (P2). Although the interface area of the Mona/Gads SH3C domain comprises mostly hydrophobic grooves (depicted in grey) where equivalent hydrophobic residues are docked, there is one prominent area of negative potential (shown in red) in the central region of the interface which promotes hydrogen bonding with the crucial Arg7* and Lys10* peptide residues (individually labelled SLP-76 peptide residues are marked with an asterisk in all structures). The first Pro of the SLP-76 peptide (shown in parentheses) is not visible in the structure.
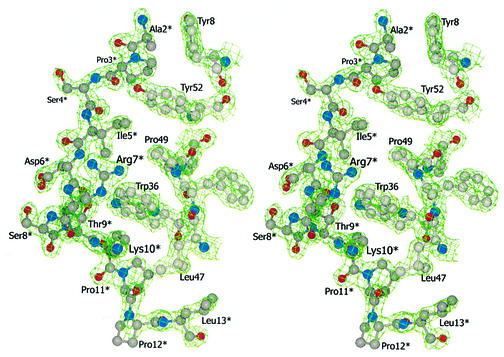
Fig. 5. Divergent stereo view of selected residues of the Mona/Gads SH3C–SLP-76 (P2) interface with the corresponding electron density contoured at 1.5 root mean square deviation from the mean density.
Fig. 6. (A and B) Close-up views of the hydrogen bonding pattern (yellow broken lines) within the core of the docking region in the two conformations present in the asymmetric unit of the crystal. All carbon atoms depicted and their bonds in the SLP-76 peptide are shown in dark grey. Carbon atoms of the SH3 domain are shown in light grey. For clarity, Glu14 is not depicted in the top view (A).
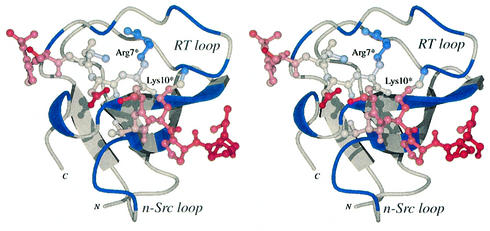
The mean square atomic displacement (<u2>) of the SLP-76 residues within the crystal structure suggests a tight binding that is in agreement with the ITC measurements described. The temperature factors (B = <u2> 8π2/3) of the peptide atoms are comparable with that of the SH3C domain with a variation between 10 and 30 Å2 (Figure 7). The lowest values, indicating the most stable regions, are exhibited by the Arg7* and Lys10* residues that hydrogen bond to residues of the RT loop. Temperature values increase gradually towards each end of the peptide chain with the highest values exhibited in the C-terminal region that contacts the n-Src loop, implying a relatively fluid local binding. Characteristically, non-essential residues such as Asp6* and Ser8* appear to have more variable atomic positions.
Fig. 7. Divergent stereo view of the Mona/Gads SH3C model with regions within the buried interface coloured in blue and a molecular model of the SLP-76 peptide coloured from red to blue relating to atomic B values from 10 to 30 Å2.
Comparisons with other SH3 domains
Grb2 is a known partner of SLP-76, attaching to the same epitope as Mona/Gads albeit with much lower affinity. The latter can be accounted for by the significant differences in the residues utilized for peptide binding between Grb2 SH3C (Kami et al., 2002) and Mona/Gads SH3C (Figure 8). In Mona/Gads SH3C, more residues from the RT loop are involved. Moreover, a significant portion of the n-Src loop, which remains mostly unengaged in the Grb2 SH3C complex, is extensively involved in peptide docking by Mona/Gads SH3C. These features probably account for the different affinities observed by ITC (Table I).
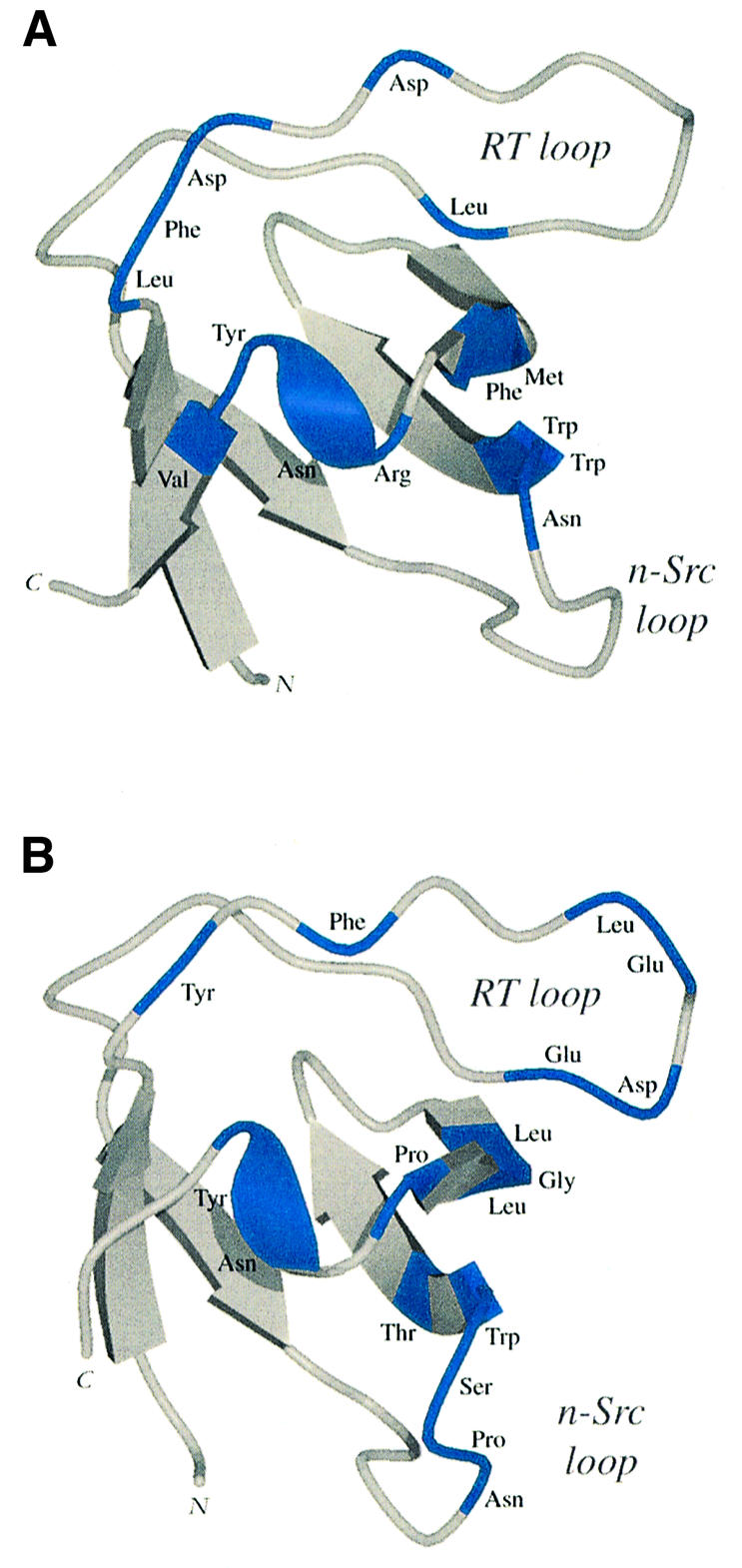
Fig. 8. Ribbon representations of the Grb2 SH3C and Mona/Gads SH3C domains with areas involved in molecular docking of an SLP-76 peptide coloured blue. (A) Grb2 SH3C–peptide interactions as previously defined by NMR experiments (Kami et al., 2002). (B) Mona/Gads SH3C–peptide contacts present in the crystal structure.
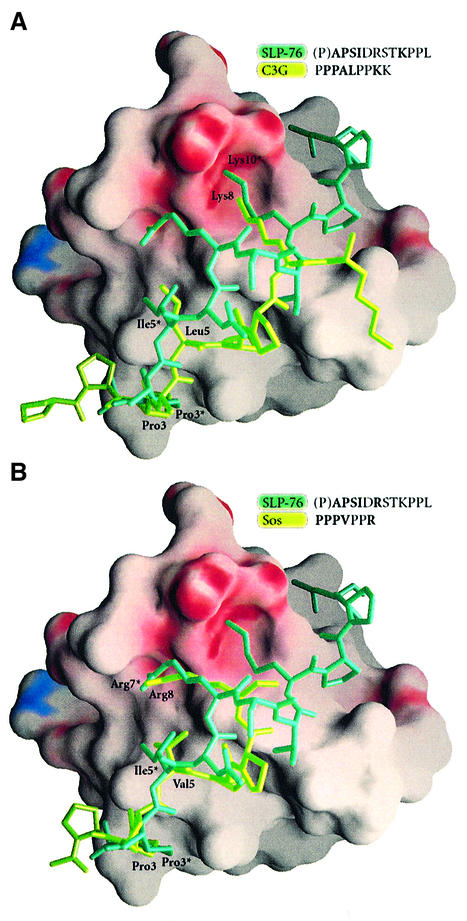
The numerous entries within the Protein Data Bank for SH3 or SH3-like domains allow extensive comparisons with the Mona/Gads SH3C. Closest structural neighbours were identified with DALI (Holm and Sander, 1994) and analysed further using the program SHP (Stuart et al., 1979). All homologous structures tested displayed positional Cα deviations of 0.7–1 Å. The structures that most closely resemble Mona/Gads SH3C are Sem-5 SH3C (1SEM.pdb), Crk SH3N (1CKB.pdb) and Grb2 SH3C (1GCQ.pdb). Of these structures, Sem-5 SH3C and Crk SH3N were in complex with peptides that share common features with the structure described here. In all three structures the peptides, although representing different motifs suited for their respective binding partners, are bound in the ‘minus’ orientation (for definition see Bork et al., 1997; Kay et al., 2000; Mayer, 2001; Musacchio, 2002) and follow convergent paths to essential binding sites (Figure 9).
Fig. 9. Electrostatic potential surface representation of Mona/Gads SH3C with molecular models of different well characterized SH3 domain binding peptides docked onto the SH3 domain. The peptides were aligned through structural superposition of their respective SH3 partners onto the present structure. (A) Comparison of the SLP-76 peptide (P2, dark green) with a C3G peptide (light green) from the mouse c-Crk SH3N–C3G complex (1CKA.pdb). (B) Comparison of the SLP-76 peptide (dark green) with an mSos peptide (light green) from the Sem-5 SH3C complex (1SEM.pdb).
The C3G peptide in the Crk SH3N complex (Wu et al., 1995) has a Pro residue in the same position as Pro3* of the SLP-76 peptide followed by a Leu two residues later, much like Ile5* (Figure 9A). The peptides then diverge up to the following Lys (Lys10* in SLP-76) which forms similar hydrogen bonds with equivalent residues on the respective SH3 domains. In the Sem-5 SH3C–mSos peptide complex (Lim et al., 1994), the peptide binds in a nearly identical fashion to SLP-76 binding to Mona/Gads SH3C from its N-terminus up to the Val position (Ile5* in the SLP-76 peptide), at which point it deviates away from the SH3 domain, only to converge back to an essentially identical position at Arg7* with its equivalent Arg residue (Figure 9B).
Characteristic residues that coincide in the three complexes are important in deciphering the interactions that promote binding. The position occupied by Pro3* seems supremely suited for just this type of residue. Two amino acids further on the nature of hydrophobic interactions required in positioning this region persists. The polypeptide chains deviate past this point away from the 310 conformation and assume a poly-Pro helical conformation. However, both peptides converge on the Arg7* or the Lys10* position. The comparatively conserved route of convergence on the domain surface is an example of the employment of different folds in molecular docking onto similar sites.
SH3 domain dimerization
Several previous reports implicate SH3 domains in oligomerization processes: the SH3 domain of PI3 kinase forms fibre-like aggregates in vitro (Guijarro et al., 1998); the EGF receptor substrate Eps8 forms intertwined SH3 homodimers which appear to regulate binding potential (Kishan et al., 1997, 2001); a Grb2 SH3C and Vav SH3N heterodimer forms the interaction basis for Grb2 and the GTPase regulator Vav (Nishida et al., 2001); the bacterial SH3-like domain of KorB forms a dimer which binds to pseudosymmetrical regulatory sites within the RP4 gene (Delbruck et al., 2002).
In this context, the non-crystallographic dimer of Mona/Gads SH3C is suggestive of a coordination potential inherent in this domain. The seven-coordination site of pentagonal bipyramid geometry displayed by the cadmium bound in the crystal (Figure 10A) is a known coordination state of Ca2+ in biological systems (Pidcock and Moore, 2001). Zn2+ can also be coordinated in a similar fashion (Rodriguez-Arguelles et al., 1995). The Mona/Gads SH3C–cadmium interaction is restricted to the molecular surface diametrically opposite the SLP-76 binding site and does not interfere with protein folding or docking potential (Figure 10A). Such an arrangement could confer higher affinities to Mona/Gads SH3C binding partners such as Gab family proteins and USP8, which have two potential Mona/Gads binding sites. Furthermore, in Drosophila, Drk SH3C has two functionally important binding sites in Dos, each of which binds only weakly by itself (Feller et al., 2002; Table I) and could require stabilization through SH3 domain dimerization or a similar mechanism.
Fig. 10. Dimerization of the Mona/Gads SH3C–peptide complex. (A) Molecular model of the Mona/Gads SH3C–peptide dimer coordinating a cadmium ion (yellow sphere) and close-up of the seven-coordinate site. The Cα trace of the two SH3C domains is light grey and the docked SLP-76 peptide residues are dark gray. Residues involved in coordination are also shown. (B) Plots of apparent whole-cell weight-average molecular weight (Mw,app) against sample concentration (mg/ml) for interference data. The same curves were also derived with the absorbance data but interference data are inherently more precisely determined. Filled symbols are for Mona/Gads SH3C–SLP-76 peptide in Ca2+, with different colours representing the different speeds as indicated. Open symbols show the complex with Zn2+. Fitted curves, shown as broken black lines, are for a linear regression of weight with concentration in Ca2+ and for a rectangularly hyperbolic regression in Zn2+, as appropriate for a dimerization process (Ikemizu et al., 2000).
To test whether the Mona/Gads SH3C domain complexed with the SLP-76 peptide can also dimerize in solution, analytical ultracentrifugation experiments were performed in the presence of the physiologically important ions Ca2+ or Zn2+. Figure 10B shows the variation of the apparent molecular weight Mw,app of the complex at different concentrations in the presence of Ca2+ and Zn2+. With Ca2+ the complex maintains virtually constant weight, with only a small statistically significant increase that indicates very weak self-association. The Mw,app of the complex in Ca2+ extrapolated to zero concentration (infinite dilution, where self-association and diffusion-limitation do not affect the observed weight) is 8703 ± 103 Da, which is close to the theoretical monomeric Mw of 8460. The Mw,app of the complex in Zn2+ shows a markedly different variation with concentration. At 26 000 r.p.m., the significant increase in Mw,app plateaus at the expected weight of a dimer. The resulting Kd of dimerization in the presence of Zn2+ is 0.73 mM (6.37 ± 0.56 mg/ml) which is within the range that may be considered significant within the cellular environment. To our knowledge, this is the first time that an ion-mediated dimerization of an SH3 domain is reported.
Zn2+ is not only crucial for a plethora of proteins involved in DNA transcription, protein–protein interactions and proteolysis but also mediates the formation of various protein dimers. Maturation and action of lymphocytes and monocytes is critically dependent on Zn2+ (Wellinghausen and Rink, 1998). A number of Zn2+-dependent proteins are known to play important roles in T-cell receptor signalling, including c-Cbl, TSAd/LAD, the SLP-76-interacting Rac activator Vav, Lck and CD4 (Lin et al., 1998; Choi et al., 1999; Zheng et al., 2000; Zugaza et al., 2002). The experiments presented here encourage further biochemical, biophysical and cell biological analyses to define the role of zinc in Mona/Gads signalling and T-cell functions.
Materials and methods
SH3 domain primary structure alignment and dendrogram
The SH3 domain consensus sequence was determined using MultAlin (Corpet, 1988). The identity matrix was calculated using MacBoxshade 2.15 (M.D.Baron, 1999; distributed at http://iubio.bio.indiana.edu/soft/molbio/mac/). Sequence alignments were generated with CLUSTAL_W (Thompson et al., 1994) and depicted through ESPript 2.0 (Gouet et al., 1999). The dendrogram was drawn using PHYLIP (Phylogeny Inference Package) (Felsenstein, 1993).
Peptide synthesis and SH3 domain construction, expression and purification
Peptides were synthesized with an amidated C-terminus using standard technology and subsequently purified by high-pressure liquid chromatography. Cloning and bacterial expression of the Drk SH3C and Grb2 SH3C domains have been described previously (Lewitzky et al., 2001; Feller et al., 2002). The Mona/Gads SH3C domain used for most ITC experiments (amino acids 256–322) was constructed by PCR and its functionality has been documented previously (Bourgin et al., 2002). Bacteria were grown in Luria broth (LB) with 100 µg/ml ampicillin and protein expression was induced with 0.1 mM ispropyl-β-d-thiogalactopyranoside (IPTG). Bacteria were lysed in cold TPE [1% (v/v) Triton X-100, 1× phosphate-buffered saline (PBS) pH 7.4, 100 mM EDTA, 10 µg/ml aprotinin, 0.7 µg/ml pepstatin A, 0.5 µg/ml leupeptin, 0.5 µg/ml antipain]. GST–SH3 fusion protein was purified using GSH-Sepharose (Amersham Biosciences). Bound GST–SH3 was eluted with 100 mM GSH pH 7.5 and dialysed against 5 mM Tris–HCl pH 7.5.
For crystallization experiments, nucleotides encoding amino acids 265–322 of mouse Mona/Gads were amplified by PCR, cloned into pGEX-6P-1 (Amersham Biosciences) using the BamHI and XhoI restriction sites, and sequenced. For MAD phasing, the protein was expressed in Escherichia coli BL21(DE3) cells grown in minimal medium (M9) supplemented with ions, vitamins, glucose, selenomethionine and all l-amino acids except methionine. Inhibition of endogenous methionine production was achieved upon induction through addition of excess of six l-amino acids (Lys, Thr, Phe, Leu, Ile and Val). The method is a combination of previously described protocols (Ramakrishnan et al., 1993; Van Duyne et al., 1993). Bacterial pellets were dissolved in 50 mM HEPES pH 7.5 with 50 mM NaCl plus protease inhibitors [0.2 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM EDTA, 1× Complete™ protease inhibitor cocktail (Roche)] and sonicated. The cleared lysates were applied to GSH–Sepharose and incubated with PreScission™ protease (2.5 U per milligram of GST–SH3 protein) in 2× PBS at 8°C for 16 h. The eluted SH3 domain was further purified by size exclusion chromatography (Sephadex-75; Amersham Biosciences) and concentrated (Vivaspin VS0611; Vivascience). The purity and monodispersity of the concentrated SH3 domain were analysed by SDS and native PAGE as well as by light scattering using a DynaPro-MS/X light scattering system (ProteinSolutions Inc.).
Isothermal titration calorimetry
Experiments were performed using a VP-ITC MicroCalorimeter (MicroCal, Northampton, MA). Peptides were used at 0.5 mM in buffer G (25 mM HEPES–KOH pH 7.5, 100 mM KAc, 5 mM MgAc) and titrated from a syringe (300 µl total volume) into a sample chamber holding 1.43 ml of 0.05 mM Mona/Gads SH3C in buffer G. The peptide concentrations were adjusted in cases of low binding affinities (P15, 1 mM; P18, 1.333 mM; P22, 1 mM). Heat generated by protein dilution was determined in separate experiments by injecting buffer G without peptide into the sample chamber filled with Gads/Mona SH3C and was negligible. Solutions used in the syringe and the sample chamber were clarified for 10 min at 20 800 g maximum prior to use. Gads/Mona SH3C solution was degassed before the measurements were made (ThermoVac; MicroCal). At the equilibrium temperature of 25°C, peptide solutions were titrated into the sample chamber by 28 injections of 10 µl each. Resulting peaks of measured deviations from the equilibrium temperature were integrated to yield the quantity of heat generated. Data were fitted using χ2 minimization on a model assuming a single set of sites to calculate the binding affinity Kd. All steps of the data analysis were performed using ORIGIN(V5.0) software provided by the manufacturer.
Crystallization and data collection
Mona/Gads SH3C protein was mixed with SLP-76 peptide (P2) at a molar ratio of 1:2. Crystallization was successful with the sitting drop vapour diffusion method at 20°C under equilibrium conditions of 20% PEG-4000, 5 mM CdCl2 and 50 mM sodium cacodylate pH 6.5 (Clear Strategy Screen; Molecular Dimensions) at a final protein concentration of 10 mg/ml. Rod-shaped crystals with approximate dimensions 0.3 × 0.2 × 0.2 mm3 appeared within 24 h. Crystals were cryoprotected through transfer into mother liquor containing 20% glycerol and vitrified.
A three-wavelength MAD experiment was performed on BM14 at the European Synchrotron Radiation Facility, Grenoble. Diffraction data to 1.7 Å were collected on an MAR CCD 133 mm detector at a crystal-to-detector distance of 100 mm (1° oscillation for each exposure). Highly redundant data were collected for each wavelength and intensities were integrated and scaled using DENZO and SCALEPACK (Otwinowski, 1993; Otwinowski and Minor, 1997) (Table II).
Structure determination and refinement
Phases were estimated with SOLVE (Terwilliger and Berendzen, 1999) using the anomalous and dispersive differences estimated from three datasets collected at wavelengths corresponding to the peak and the inflection point of the anomalous scattering of the selenium atoms as well as at a higher-energy remote wavelength (see Table II). Phases were further modified with RESOLVE (Terwilliger, 2001). The structure was autotraced with WarpNtrace (Perrakis et al., 1999) (133 out of 136 structured amino acids in the asymmetric unit were traced) with a resulting R factor over all reflections of 19.3%. Several additional cycles of manual building in O (Jones et al., 1991) and refinement in REFMAC (Murshudov et al., 1997) resulted in full structure assignment. Water molecules were assigned automatically with ARP (Lamzin and Wilson, 1993) and further refined in REFMAC. Phasing, refinement and model quality statistics are shown in Table II.
Structure and density representations
Ball-and-stick, ribbon and electron density representations were created using BOBSCRIPT (Esnouf, 1997, 1999) and rendered with Raster3D (Merritt and Murphy, 1994).
Calculation of buried surface areas and surface representations
Surface area calculations were carried out in CNS (Brunger et al., 1998) and surface representations and electrostatic potential calculations were performed with GRASP (Nicholls et al., 1991).
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-I analytical ultracentrifuge, essentially as previously described (Ikemizu et al., 2000). Briefly, Mona/Gads SH3C and SLP-76 peptide (molar ratio 1:2) in 50 mM Tris–HCl pH 6.8, 50 mM NaCl and 2 mM metal ions (ZnCl2 or CaCl2) were used at SH3C concentrations ranging from 2 to 9 mg/ml, centrifuged at 15 000, 18 000 or 26 000 r.p.m., respectively, at 20°C and imaged using absorbance optics at 305 and 310 nm wavelengths and interference optics. The sample distributions measured at equilibrium were fitted with the program ULTRASPIN (Altamirano et al., 2001) using a single-species equation. Any non-ideal behaviour, such as self-association, manifests itself as increasing apparent whole-cell molecular weight-average weights (Mw,app) with increasing concentration. The values for Mw,app were plotted together against sample concentration over the full range studied and fitted with either a straight line or the equation:
for a dimerization, as appropriate. Here, M1 is the mass of a monomer, c is the sample concentration and Kd is the equilibrium constant of dissociation.
PDB accession code
The coordinates and structure factors have been deposited in the Protein Data Bank with ID code 1oeb.pdb.
Acknowledgments
Acknowledgements
Erika Mancini and Geoff Sutton kindly helped with synchrotron data collection, Jonathan Grimes with the calculations and a critical reading of the manuscript, and Robert Esnouf and Karl Harlos with computing and in-house data collection. We also thank Martin Walsh at ESRF Grenoble, BM14, for help with the MAD experiment. The analytical ultracentrifugation experiments were performed in the AUC facility established by the BBSRC and the Wellcome Trust in the Glycobiology Institute of the University of Oxford and managed by Russell Wallis. This work has been funded by Cancer Research UK and the European Union. E.Y.J. is a Cancer Research UK Principal Research Fellow.
References
- Altamirano M.M. et al. (2001) Ligand-independent assembly of recombinant human CD1 by using oxidative refolding chromatography. Proc. Natl Acad. Sci. USA, 98, 3288–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada H. et al. (1999) Grf40, a novel Grb2 family member, is involved in T cell signaling through interaction with SLP-76 and LAT. J. Exp. Med., 189, 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett P., Bottger,G., Klein,A.T., Tabak,H.F. and Distel,B. (2000) The peroxisomal membrane protein Pex13p shows a novel mode of SH3 interaction. EMBO J., 19, 6382–6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D.M., Nash,P., Liu,S.K., Pawson,T. and McGlade,C.J. (2002) A high-affinity Arg–X–X–Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr. Biol., 12, 1336–1341. [DOI] [PubMed] [Google Scholar]
- Bork P., Schultz,J. and Ponting,C.P. (1997) Cytoplasmic signalling domains: the next generation. Trends Biochem. Sci., 22, 296–298. [DOI] [PubMed] [Google Scholar]
- Bourette R.P., Arnaud,S., Myles,G.M., Blanchet,J.P., Rohrschneider,L.R. and Mouchiroud,G. (1998) Mona, a novel hematopoietic-specific adaptor interacting with the macrophage colony-stimulating factor receptor, is implicated in monocyte/macrophage development. EMBO J., 17, 7273–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgin C., Bourette,R.P., Arnaud,S., Liu,Y., Rohrschneider,L.R. and Mouchiroud,G. (2002) Induced expression and association of the Mona/Gads adapter and Gab3 scaffolding protein during monocyte/macrophage differentiation. Mol. Cell. Biol., 22, 3744–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burack W.R., Cheng,A.M. and Shaw,A.S. (2002) Scaffolds, adaptors and linkers of TCR signaling: theory and practice. Curr. Opin. Immunol., 14, 312–316. [DOI] [PubMed] [Google Scholar]
- Cestra G., Castagnoli,L., Dente,L., Minenkova,O., Petrelli,A., Migone,N., Hoffmuller,U., Schneider-Mergener,J. and Cesareni,G. (1999) The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity. J. Biol. Chem., 274, 32001–32007. [DOI] [PubMed] [Google Scholar]
- Choi Y.B., Kim,C.K. and Yun,Y. (1999) Lad, an adapter protein interacting with the SH2 domain of p56lck, is required for T cell activation. J. Immunol., 163, 5242–5249. [PubMed] [Google Scholar]
- Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbruck H., Ziegelin,G., Lanka,E. and Heinemann,U. (2002) An Src homology 3-like domain is responsible for dimerization of the repressor protein KorB encoded by the promiscuous IncP plasmid RP4. J. Biol. Chem., 277, 4191–4198. [DOI] [PubMed] [Google Scholar]
- Ellis J.H., Ashman,C., Burden,M.N., Kilpatrick,K.E., Morse,M.A. and Hamblin,P.A. (2000) GRID: a novel Grb-2-related adapter protein that interacts with the activated T cell costimulatory receptor CD28. J. Immunol., 164, 5805–5814. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134, 112–133. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Feller S.M., Wecklein,H., Lewitzky,M., Kibler,E. and Raabe,T. (2002) SH3 domain-mediated binding of the Drk protein to Dos is an important step in signaling of Drosophila receptor tyrosine kinases. Mech. Dev., 116, 129–139. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1993) PHYLIP (Phylogeny Inference Package) Version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle. http://evolution.genetics.washington.edu/phylip.html
- Ghose R., Shekhtman,A., Goger,M.J., Ji,H. and Cowburn,D. (2001) A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat. Struct. Biol., 8, 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Guijarro J.I., Sunde,M., Jones,J.A., Campbell,I.D. and Dobson,C.M. (1998) Amyloid fibril formation by an SH3 domain. Proc. Natl Acad. Sci. USA, 95, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1994) Searching protein structure databases has come of age. Proteins, 19, 165–173. [DOI] [PubMed] [Google Scholar]
- Ikemizu S., Gilbert,R.J., Fennelly,J.A., Collins,A.V., Harlos,K., Jones,E.Y., Stuart,D.I. and Davis,S.J. (2000) Structure and dimerization of a soluble form of B7-1. Immunity, 12, 51–60. [DOI] [PubMed] [Google Scholar]
- Jackman J.K., Motto,D.G., Sun,Q., Tanemoto,M., Turck,C.W., Peltz,G.A., Koretzky,G.A. and Findell,P.R. (1995) Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J. Biol. Chem., 270, 7029–7032. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kami K., Takeya,R., Sumimoto,H. and Kohda,D. (2002) Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. EMBO J., 21, 4268–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Freund,C., Duke-Cohan,J.S., Musacchio,A., Wagner,G. and Rudd,C.E. (2000) SH3 domain recognition of a proline-independent tyrosine-based RKxxYxxY motif in immune cell adaptor SKAP55. EMBO J., 19, 2889–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Miyazawa,K. and Kitamura,N. (2000) A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem., 275, 37481–37487. [DOI] [PubMed] [Google Scholar]
- Kay B.K., Williamson,M.P. and Sudol,M. (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J., 14, 231–241. [PubMed] [Google Scholar]
- Kishan K.V., Scita,G., Wong,W.T., Di Fiore,P.P. and Newcomer,M.E. (1997) The SH3 domain of Eps8 exists as a novel intertwined dimer. Nat. Struct. Biol., 4, 739–743. [DOI] [PubMed] [Google Scholar]
- Kishan K.V., Newcomer,M.E., Rhodes,T.H. and Guilliot,S.D. (2001) Effect of pH and salt bridges on structural assembly: molecular structures of the monomer and intertwined dimer of the Eps8 SH3 domain. Protein Sci., 10, 1046–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin V.S. and Wilson,K.S. (1993) Automated refinement of protein models. Acta Crystallogr. D, 49, 129–147. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Law C.L., Ewings,M.K., Chaudhary,P.M., Solow,S.A., Yun,T.J., Marshall,A.J., Hood,L. and Clark,E.A. (1999) GrpL, a Grb2-related adaptor protein, interacts with SLP-76 to regulate nuclear factor of activated T cell activation. J. Exp. Med., 189, 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I. et al. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitzky M., Kardinal,C., Gehring,N.H., Schmidt,E.K., Konkol,B., Eulitz,M., Birchmeier,W., Schaeper,U. and Feller,S.M. (2001) The C-terminal SH3 domain of the adapter protein Grb2 binds with high affinity to sequences in Gab1 and SLP-76 which lack the SH3-typical P–x–x–P core motif. Oncogene, 20, 1052–1062. [DOI] [PubMed] [Google Scholar]
- Lim W.A. (1996) Reading between the lines: SH3 recognition of an intact protein. Structure, 4, 657–659. [DOI] [PubMed] [Google Scholar]
- Lim W.A., Richards,F.M. and Fox,R.O. (1994) Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature, 372, 375–379. [DOI] [PubMed] [Google Scholar]
- Lin R.S., Rodriguez,C., Veillette,A. and Lodish,H.F. (1998) Zinc is essential for binding of p56(lck) to CD4 and CD8alpha. J. Biol. Chem., 273, 32878–32882. [DOI] [PubMed] [Google Scholar]
- Liu S.K. and McGlade,C.J. (1998) Gads is a novel SH2 and SH3 domain-containing adaptor protein that binds to tyrosine-phosphorylated Shc. Oncogene, 17, 3073–3082. [DOI] [PubMed] [Google Scholar]
- Liu S.K., Fang,N., Koretzky,G.A. and McGlade,C.J. (1999) The hematopoietic-specific adaptor protein gads functions in T-cell signaling via interactions with the SLP-76 and LAT adaptors. Curr. Biol., 9, 67–75. [DOI] [PubMed] [Google Scholar]
- Liu S.K., Smith,C.A., Arnold,R., Kiefer,F. and McGlade,C.J. (2000) The adaptor protein Gads (Grb2-related adaptor downstream of Shc) is implicated in coupling hemopoietic progenitor kinase-1 to the activated TCR. J. Immunol., 165, 1417–1426. [DOI] [PubMed] [Google Scholar]
- Liu S.K., Berry,D.M. and McGlade,C.J. (2001) The role of Gads in hematopoietic cell signalling. Oncogene, 20, 6284–6290. [DOI] [PubMed] [Google Scholar]
- Lock L.S., Royal,I., Naujokas,M.A. and Park,M. (2000) Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J. Biol. Chem., 275, 31536–31545. [DOI] [PubMed] [Google Scholar]
- Manser E., Loo,T.H., Koh,C.G., Zhao,Z.S., Chen,X.Q., Tan,L., Tan,I., Leung,T. and Lim,L. (1998) PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell, 1, 183–192. [DOI] [PubMed] [Google Scholar]
- Mayer B.J. (2001) SH3 domains: complexity in moderation. J. Cell Sci., 114, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3d Version-2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Mongiovi A.M., Romano,P.R., Panni,S., Mendoza,M., Wong,W.T., Musacchio,A., Cesareni,G. and Di Fiore,P.P. (1999) A novel peptide–SH3 interaction. EMBO J., 18, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Musacchio A. (2002) How SH3 domains recognize proline. Adv. Protein Chem., 61, 211–268. [DOI] [PubMed] [Google Scholar]
- Naviglio S., Mattecucci,C., Matoskova,B., Nagase,T., Nomura,N., Di Fiore,P.P. and Draetta,G.F. (1998) UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J., 17, 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nishida M., Nagata,K., Hachimori,Y., Horiuchi,M., Ogura,K., Mandiyan,V., Schlessinger,J. and Inagaki,F. (2001) Novel recognition mode between Vav and Grb2 SH3 domains. EMBO J., 20, 2995–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K., Nagata,K., Horiuchi,M., Ebisui,E., Hasuda,T., Yuzawa,S., Nishida,M., Hatanaka,H. and Inagaki,F. (2002) Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain. J. Biomol. NMR, 22, 37–46. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1993) In Proceedings of the CCP4 Study Weekend: Data Collection and Processing. (Sawyer,L., Isaacs,N. and Bailey,S. (eds), pp. 56–62, SERC Daresbury Laboratory, Daresbury, England.
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Pidcock E. and Moore,G.R. (2001) Structural characteristics of protein binding sites for calcium and lanthanide ions. J. Biol. Inorg. Chem., 6, 479–489. [DOI] [PubMed] [Google Scholar]
- Qiu M., Hua,S., Agrawal,M., Li,G., Cai,J., Chan,E., Zhou,H., Luo,Y. and Liu,M. (1998) Molecular cloning and expression of human grap-2, a novel leukocyte-specific SH2- and SH3-containing adaptor-like protein that binds to gab-1. Biochem. Biophys. Res. Commun., 253, 443–447. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch,J.T., Graziano,V., Lee,P.L. and Sweet,R.M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Arguelles M.C., Belicchi Ferrari,M., Gasparri Fava,G., Pelizzi,C., Tarasconi,P., Albertini,R., Dall’Aglio,P.P., Lunghi,P. and Pinelli,S. (1995) 2,6-Diacetylpyridine bis(thiosemicarbazones) zinc complexes: synthesis, structure and biological activity. J. Inorg. Biochem., 58, 157–175. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D.I., Levine,M., Muirhead,H. and Stammers,D.K. (1979) Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol., 134, 109–142. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2001) Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr. D, 57, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson M.G., Lin,J. and Weiss,A. (2000) Lymphocytes with a complex: adapter proteins in antigen receptor signaling. Immunol. Today, 21, 584–591. [DOI] [PubMed] [Google Scholar]
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Wellinghausen N. and Rink,L. (1998) The significance of zinc for leukocyte biology. J. Leukoc. Biol., 64, 571–577. [DOI] [PubMed] [Google Scholar]
- Wu X., Knudsen,B., Feller,S.M., Zheng,J., Sali,A., Cowburn,D., Hanafusa,H. and Kuriyan,J. (1995) Structural basis for the specific interaction of lysine-containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure, 3, 215–226. [DOI] [PubMed] [Google Scholar]
- Xia C., Bao,Z., Tabassam,F., Ma,W., Qiu,M., Hua,S. and Liu,M. (2000) GCIP, a novel human grap2 and cyclin D interacting protein, regulates E2F-mediated transcriptional activity. J. Biol. Chem., 275, 20942–20948. [DOI] [PubMed] [Google Scholar]
- Yankee T.M., Solow,S.A., Draves,K.D. and Clark,E.A. (2003) Expression of the grb2-related protein of the lymphoid system in B cell subsets enhances B cell antigen receptor signaling through mitogen-activated protein kinase pathways. J. Immunol., 170, 349–355. [DOI] [PubMed] [Google Scholar]
- Yoder J., Pham,C., Iizuka,Y.M., Kanagawa,O., Liu,S.K., McGlade,J. and Cheng,A.M. (2001) Requirement for the SLP-76 adaptor GADS in T cell development. Science, 291, 1987–1991. [DOI] [PubMed] [Google Scholar]
- Zheng N., Wang,P., Jeffrey,P.D. and Pavletich,N.P. (2000) Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin–protein ligases. Cell, 102, 533–539. [DOI] [PubMed] [Google Scholar]
- Zugaza J.L., Lopez-Lago,M.A., Caloca,M.J., Dosil,M., Movilla,N. and Bustelo,X.R. (2002) Structural determinants for the biological activity of Vav proteins. J. Biol. Chem., 277, 45377–45392. [DOI] [PubMed] [Google Scholar]